Abstract
Transcription factors that regulate quiescence, proliferation, and homing of lymphocytes are critical for effective immune system function. In the present study, we demonstrated that the transcription factor ELF4 directly activates the tumor suppressor KLF4 downstream of T cell receptor (TCR) signaling to induce cell-cycle arrest in naïve CD8+ T cells. Elf4- and Klf4-deficient mice accumulated CD8+CD44hi T cells during steady-state conditions and generated more memory T cells after immunization. The homeostatic expansion of CD8+CD44hi T cells in Elf4-null mice resulted in a redistribution of cells to non-lymphoid tissue due to reduced expression of the transcription factor KLF2, and the surface proteins CCR7 and CD62L. This work describes the combinatorial role of lymphocyte-intrinsic factors on T cell homeostasis, activation and homing.
INTRODUCTION
A delicate balance between quiescence and homeostatic proliferation maintains the T cell pool. An emerging paradigm suggests that quiescence is an actively regulated state, rather than the default state, in the absence of proliferation-inducing signals 1. Homeostatic proliferation of CD8+ T cells is an efficient mechanism to maintain the T cell pool during aging 2. However, neither the molecular mechanisms regulating CD8+ T cell homeostasis nor its impact on T cell activation and homing are completely understood. The identification of new negative regulators of T cell proliferation is vital to further understand lymphocyte homeostasis and immune responses, as well as to develop protocols of gene modulation to enhance the immunological memory generated after vaccination.
The transcription factor ELF4 (also known as MEF) is a member of the ETS family of proteins 3–5. Recent reports suggest that ELF4 can function as a tumor suppressor in hematopoietic cells. ELF4 expression is downregulated in acute myelogenous leukemia, either by oncoproteins or by chromosome translocation with the ERG gene 6–8. Using ELF4-deficient mice, we previously demonstrated that ELF4 induces a state of readiness in natural killer (NK) cells by activating perforin gene expression 3, 9. We also demonstrated that ELF4 controls the balance between activation and quiescence in hematopoietic stem cells (HSCs) during steady state conditions but not during regenerative hematopoiesis 10. Therefore, we hypothesized that ELF4 may regulate T cell entry into the cell cycle by activating an inhibitor of proliferation.
The Krüppel-like factor 4 (KLF4) is a zinc-finger transcriptional regulator that acts as a tumor suppressor and oncogene in the gastrointestinal tract and breast tissue, respectively 11–13. In the hematopoietic system, Klf4 gene was found silenced in patients with adult T cell leukemia 14. The recent finding that KLF4 can reprogram adult somatic cells into pluripotent stem cells in combination with Sox2, Oct3/4 and c-Myc suggests a role for this transcription factor in the prevention of cellular proliferation and differentiation 15–17.
KLF2 (http://www.signaling-gateway.org/molecule/query?afcsid=A004085) was the first transcription factor described as capable of programming T cell quiescence by inhibiting the c-Myc pathway 18. Other cell-intrinsic inhibitors of T cell proliferation include the FOXO transcription factors, the Tob anti-proliferative protein, dipeptidyl peptidase 2 (Dpp2), and the transcription factor Nfat2c 19–22. Gene products involved in controlling the cell cycle can also influence the number of antigen-experienced memory CD8+ T cells generated at the end of an immune response 23–26. In addition, the transcription factor T-bet promotes the differentiation of effector T cells over memory precursor cells, whereas the basic helix loop helix protein Id2 regulates the survival of effector T cells via the proteinase inhibitor 6 (Spi6) 27–29.
Memory-like T cells are generated during lymphopenia- or cytokine-induced homeostatic proliferation of naïve T cells2, 30. A similar gene expression profile and the same cell of origin suggests that antigen-experienced and memory-like T cells use common mechanisms to control proliferation 31. Gene products that influence T cell proliferation can also regulate homing receptors and thereby direct T cell trafficking. In contrast to CD62LloCCR7− effector memory T cells (Tem) that home to non-lymphoid tissues and the spleen, CD62LhiCCR7+ central memory T cells (Tcm) home to the lymph nodes (LN) 32, 33. KLF2 promotes the circulation of naïve T cells through lymphoid organs by regulating the expression of CD62L, sphingosine 1 phosphate 1 receptor, CCR7, and β7-integrin 34, 35. Conversely, the suppression of KLF2 expression induced by T cell receptor (TCR) stimulation results in the mobilization of effector T cells to inflamed tissues because KLF2 represses CCR3 and CCR5 expression 36. Although the role of KLF2 has been established in naïve T cells, it has not been defined in memory T cells or in the maintenance of the size of the T cell pool. Neither the transcriptional mechanisms controlling the homeostatic and antigen-driven proliferation of naïve T cells and the differentiation of naïve T cells into memory T cells, nor the lineage relationship between memory T cell subsets have been identified, in spite of the importance of these mechanisms in vaccination and immune protection.
Here we demonstrated that ELF4 regulates the proliferation of naïve CD8+ T cells and the tissue distribution of CD8+CD44hi T cells. ELF4 activated KLF4 downstream of TCR signaling to induce cell cycle arrest in naïve CD8+ T cells. Naïve CD8+ T cells from mice lacking ELF4 or KLF4 were ‘primed’ to proliferate during homeostasis or after immunization, due to accumulation of cyclin D in the cytosol and repression of cyclin-dependent kinase inhibitors. In addition, in ELF4-deficient mice, CD8+CD44hi T cell populations gradually accumulated and redistributed to non-lymphoid tissues due to reduced expression of KLF2, CCR7 and CD62L. In addition to KLF2, ELF4 and KLF4 tumor suppressors regulate T cell proliferation and homing.
RESULTS
ELF4 suppresses homeostatic and antigen-driven proliferation
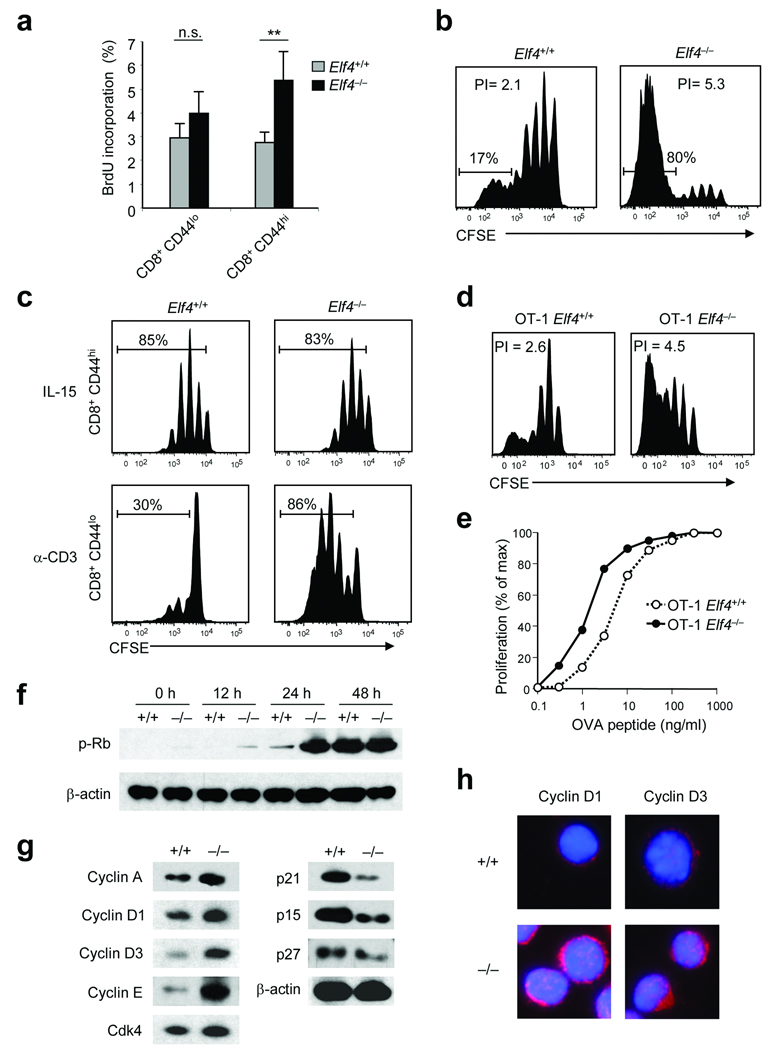
To test whether ELF4 regulates cell cycle entry in naïve CD8+ T cells similarly to HSC, we measured proliferation in response to homeostatic and antigen-driven stimuli. We used two systems to test homeostatic proliferation of Elf4−/− CD8+ T cells: in vivo BrdU incorporation and adoptive transfer of naïve CD8+ T cells into lymphopenic hosts. Although administration of BrdU revealed a low rate of T cell proliferation, activated Elf4−/− CD8+CD44hi T cells incorporated significantly more BrdU than did Elf4+/+ CD8+CD44hi T cells (Fig. 1a). Next, CD8+ CD44loCD45.2+ T cells were purified from Elf4+/+ and Elf4−/− mice, labeled with CFSE and adoptively transferred into sublethally irradiated B6.SJL mice to induce homeostatic proliferation. At least 80% of the transferred Elf4−/− CD8+ T cells underwent more than 5 divisions compared to only 17% of the Elf4+/+ CD8+ T cells (Fig. 1b). Furthermore, the Elf4−/− naïve CD8+ T cells out-competed the Elf4+/+ CD8+ T cells when the two populations were co-injected into irradiated B6.SJL mice (Supplementary Fig. 1 online). To determine whether self-renewal of memory-like T cells is also increased, we purified CD8+CD44lo and CD8+CD44hi T cells and measured their proliferation in vitro. IL-15 induced similar proliferation of Elf4+/+ and Elf4−/− CD8+CD44hi T cells (Fig. 1c). In contrast, CD8+CD44lo T cells isolated from Elf4−/− mice proliferated more than Elf4+/+ CD8+CD44lo cells in response to TCR activation (Fig. 1c). Similar results were observed when adoptively transferred Elf4+/+ OT-1 CD8+CD44lo T cells or Elf4−/− OT-1 CD8+CD44lo CD8+ T cells were activated in vivo by intravenous administration of OVA257–264 peptide (Fig. 1d). This readiness to proliferate suggests a lower threshold of activation. In support of this hypothesis, Elf4−/− OT-1 CD8+ T cells underwent more proliferation than Elf4+/+ OT-1 CD8+ T cells in response to low concentrations of OVA257–264 peptide in vitro (Fig. 1e).
Figure 1. ELF4 negatively regulates the proliferation of naïve CD8+ T cells.
(a) Elf4+/+ and Elf4−/− mice were injected intraperitoneally with bromodeoxyuridine (BrdU). BrdU incorporation by indicated CD8+ T cell subsets was analyzed by flow cytometry (n=4, ** P < 0.01). A representative experiment is shown with four mice per group (mean and s.d.). (b) Proliferation of CFSE-labeled naïve Elf4+/+ and Elf4−/− CD8+ T cells was assessed 9 days after adoptive transfer into irradiated B6.SJL mice. The percentage of divided cells and the proliferation indices (PI) are indicated. Data are representative of three independent experiments. (c) Proliferation of CFSE-labeled Elf4+/+ and Elf4−/− CD8+CD44hi and CD8+CD44lo T cells stimulated with IL-15 in vitro for 7 days was measured by flow cytometry. Data are representative of at least three independent experiments. (d) OT-1 Elf4+/+ and Elf4−/− CD8+ T cells were labeled with CFSE and adoptively transferred into B6.SJL mice. One day later, recipient mice were injected with OVA257–264 peptide. Three days after OVA257–264 injection, CFSE dilution was assessed by flow cytometry. Data are representative of three independent experiments. (e) OT-1 Elf4+/+ and Elf4−/− CD8+ T cells were labeled with CFSE and incubated with B6.SJL splenocytes pulsed with the indicated concentrations of OVA257–264 peptide. CFSE dilution was measured by flow cytometry. Data are representative of two experiments. (f) Elf4+/+ and Elf4−/− CD8+ T cells were stimulated with anti-CD3 and anti-CD28 for the indicated time periods. Rb phosphorylation was assessed by immunoblot. β-actin, loading control. Data are representative of two experiments. (g) Expression of the indicated cyclins and CDK inhibitors in unstimulated Elf4+/+ and Elf4−/− CD8+ T cells was measured by immunoblot. Data are representative of two experiments. (h) Expression of cyclin D1 and cyclin D3 (magenta) in Elf4+/+ and Elf4−/− CD8+ T cells was assessed by immunofluorescence. DAPI (blue) staining marks the nuclei. Data are representative of two experiments.
Aberrant expression of factors that facilitate the G1- to S-phase transition can prime T cells to rapidly proliferate upon activation. TCR crosslinking induced faster phosphorylation of Rb in Elf4−/− CD8+ T cells than Elf4+/+ CD8+ T cells (Fig. 1f). Additionally, the expression of cyclins D1, D3 and E was increased, whereas the expression of the p21, p15 and p27 cell cycle inhibitors was reduced in Elf4−/− compared to Elf4+/+ CD8+ T cells (Fig. 1g). This finding, together with the accumulation of cyclins D1 and D3 in the cytosol of Elf4−/− CD8+ T cells, is consistent with the notion that Elf4−/− CD8+ T cells exist in a late G1 stage (Fig. 1h). Although we did not detect a substantial amount of phosphorylated Rb in unstimulated Elf4−/− CD8+ T cells, the BrdU incorporation experiments suggested a higher steady-state proliferation rate of this population (Fig. 1a). Collectively, we propose that the transcription factor ELF4 negatively regulates proliferation of naïve CD8+ T cells by raising the activation threshold. Thus, we investigated the effect of Elf4 deletion on the generation of memory-like and antigen-experienced memory T cells during homeostasis and antigen driven presentation, respectively.
ELF4 regulates naïve T cell quiescence
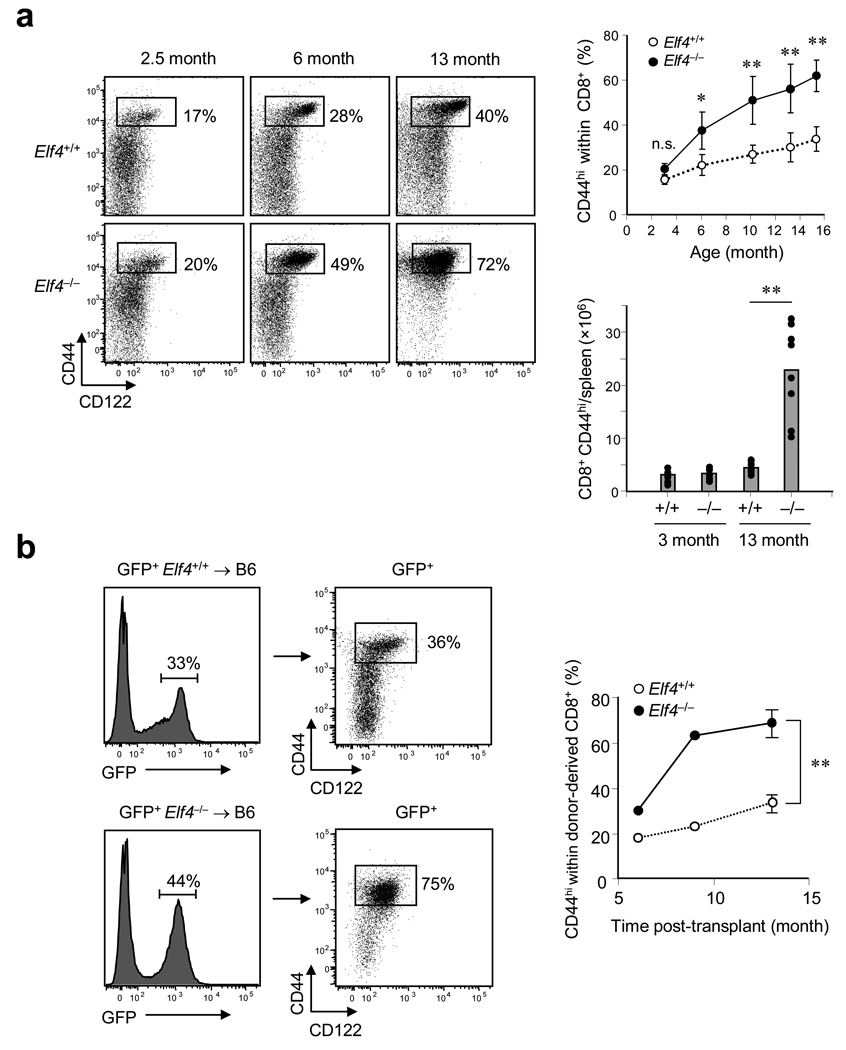
Homeostatic proliferation of naïve T cells contributes to the maintenance of the T cell pool, especially as thymic production wanes during aging, leading to accumulation of T cells with a memory-like phenotype 30, 37, 38. These memory-like CD8+CD44hiCD122+ T cells are detected even in mice kept in a pathogen-free environment. The percentage of CD44hiCD122+ cells within the CD8+ T cell pool gradually increases with age to constitute 30–40% of CD8+ T cells in the spleens of wild-type mice (Fig. 2a). The expansion of the CD8+CD44hiCD122+ T cell population was significantly accelerated in Elf4−/− mice, such that up to 70% of the total CD8+ T cell compartment in 13-month-old mice was CD44hiCD122+ (Fig. 2a). This accumulation correlated with a reduction in the number of naïve CD8+CD44lo T cells, suggesting that CD8+ T cells in Elf4−/− mice are spontaneously activated. In addition, Elf4−/− mice displayed splenomegaly and hyperplasia of the white pulp, accumulation of CD4+ and CD8+ T cells in the spleen, and lymphocytic infiltration of the liver, with age (Supplementary Fig. 2 online). To determine whether this T cell population expansion was due to T cell-intrinsic factors, we generated bone marrow (BM) chimeras using GFP+ Elf4+/+ or GFP+ Elf4−/− BM cells as donors and wild-type mice as recipients. The percentage of CD44hiCD122+ cells in the CD8+ T cell pool derived from Elf4−/− donor cells increased over time at a rate significantly greater than that of Elf4+/+ donor-derived cells, suggesting that the accelerated expansion of Elf4−/− T cell populations is T cell intrinsic and not due to an altered environment in Elf4−/− mice (Fig. 2b).
Figure 2. ELF4 restricts the homeostatic expansion of CD8+ T cell populations with a memory-like phenotype.
(a) The percentage of CD44hiCD122+ cells within the CD8+ T cell pool in Elf4+/+ and Elf4−/− spleens from mice of different ages (left) was determined by flow cytometry. The proportion and number of CD8+CD44hi T cells in the spleen is shown on the right (at least 3 mice were analyzed for each time point). A two-tailed Student’s t-test was used to compare Elf4+/+ and Elf4−/− mice (* P < 0.05, ** P < 0.01). Data are representative of three independent experiments. (b) BM cells from GFP-transgenic Elf4+/+ or Elf4−/− mice were transplanted into irradiated B6 mice. Expansion of the donor-derived CD8+CD44hiCD122+ T cell populations in the spleen was monitored at different times post-transplantation (n=3, ** P < 0.01). Data are representative of two independent experiments.
Outgrowth of autoreactive T cell clones could potentially account for the T cell population expansion observed in Elf4−/− mice. However, we noted a similar distribution of TCR Vβchains on CD8+CD44hi T cells in Elf4−/− mice compared to age-matched Elf4+/+ controls, suggesting that the T cell population was polyclonal rather than oligoclonal (Supplementary Fig. 3a online). We also examined the homeostatic expansion of Elf4−/− T cell populations specific for a foreign antigen by crossing Elf4−/− mice to OT-1 transgenic mice. OT-1 Elf4−/− mice accumulated CD8+CD44hiCD122+ T cells at a rate and to an extent similar to Elf4−/− mice (Supplementary Fig. 3b online). In addition, whereas the majority (77%) of CD8+CD44hi CD122+ T cells in OT-1 Elf4−/− mice stained with the H2-Kb-OVA257–264 tetramer, only 16% of CD8+CD44hiCD122+ T cells in OT-1 Elf4+/+ mice bound this tetramer. Therefore, ELF4 regulates T cell homeostasis, and Elf4−/− mice exhibit polyclonal T cell population expansion due to T cell-intrinsic defect(s).
ELF4 regulates the expression of KLF2
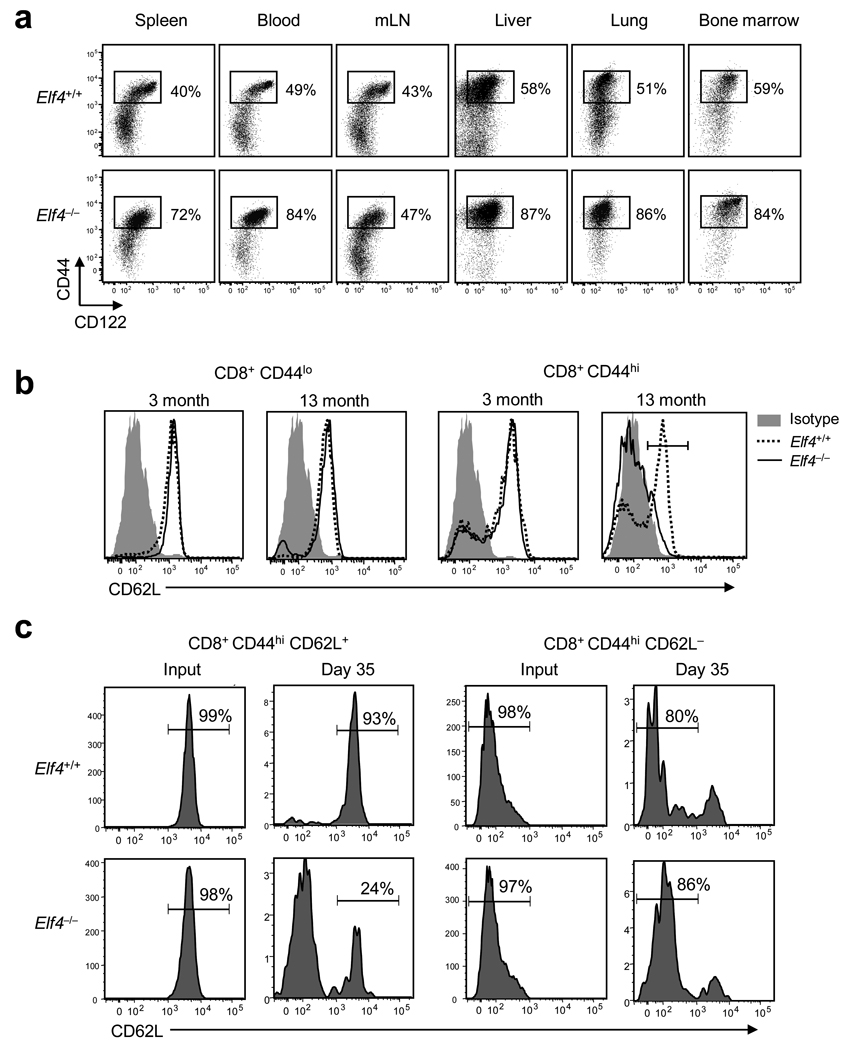
Next we analyzed T cell distribution in different tissues of aged Elf4+/+ and Elf4−/− mice because a continuously expanding T cell pool could induce a redistribution of T cells. In 13-month-old Elf4−/− mice, CD8+CD44hi T cells accumulated in the spleen, blood, liver, lung, and BM but not in the LNs (Fig. 3a). Even though there was no accumulation of cells in the LNs, the presence of some Elf4−/− memory-like T cells in this tissue could be due to entry via afferent lymphatics. Next, we examined the cell surface expression of CD62L—an indicator of LN homing potential—on Elf4−/− CD8+ T cells (Fig. 3b). Although CD8+CD44lo T cells demonstrated similar CD62L expression in 3-month and 13-month-old Elf4+/+ and Elf4−/− mice, the expression of CD62L was substantially downregulated on CD8+CD44hi T cells from 13-month-old Elf4−/− mice (Fig. 3b). This downregulation of CD62L was a gradual process that correlated with T cell expansion (Supplementary Fig. 4a online). CCR7 was also downregulated in CD8+CD44hi T cells from 13-month-old Elf4−/− mice (Supplementary Fig. 5 online). Expansion of CD44hi T cells in Elf4−/− is not exclusive of CD8+ T cells, CD4+CD44hi T cells also accumulated and lost CD62L and CCR7 expression over time (Supplementary Fig. 6 online). Loss of CD62L and CCR7 expression on CD8+CD44hi T cells suggests an age-dependent conversion of Tcm to Tem phenotype. To evaluate the relationship between these subsets, we tracked the cell fate of adoptively transferred memory T cell populations from Elf4+/+ and Elf4−/− mice. Tem cells purified from Elf4+/+ and Elf4−/− mice did not significantly acquire CD62L expression upon adoptive transfer (Fig. 3c). However, unlike Elf4+/+ Tcm cells, approximately 80% of the Elf4−/− Tcm cells lost CD62L from the cell surface by 35 days after adoptive transfer (Fig. 3c).
Figure 3. The gradual expansion of Elf4−/− CD8+CD44hi T cell populations leads to reduced CD62L expression.
(a) Distribution of CD8+CD44hiCD122+ T cells in lymphoid and non-lymphoid tissues of 13-month-old Elf4+/+ and Elf4−/− mice (mLN, mesenteric lymph node). Data are representative of two experiments. (b) Cell surface expression of CD62L on naïve (CD8+CD44lo) and memory-like (CD8+CD44hi) T cells from 3 and 13-month-old mice (54±2% CD62L+ in Elf4+/+ T cells versus 24±3% in Elf4−/− T cells, P < 0.01, n=4). Data are representative of five independent experiments. (c) The lineage relationship between memory T cell subsets was examined following adoptive transfer of Elf4+/+ and Elf4−/− CD8+CD44hiCD62L+ and CD8+CD44hiCD62L− cells into B6.SJL (CD45.1+) mice. CD62L expression in CD45.2+ donor-derived cells was analyzed after 35 days. Data are representative of at least two independent experiments.
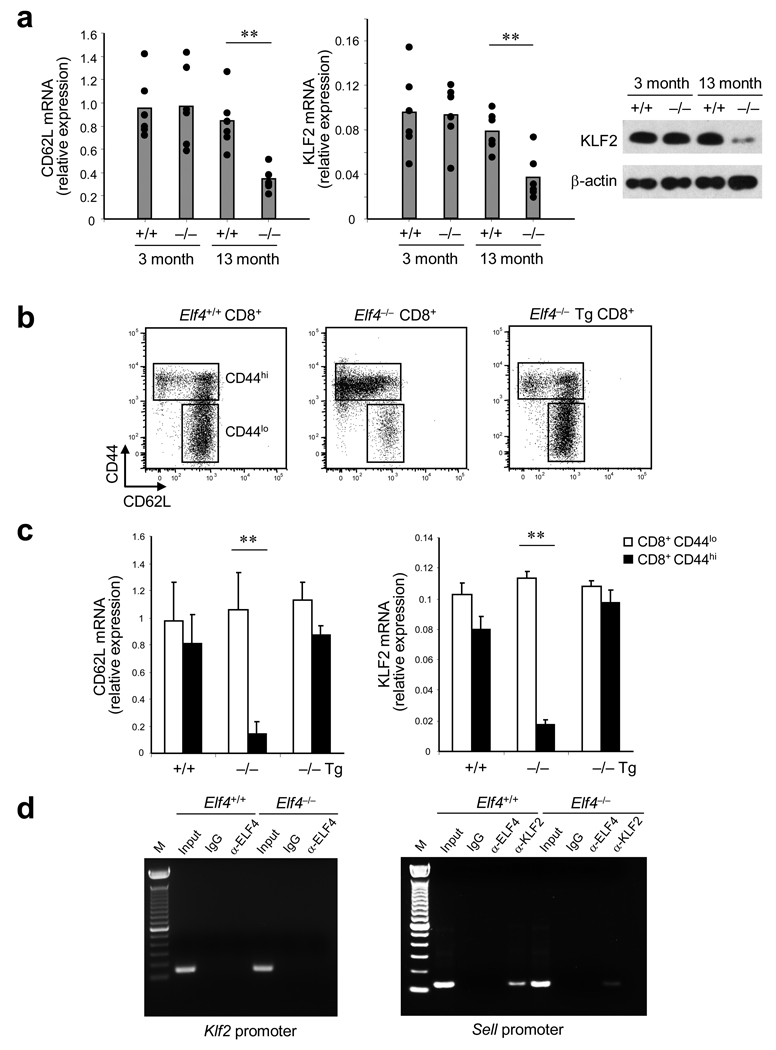
Homeostatic expansion of CD8+ T cell populations in Elf4−/− mice leads to the continuous accumulation of CD8+CD44hiCD62Llo T cells, suggesting that ELF4 may be regulating the expression of inducers of CD62L expression. Hence, we examined expression of KLF2—the main activator of CD62L expression in naïve T cells—in Elf4−/− CD8+ T cells 34, 35. CD62L and KLF2 mRNA and protein expression was reduced in CD8+ T cells isolated from 13-month-old Elf4−/− mice (Fig. 4a). To confirm that this effect was due to ELF4, Elf4−/− mice were crossed to Vav-ELF4 transgenic mice to restore ELF4 activity in the hematopoietic system. As predicted, the ectopic expression of ELF4 restored CD62L expression on the surface of CD8+CD44hi T cells of 13-month-old Elf4−/− mice (Fig. 4b). Reduced CD62L mRNA was observed in CD8+CD44hi T cells from 13-month-old Elf4−/− mice, and this reduction was prevented by transgenic expression of ELF4 (Fig. 4c). Even though ELF4 can activate the Klf2 promoter in vitro (Supplementary Fig. 4b online), we were unable to detect ELF4 bound to the Klf2 regulatory sequences in CD8+ T cells isolated from Elf4+/+ 13-month-old mice (Fig. 4d). However, the chromatin immunoprecipitation analyses demonstrated that KLF2, but not ELF4, bound to the Sell promoter in Elf4+/+ but not Elf4−/− CD8+ T cells (Fig. 4d). Of note, normal levels of Foxo1, another activator of CD62L and CCR7 expression in naïve T cells 39, were found in Elf4−/− CD8+CD44hi T cells (data not shown). Collectively, this suggests that the homeostatic expansion of CD8+ T cell populations with a memory-like phenotype results in downregulation of CD62L and CCR7, which diverts the excess CD8+CD44hi T cells from the LNs to the spleen and non-lymphoid tissues.
Figure 4. ELF4 regulates KLF2 and CD62L expression solely in CD8+CD44hi T cells from aged mice.
(a) KLF2 and CD62L expression relative to β-actin was measured by qPCR (left) and by immunoblot (right, KLF2 only) in CD8+ T cells from 3-and 13-month-old Elf4+/+ and Elf4−/− mice. Data are representative of three independent experiments. (b) The cell surface expression of CD62L was measured in CD8+ T cells from 13-month-old Elf4+/+ and Elf4−/− mice, and from Elf4−/− mice expressing a Vav-ELF4 transgene (Tg) by flow cytometry. The sorting gates of CD44lo and CD44hi CD8+ T cells used for the next experiment are indicated. Data are representative of two independent experiments in triplicates. (c) CD62L and KLF2 expression in purified CD8+CD44lo and CD8+CD44hi cells from 13-month-old Elf4+/+, Elf4−/−, and Elf4−/− Tg mice was measured using qPCR. Expression relative to β-actin is shown. Data are representative of two experiments in triplicates (mean and s.d.). (d) Promoter occupancy in the Klf2 (left) and Sell (right) regulatory sequences. Sheared DNA from Elf4+/+ and Elf4−/− CD8+ T cells isolated from 13-month-old mice was immunoprecipitated with anti-ELF4 and anti-KLF2 antibodies. The input DNA and immunoprecipitation with IgG control are also shown. Data are representative of at least three independent experiments.
ELF4 restricts memory after vaccination
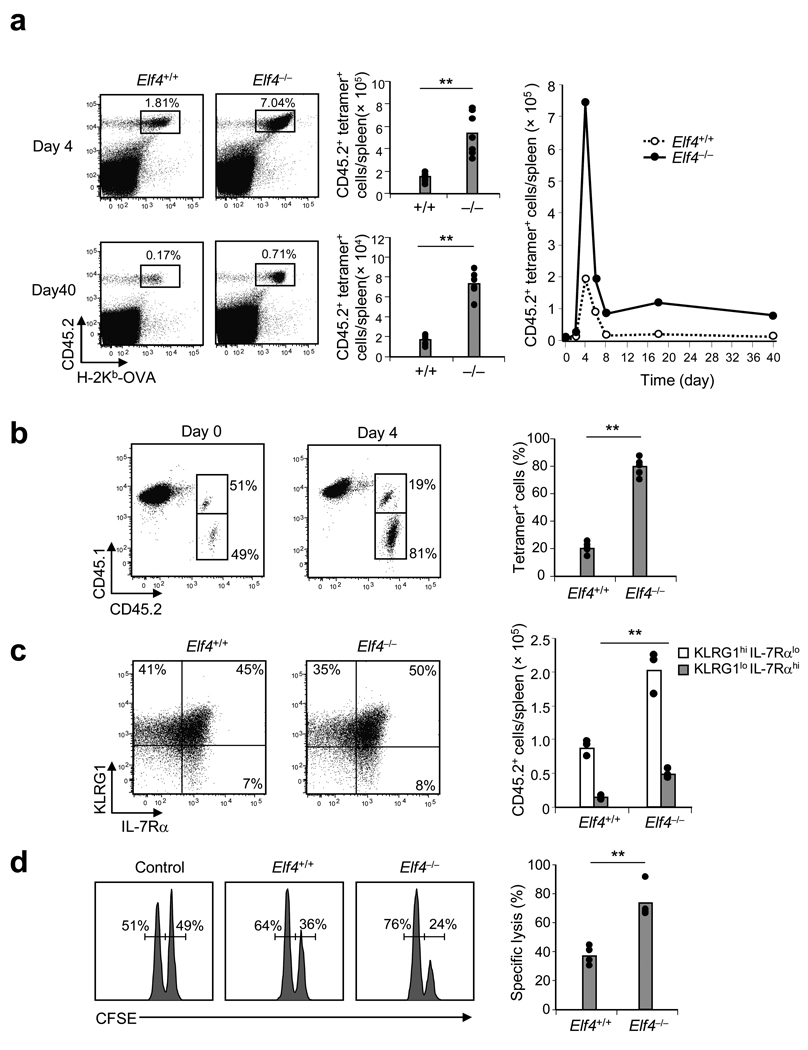
Cell cycle regulation plays a critical role in the clonal expansion of antigen-specific naïve T cell populations during infection or vaccination. To test whether ELF4 also regulates proliferation upon priming, we used dendritic cells (DC) pulsed with OVA257–264 peptide (DC-OVA) to immunize B6.SJL mice that had been adoptively transferred 24 hours earlier with CD8+ T cells isolated from either OVA-specific OT-1 Elf4+/+ or OT-1 Elf4−/− mice. H-2Kb-OVA tetramer staining demonstrated a significantly higher proportion and number of Elf4−/− compared to Elf4+/+ OVA-specific CD8+ T cells at the peak of expansion (day 4) and at day 40 (Fig. 5a and Supplementary Fig. 7 online). To rule out microenvironmental effects, we transferred a 1:1 mixture of Elf4+/+ and Elf4−/− OT-1 CD8+ T cells into B6.SJL recipients. Elf4+/+ CD8+ T cells were outcompeted by Elf4−/− CD8+ T cells following immunization in the same environment (Fig. 5b).
Figure 5. Elf4−/− CD8+ T cells generate more effector and memory T cells following immunization.
(a) Purified CD8+ T cells from OT-1 Elf4+/+ or Elf4−/− mice (CD45.2+) were transferred into B6.SJL mice (CD45.1+) and activated 24 h later with DCs pulsed with OVA257–264 peptide. The percent and number of donor-derived tetramer-positive cells 4 and 40 days after immunization is shown in the dot plots and bar graphs. The kinetics of OT-1 CD8+ T cell population expansion and contraction are also shown. Data are representative of at least three independent experiments. (b) CD8+ T cells from Elf4+/+ (CD45.1+CD45.2+) and Elf4−/− (CD45.1− CD45.2+) mice were co-transferred into B6.SJL mice (CD45.1+CD45.2−). One day later, recipient mice were injected with OVA257–264 -pulsed DCs and donor CD8+ T cells were examined at day 4 (n=5, ** P < 0.01). Data are representative of two experiments. (c) The number of effectors (KLRG1hi IL-7Rαlo) and memory precursor cells (KLRG1lo IL-7Rαhi) in mice adoptively transferred with OT-1 Elf4+/+ or OT-1 Elf4−/− CD8+ T cells was determined by flow cytometry at day 4 after immunization with OVA-pulsed DCs (n=3, ** P < 0.01). Data are representative of two experiments. (d) OT-1 Elf4+/+ or Elf4−/− mice were immunized or not (Control) 40 days prior to injection with a 1:1 mixture of CFSEdim-labeled control cells and CFSEbright-labeled target cells. The specific lysis was calculated on the basis of flow cytometric enumeration of the CFSEdim and CFSEbright cells in the spleen (n=4, ** P < 0.01). Data are representative of two independent experiments.
Our data suggest that the naïve Elf4−/− CD8+ T cell populations did not exhaust the memory precursor cells (MPEC) during differentiation to effector T cells. Even though the distribution of short-lived effector (SLEC) and MPEC did not differ significantly between Elf4+/+ and Elf4−/− donor-derived CD8+ T cells, the overall expansion of both subsets was greater for Elf4−/− CD8+ T cells (Fig. 5c). A caveat to this determination is that not all MPEC identified in this way become memory T cells following DC vaccination 40, 41. Despite this limitation, our MPEC accumulation findings correlated with the increased number of tetramer positive CD8+ T cells detected at late time points after vaccination. We next performed in vivo cytolytic assays to evaluate whether Elf4−/− memory CD8+ T cells generated after vaccination can recognize and lyse antigen-bearing target cells (CFSEbright OVA-pulsed splenocytes). Compared to the OVA-specific memory CD8+ T cells generated from Elf4+/+ OT-1 cells, those generated from Elf4−/− OT-1 CD8+ T cells more efficiently lysed antigen-bearing target cells in vivo (Fig. 5d). However, this assay was not analyzed on a cell-to-cell basis; thus the apparent higher specific lysis by Elf4−/− memory T cells was due to their increased frequency in the spleen. Therefore, our data indicate that ELF4 negatively regulates the clonal proliferation of naïve CD8+ T cells and the generation of functional memory T cells after immunization.
KLF4 inhibits proliferation of CD8+ T cells
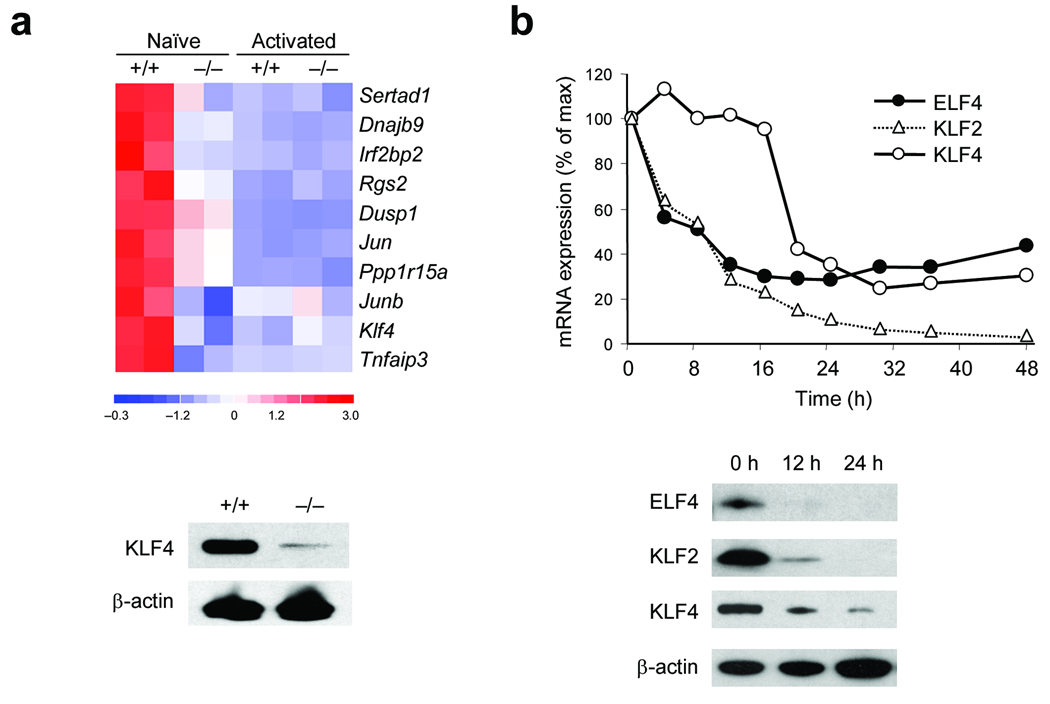
We hypothesized that ELF4 most likely activates the expression of an inhibitor of proliferation, because ELF4 does not have a known direct role in the progression of the cell cycle. In an effort to identify ELF4 target genes, we performed global gene expression analysis using RNA isolated from resting and in vitro activated CD8+ T cells. Most of the differences Elf4+/+ and Elf4−/− CD8+ T cells were detected in the non-activated populations (Fig. 6a). Naïve Elf4−/− CD8+ T cells expressed higher amounts of factors that promote passage through the G1-to -S-phase transition and lower amounts of factors that inhibit cell cycle progression (Supplementary Fig. 8 online). In addition, the expression of KLF4 was significantly downregulated in Elf4−/− CD8+ T cells. We decided to further investigate KLF4 given its homology to KLF2 and its dual function as tumor suppressor and oncogene 11, 42.
Figure 6. ELF4 inhibits T cell proliferation by activating the tumor suppressor KLF4 downstream of TCR signaling.
(a) Heat map of the 10 most downregulated genes comparing duplicates of naïve and activated CD8+ T cells from Elf4+/+ and Elf4−/− mice by microarray analysis (top). KLF4 expression in naïve CD8+ T cells was confirmed by immunoblot (bottom). (b) Expression of ELF4, KLF2, and KLF4 mRNA (top) and protein (bottom) at indicated times after TCR activation. mRNA was measured by qPCR and protein was measured by immunoblot. Data are representative of three independent experiments.
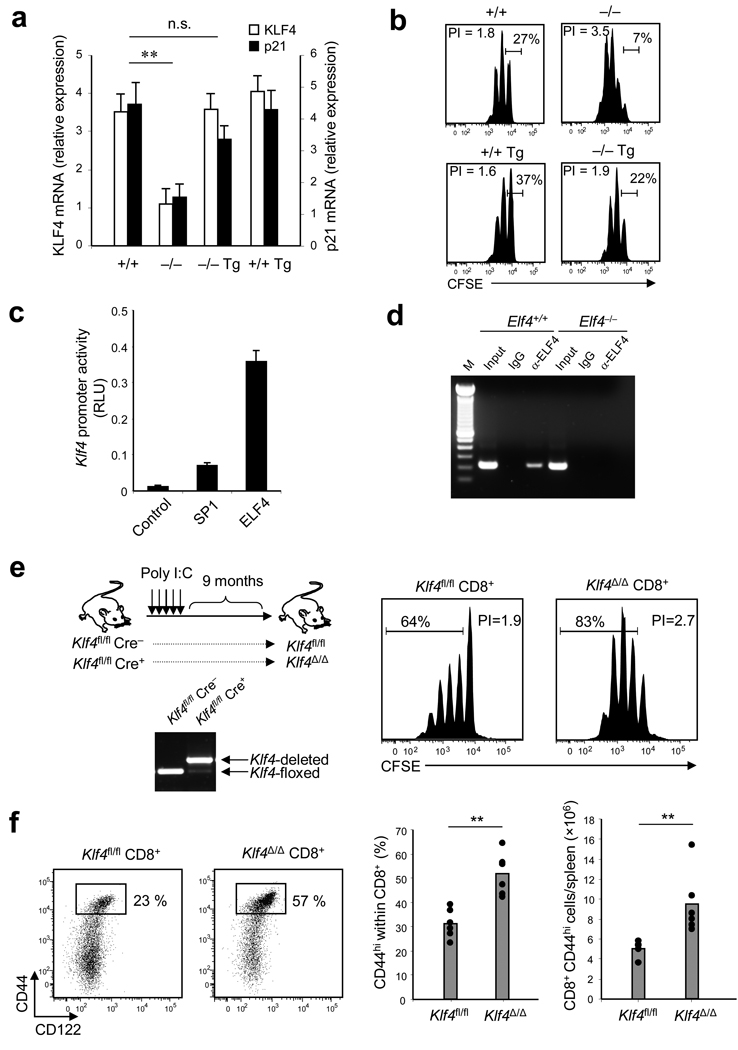
If ELF4 negatively regulates T cell proliferation by directly inducing expression of KLF4, this ELF4-mediated suppression of proliferation must be blocked during T cell activation. In agreement with this hypothesis, the expression of ELF4 and KLF2 decreased immediately after TCR activation, whereas expression of KLF4 decreased 16–20 hours later (Fig. 6b). In further support of this model, the expression of KLF4 and p21 inversely correlated with the extent of proliferation of CD8+ T cells from Elf4−/− and Elf4−/− Vav-ELF4 mice (Fig. 7a). Ectopic expression of ELF4 restored the expression of KLF4 and p21 and normalized the proliferation of Elf4−/− CD8+ T cells by increasing the percentage of non-proliferative T cells (Fig.7b). This finding suggests that the hyperproliferation of Elf4−/− CD8+ T cells is due to defective expression of KLF4 and p21 (Fig. 7a). Reporter assays demonstrated that Klf4 transcription is induced by ELF4, as well as by the known activator SP1 (Fig. 7c)43. To further confirm the direct regulation of Klf4 by ELF4 in vivo, we performed ChIP assays to evaluate Klf4 promoter occupancy. We detected Klf4 regulatory sequences by PCR using primers that span the Klf4 promoter among the DNA immunoprecipitated with anti-ELF4 (Fig. 7d). These data indicate that KLF4 maintains T cell quiescence downstream of ELF4, perhaps by directly activating p21 expression in CD8+ T cells.
Figure 7. KLF4, a direct target of ELF4, negatively regulates the proliferation of naïve CD8+ T cells.
(a) CD8+ T cells were isolated from Elf4+/+, Elf4−/−, Vav-ELF4 transgenic (Tg), and Elf4−/− Tg mice. KLF4 and p21 transcripts were measured by real-time PCR and expressed as relative expression to β-actin. Data are representative of at least two experiments in triplicates (mean and s.d.). (b) CD8+ T cells from (a) were labeled with CFSE and stimulated with anti-CD3 and anti-CD28 in vitro for 3 days. CFSE dilution was measured by flow cytometry. The percentage of non-proliferative T cells is indicated. Data are representative of two experiments. (c) COS-7 cells expressing a luciferase construct driven by the Klf4 promoter were transfected with empty vector (Control) or with vectors encoding SP1 or ELF4 proteins. Data are representative of two independent experiments. (d) Sheared DNA from Elf4+/+ and Elf4−/− CD8+ T cells was immunoprecipitated with an ELF4-specific antibody or an immunoglobulin (IgG) control, and DNA was amplified with primers spanning the Klf4 proximal promoter. Input, total DNA without immunoprecipitation. Results are representative of three experiments. (e) Klf4fl/fl Cre+ and Klf4fl/fl Cre− mice were injected with polyI:C to induce gene deletion. Deletion was confirmed by PCR of genomic DNA isolated from CD8+ T cells after 9 months of treatment. CD8+CD44lo T cells from these mice were labeled with CFSE and stimulated in vitro with anti-CD3 and anti-CD28 for 3 days. CFSE dilution was measured by flow cytometry. (f) Thirteen months after polyI:C-induced Klf4 gene deletion, expression of CD44 and CD122 on splenic CD8+ T cells was measured by flow cytometry (n=6, **P < 0.01). Data are representative of at least three independent experiments.
To directly assess the influence of KLF4 on T cell proliferation, we generated Klf4-deficient mice to test the effect of KLF4 in T cell proliferation. We crossed Klf4-floxed mice (Klf4fl/fl) to Mx1-Cre (Cre) transgenic mice to induce deletion of the Klf4 gene in adult mice with polyI:C. The efficacy of Klf4 gene deletion was confirmed by PCR using genomic DNA isolated from purified CD8+ T cells (Fig. 7e). Klf4fl/+ Mx1-Cre+ mice were not used as controls because Elf4 heterozygosis leads to expansion of CD8+CD44hiCD122+ T cells (Supplementary Fig. 9 online). The effect of transient Cre expression on T cell expansion was indirectly evaluated using CD8+ T cells from CD4-Cre transgenic mice (Supplementary Fig. 9 online). In vitro TCR cross-linking experiments demonstrated that CD8+ T cells from Klf4Δ/Δ mice proliferated more than the Klf4fl/fl controls (Fig. 7e). Furthermore, Klf4-deficient mice exhibited increased homeostatic expansion of CD8+CD44+ T cells in the spleen, similar to that observed in aged Elf4−/− mice (Fig. 7f). Our data collectively indicate that circulating CD8+ T cells remain quiescent due to the coordinated expression of ELF4 and KLF4, and that TCR activation disrupts this inhibition of proliferation by repressing ELF4 expression.
DISCUSSION
Here we demonstrated that ELF4 controls homeostatic and antigen-driven T cell proliferation and T cell homing by inducing expression of KLF4 and KLF2, respectively. ELF4 transactivates the tumor suppressor Klf4 in naïve CD8+ T cells, which is needed to maintain the quiescent state. Loss of ELF4 leads to deregulated T cell population expansion during homeostasis and after immunization, likely due to enrichment of factors that promoting progression through the G1-to S-phase transition. Similarly, deletion of Klf4 results in increased homeostatic expansion of CD8+CD44hi T cell populations and deregulated proliferation of CD8+CD44lo T cells upon activation. This inhibition of proliferation is disrupted upon T cell activation by inhibition of the Elf4 and Klf4 genes. In addition, we observed a progressive shift in the tissue distribution of CD8+CD44hi T cells due to loss of KLF2, CCR7 and CD62L expression; KLF2-deficient cells homed to non-lymphoid tissues, presumably in order to accommodate a growing pool of memory-like T cells.
This work suggests that the tumor suppressor KLF4 is a negative regulator of naïve CD8+ T cell proliferation downstream of ELF4. These intrinsic regulatory factors not only prevent homeostatic proliferation in response to weak self-MHC signals but also prevent clonal expansion of naïve CD8+ T cells and the generation of immunological memory. Adoptive transfer experiments demonstrated that ELF4-dependent homeostatic expansion of CD8+ T cells is independent of environmental factors, some of which (e.g. IL-7 and IL-15) are important extrinsic regulators of homeostatic T cell proliferation 2. The enhanced antigen-driven proliferation of Elf4−/− CD8+ T cells is also cell autonomous, as immunization of recipients of both Elf4−/− and Elf4+/+ OT-1 CD8+ T cells resulted in Elf4−/− OT-1 CD8+ T cells out-competing Elf4+/+ OT-1 CD8+ T cells in the same environment.
In addition to CD4+ and CD8+ T cells, ELF4 regulates proliferation of B cells 9, ovarian epithelial cancer cell lines 44, and HSCs 10. ELF4 and c-Myb are the only known negative regulators of HSC quiescence during homeostasis 10, 45. In contrast to HSCs, Elf4−/− naïve T cells remain in the late-G1 stage poised to proliferate. We demonstrated that the tumor suppressor Klf4 is directly regulated by ELF4, and that KLF4 expression correlates with expression of the p21 cell cycle inhibitor in naïve T cells. Interestingly, the ‘pre-activated’ state of Elf4−/− CD8+ T cells is reminiscent of memory T cells 46. There are no previous reports describing a role for KLF4 in T cell proliferation, although Klf4 has been shown to suppress B cell proliferation as a target of the FOXO transcription factor 47.
Recent reports have identified cell surface molecules that are required for trafficking of naïve T cells to the LNs and inflamed tissues 34, 35, 36, 48. Even though KLF2 is the best-known trafficking ‘controller’ in naïve T cells, the role of KLF2 in tissue homing and the relationship among different subsets of memory T cells was unknown. Deletion of Elf4 led to the loss of KLF2, CCR7 and CD62L expression in CD8+CD44hi T cells. We demonstrated that even though KLF2 regulates CD62L in CD8+ T cells, ELF4 does not activate KLF2 expression in spite of its ability to activate the Klf2 promoter in vitro. Genes activated by ETS proteins in vitro often do not correlate with their role in vivo due to cell specific transcriptional machinery and functional redundancy. Therefore, the increased homeostatic proliferation of CD8+ T cells triggers downregulation of KLF2 and accumulation of CD8+CD44hiCD62Llo T cells, presumably in order to prevent LN swelling. In fact, it was recently shown that exacerbated expansion of memory CD8+ T cell populations results in an increase in the numbers of CD8+CD44hiCD62Llo T cells in the spleen and non-lymphoid tissues but not in the LNs 49. Interestingly, loss of CD62L expression has been detected in T lymphocytes from elderly humans, and therefore Elf4 deletion may accelerate the normal process of aging in the immune system 50. The signals that regulate the size of the CD8+ T cell pool in the LNs are largely unknown, although data obtained in the present study support the argument that ELF4 prevents CD8+ T cells from reaching the size limit of the T cell pool. Our findings also support the hypothesis that the size of the CD8+ T cell pool is flexible rather than invariable as was previously thought 49. Our findings also raise the question of whether ELF4 regulates the relationship between Tcm and Tem cells, and the retention of Tcm cells in the LNs. However, it is possible that this apparent conversion of Tcm into Tem cells actually represents Tcm cells that have lost CD62L and CCR7 expression but have retained all other features of Tcm cells.
In summary, our study identified two important functions of the transcription factor ELF4. First, ELF4 acts as a negative regulator of naïve CD8+ T cell proliferation by directly activating expression of Klf4. Second, by inducing expression of KLF2, ELF4 maintains CD62L and CCR7 expression on memory T cells. In this way, ELF4 regulates proliferation in naïve T cells and homing to the LN in memory T cells. A detailed map of the transcriptional machinery involved in the generation of antigen-specific memory CD8+ T cells is needed to improve long-lasting immune protection. Hence, modulation of ELF4 or KLF4 expression could be used in the future to heighten immune response and immunological memory by augmenting the clonal expansion of antigen-specific cells without promoting their terminal differentiation into effector T cells.
METHODS
Mice
Elf4−/− and Vav-ELF4 mice were obtained from S. Nimer (Memorial Sloan-Kettering Cancer Center, New York) (backcrossed for more than 12 generations to C57BL/6 background). Klf4fl/+ mice were obtained from K. Kaestner (University of Pennsylvania, Philadelphia) 51. C57BL/6 (B6), B6.SJL, OT-1 TCR transgenic and GFP transgenic mice were purchased from Jackson Laboratories. Klf4fl/fl mice were crossed with Mx1-Cre transgenic mice (Jackson Laboratories) to induce deletion. The OT-1 and GFP transgenic mice were crossed with Elf4−/− mice to generate OT-1 Elf4−/− and GFP+ Elf4−/− mice. For adoptive transfers, B6 mice were crossed with B6.SJL mice to generate Elf4+/+ CD45.1+ CD45.2+ mice. Mice were irradiated using a Gammacell 40 Exactor (Nordion) irradiator for BM transplantation (9.5 Gy) or to induce lymphopenia (4.5 Gy). All mice were bred and maintained under specific pathogen-free conditions at the Baylor College of Medicine. All experiments were performed with the approval of the Institutional Animal Care and Usage Committee of Baylor College of Medicine.
Flow cytometry
The following fluorescence- or biotin-labeled antibodies were used: anti-CD44-phycoerythrin (PE) (IM7), anti-CD45.1-PE (A20), anti-CD45.2-FITC (104), anti-CD122-FITC (TM-β1), anti-CD8-APC (53-6.7), anti-KLRG1-biotin (2F1), anti-CD127-PE (SB/199) and anti-CD62L-APC (MEL-14). All antibodies and streptavidin-APC used in this study were purchased from BD Biosciences. CCL19-Fc and PE-labeled anti-human IgG Fcγ fragment were used for CCR7 staining (eBioscience). TCR Vβ repertoire was analyzed using a panel of mouse TCR Vβ chain-specific antibodies (BD Biosciences). PE-labeled H2-Kb-OVA257– 264 tetramer was obtained from the MHC Tetramer Core Laboratory (Baylor College of Medicine). Samples were analyzed by flow cytometry using a FACSCanto instrument (BD Biosciences) and FlowJo software (Tree Star). CFSE dilution was analyzed using ModFit software (Verity).
Purification of CD8+ T cells
CD8+ T cells were purified from spleen using the negative selection BD-IMag magnetic-bead separation system (BD Biosciences) following the manufacturer’s instructions. Purified CD8+ T cells were stained with PE-labeled anti-CD44 to purify CD8+CD44hi (memory-like) and CD8+CD44lo (naïve) T cells using a MoFlo cell sorter (Cytomation Inc.).
Adoptive transfer
CD8+CD44hiCD62L+ and CD8+CD44hiCD62L− T cells were purified by cell sorting from CD45.2+ mice. Purified cells (1 × 104) were adoptively transferred into non-irradiated B6.SJL mice (CD45.1+). Cell fate was analyzed 35 days later based on the expression of CD62L in donor-derived T cells (CD45.2+).
T cell proliferation
CD8+ T cells isolated from spleen were labeled with 4 µM of carboxyfluorescein diacetate succinimidyl ester (CFSE, Invitrogen) in PBS with 0.1% BSA at 37°C for 10 min. CFSE was removed by washing cells with ice-cold RPMI containing 10% fetal bovine serum. For in vitro stimulation, CD8+CD44lo T cells were isolated from 3-month-old mice, labeled with CFSE and cultured in anti-CD3 coated 96-well plates (Bio-Coat, BD Biosciences) in the presence of 2 µg/ml of anti-CD28 (BD Biosciences) at 1 × 105 cells/well in RPMI containing 10% FBS and 5% T-Stim (BD Biosciences). CD8+CD44hi T cells were stimulated with rmIL-15 (20 ng/ml) (R&D Systems) for 7 days. For in vivo stimulation, 5 × 106 CFSE-labeled CD8+ T cells from 3-month-old OT-1 Elf4+/+ or OT-1 Elf4−/− mice (CD45.2+) were adoptively transferred intravenously into recipient B6.SJL (CD45.1+) mice followed by activation with 50 ng of OVA257– 264 peptide (SIINFEKL) (ANASpec) the following day. CFSE dilution was examined by flow cytometry after 3 days of activation. To evaluate homeostatic proliferation of naïve T cells in lymphopenic hosts, CD8+CD44lo T cells (2 × 106) from young CD45.2+ Elf4+/+ or CD45.2+ Elf4−/− mice were labeled with CFSE and transferred into lymphopenic B6.SJL (CD45.1+) mice (irradiated with 4.5 Gy). After 9 days, CFSE dilution was measured in donor-derived cells (CD45.2+) in the spleen. For in vitro stimulation with OVA257–264 peptide, splenocytes from B6.SJL mice were pulsed with different concentrations of OVA257–264 peptide at 37°C for 1 h. CFSE-labeled OT-1 CD8+ T cells were then stimulated with OVA257–264 -pulsed splenocytes. CFSE dilution in CD45.2+ cells was analyzed by flow cytometry 3 days later. Percentage of proliferation was calculated respect to the maximum proliferation induced by 1,000 ng/ml OVA257–264 peptide.
Immunization with peptide-pulsed DCs
DCs were generated from BM cells cultured with 10 ng/ml of mGM-CSF (Peprotech Inc.). Media was hemi-replenished every 2 days and non-adherent cells were transferred to a new plate at day 6. DCs were activated with 100 ng/ml of LPS (Sigma) for 18 hours and activation (CD11c+ CD11b+ MHC-class IIhi phenotype) was confirmed by flow cytometry. Finally, DCs were pulsed with 5 µg/ml of OVA257–264 peptide at 37°C for 1 hour and washed twice with PBS. CD8+ T cells were purified from OT-1 Elf4+/+ and OT-1 Elf4−/− mice and adoptively transferred into B6.SJL mice (1 × 105 cells/mouse). Twenty-four hours later, mice were immunized with 1 × 106 peptide-pulsed DCs. Expansion of CD8+ T cells in spleen was monitored at different times following immunization by flow cytometry using anti-CD8-APC, anti-CD45.2-FITC and PE-labeled H2-Kb-OVA tetramer.
In vivo cytotoxicity assay
The in vivo killing assay was performed as previously described 52. Briefly, splenocytes were labeled with two concentrations of CFSE (2 µM or 8 µM) to generate dim and bright populations. To generate target cells, CFSE-bright cells were pulsed with 5 µg/ml of OVA peptide for 1 h at 37 °C and washed twice with 10-fold PBS. The CFSE-dim population was not pulsed with OVA peptide. A mixture of CFSE-dim cells (5 × 106) and CFSE-bright cells (5 × 106) was injected intravenously into mice that were immunized with OVA peptide-pulsed DCs 40 days earlier. CFSE profiles were analyzed in spleen 16 hours after transfer. Specific lysis was calculated by the following formula: Specific lysis = [(% of CFSEbright - % of CFSEdim)/ % of CFSEdim×100].
Isolation of lymphocytes from liver and lung
Livers and lungs were perfused after dissection by injecting 5–10 ml of ice-cold PBS into the hepatic artery or into the ventricle of lung, respectively. Both tissues were homogenized using a pestle motor mixer (VWR) in PBS. After washing with PBS, homogenized tissues were incubated in RPMI containing 5% FBS, 0.5 mg/ml of collagenase IV (Sigma-Aldrich) and 1.5 U/ml of DNase I (Qiagen) at 37 °C for 45–60 min. Then, digested tissues were applied to a Ficoll type 40 (Sigma-Aldrich) and centrifuged at 1,900 r.p.m. at 20 °C for 20 min. Lymphocytes were harvested from the interface.
BM transplantation
Bone marrow cells were extracted from femurs and tibias and placed in DMEM (Lonza) with 2% FBS. Cell suspension was filtered through 40-micron cell strainers (BD Falcon). Five million BM cells were injected intravenously into the lateral tail vein of recipient mice that were previously irradiated (4.5 Gy).
Immunoblot
CD8+ T cells were lysed directly with LDS sample buffer (Invitrogen) at 1 × 104 cells /µl and sonicated to shear DNA. 10 µl of cell lysates were loaded onto NuPAGE Bis-Tris gels (Invitrogen) and proteins were transferred to PVDF membrane. Specific proteins were detected using rabbit polyclonal anti-KLF2 (Aviva Systems Biology) and anti-actin (Sigma-Aldrich). For the analysis of cell cycle regulators, rabbit polyclonal anti-cyclin E, anti-cyclin A, and anti-p21 were purchased from eBioscience. Rabbit polyclonal anti-p27 and mouse anti-cyclin D1, anti-cyclin D3, and anti-cdk4 were purchased from Cell Signaling Technology. To analyze phosphorylation of Rb protein, CD8+ T cells were activated in anti-CD3 coated plates with soluble anti-CD28 and lysates were analyzed using rabbit polyclonal anti-phospho-Rb (Cell Signaling Technology). Goat anti-rabbit IgG and goat anti-mouse IgG conjugated with peroxidase (Pierce) were used as secondary antibodies. X-ray films were developed using immunoblotting detection reagent (Westdura).
Quantitative real-time PCR
Total RNA was extracted from CD8+ T cells by RNeasy Mini kit (Qiagen). Then cDNA was syntyhesized from 100–500 ng RNA with random hexamer primers using SuperScript III kit (Invitrogen). Quantitative real time PCR was performed using LightCycler FastStart DNA Master SYBR Green I (Roche) as specified by the manufacturer. Primer sequences for PCR were as follows: β-actin forward (5’-GTGGGCCGCTCTAGGCACCA-3’) and reverse (5’-CGGTTGGCCTTAGGGTTCAGGGG-3’), Elf4 forward (5’-CGGAAGTGCTTTCAGACTCC-3’) and reverse (5’-GGTCAGTGACAGGTGAGGTA-3’), CD62L forward (5’-CATTCCTGTAGCCGTCATGG-3’) and reverse (5’-AGGAGGAGCTGTTGGTCATG-3’), Klf2 forward (5’-CCAACTGCGGCAAGACCTAC-3’) and reverse (5’-AGTCGACCCAGGCTACATGTG-3’), Klf4 forward (5’-CTGAACAGCAGGGACTGTCA-3’) and reverse (5’-GTGTGGGTGGCTGTTCTTTT-3’), p21 forward (5’-CTGTCTTGCACTCTGGTGTCTGAG-3’) and reverse (5’-TTTCTCTTGCAGAAGACCAATCTG-3’). Reactions were run on a Smart Cycler (Cepheid). Thermocycler conditions consisted of an initial step at 95 °C for 10 min, followed by 35 cycles of a 3-step PCR program consisting of 95° C for 15 sec, 55 °C for 30 sec and 72 °C for 30 sec. Relative expression was calculated by normalizing with β-actin expression.
Chromatin Immunoprecipitation
To examine DNA binding of ELF4 or KLF2 protein, ChIP assay was performed using the magnetic chromatin immunoprecipitation kit, ChIP-IT express (Active Motif). Fixed and sheared DNA from wild type and Elf4−/− CD8+ T cells was prepared and ELF4 or KLF2 was immunoprecipitated. Immunoprecipitated chromatin was amplified by PCR using primers spanning the Sell (CD62L), Klf2 or Klf4 promoters. Primer sequences for PCR were as follows: Sell promoter forward (5’-CTCTCCCTTTCCTTTCCTCCTTCC-3’) and reverse (5’-CTCTCCCCACCCCTTTCC-3’), Klf2 promoter forward (5’-AGTTCGCCAATGTTTCTGCT-3’) and reverse (5’-GCATTGAAGCTGTGACCTGA-3’), Klf4 promoter forward (5’-CCCACTTCATTGGGAAGAGA-3’) and reverse (5’-GGACTGAAGAAAGGCACTCG-3’). Thermocycler conditions consisted of an initial step at 94 °C for 10 min, followed by 32 cycles of a 3-step PCR program consisting of 94 °C for 20 sec, 59 °C for 30 sec and 72 °C for 30 sec.
Promoter assay
Elf4, Mef2c and Sp1 cDNAs were cloned into the expression vector pcDNA 3.1/V5-His A (Invitrogen). Klf2 and Klf4 promoter regions were cloned from mouse genomic DNA into pGL3-Basic vector (Promega). pGL3-Klf2 consists of 1.8 kb containing 43 bp of the 5’ non-coding region and pGL3-Klf4 consists of 2.1 kb containing 542 bp of the 5’ non-coding region. COS-7 cells were transfected with a mixture of expression vector (200ng), promoter plasmid (20ng) and 4ng of renilla luciferase expression plasmid (pRL-CMV, Promega) using Lipofectamine 2000 (Invitrogen). After 48 hours, all transfectants were analyzed by dual-luciferase reporter assay system (Promega) with luminometer (Sirius, Berthold). Transfection efficiency was normalized with renilla luciferase. The expression of exogeneous ELF4, SP1 and MEF2C was confirmed by immunoblot.
Induction of Klf4 deletion
Klf4fl/fl Mx1-Cre+ mice (2–5 week old) were injected i.p. with poly I:C (300 µg every other day for a total of five times) as described 53. Gene deletion was confirmed by PCR using genomic DNA from CD8+ T cells and three primers: Klf4 exon1, CTGGGCCCCCACATTAATGAG; Klf4 exon2, CGCTGACAGCCATGTCAGACT; Klf4 intron3, CAGAGCCGTTCTGGCTGTTTT as described 51.
Immunocytochemistry
Naïve CD8+ T cells from mice were spun down onto glass slides, fixed with 1% paraformaldehyde and permeabilized with 0.1% Triton X-100. Slides were stained with cyclin D1-or cyclin D3-specific antibodies and mouse-specific antibody conjugated with Alexa-594. Nuclei were stained with DAPI.
BrdU incorporation
BrdU (2mg) was intraperitonealy injected into 2-month-old Elf4+/+ and Elf4−/− mice every other day for a total 8 times. Splenocytes were stained with anti-CD8-PE, anti-CD44-PerCP-Cy5.5. Then, BrdU incorporation was measured by flow cytometry using the APC BrdU flow kit (BD).
Microarray analysis
Total RNA was extracted from CD8+ T cells purified from Elf4+/+ and Elf4−/− mice using RNeasy Mini kit (QIAGEN). RNA probes were used to hybridize Affymetrix gene chips (mouse 430 version 2.0) in the Microarray Core Facility at Baylor College of Medicine. Data was analyzed using Bioconductor to compare mRNA expression level.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Finegold for his support, T. Matano (Tokyo, Japan) for his support from the Japanese Foundation for AIDS prevention (to T.Y.), S. Nimer for kindly providing Elf4−/− and Vav-ELF4 mice (New York, NY), and K. Kaestner for providing Klf4-floxed mice (Philadelphia, PA). We are grateful to M. Puppi and L. Bae for their valuable assistance, M. Cubbage, C. Threeton and T. Goltsova for cell sorting, the Gulf Coast Digestive Disease Morphology Core, and T-A. Mistretta for microarray analysis. We are also indebted to C. Rooney for critical review of the manuscript. This work was funded by a Howard Temin Award from NCI/ NIH (KO1 CA099156-01 to H.D.L.), a Pilot Project of the Dan Duncan Cancer Center in Baylor College of Medicine to H.D.L, the Curtis Hankamer Basic Research Fund (Junior-Faculty Seed Fund Award, BCM) to H.D.L., and 1RO1AI077536-01 (NIAID) to H.D.L.
Footnotes
AUTHOR CONTRIBUTIONS
T.Y. designed and performed most experiments, analyzed data, and assisted in writing of the manuscript, C.S.P. contributed to promoter assays, deletion of Klf4 gene, and reviewed manuscript, M.M. contributed to promoter assays, performed flow cytometric detection of CCR7 in CD4+ and CD8+ T cells and reviewed manuscript, and H.D.L. conceptualized the research, designed experiments, directed the project as principal investigator, wrote the manuscript and funded the research.
ACCESSION CODES. GEO: microarray data, GSE15324
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Tzachanis D, Lafuente EM, Li L, Boussiotis VA. Intrinsic and extrinsic regulation of T lymphocyte quiescence. Leuk Lymphoma. 2004;45:1959–1967. doi: 10.1080/1042819042000219494. [DOI] [PubMed] [Google Scholar]
- 2.Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29:848–862. doi: 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Lacorazza HD, Nimer SD. The emerging role of the myeloid Elf-1 like transcription factor in hematopoiesis. Blood Cells Mol Dis. 2003;31:342–350. doi: 10.1016/s1079-9796(03)00162-1. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y, Hedvat CV, Mao S, Zhu XH, Yao J, Nguyen H, Koff A, Nimer SD. The ETS protein MEF is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCFSkp2. Mol Cell Biol. 2006;26:3114–3123. doi: 10.1128/MCB.26.8.3114-3123.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyazaki Y, Boccuni P, Mao S, Zhang J, Erdjument-Bromage H, Tempst P, Kiyokawa H, Nimer SD. Cyclin A-dependent Phosphorylation of the ETS-related Protein, MEF, Restricts Its Activity to the G1 Phase of the Cell Cycle. J Biol Chem. 2001;276:40528–40536. doi: 10.1074/jbc.M103051200. [DOI] [PubMed] [Google Scholar]
- 6.Alcalay M, Meani N, Gelmetti V, Fantozzi A, Fagioli M, Orleth A, Riganelli D, Sebastiani C, Cappelli E, Casciari C, Sciurpi MT, Mariano AR, Minardi SP, Luzi L, Muller H, Di Fiore PP, Frosina G, Pelicci PG. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J Clin Invest. 2003;112:1751–1761. doi: 10.1172/JCI17595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park DJ, Vuong PT, de Vos S, Douer D, Koeffler HP. Comparative analysis of genes regulated by PML/RAR alpha and PLZF/RAR alpha in response to retinoic acid using oligonucleotide arrays. Blood. 2003;102:3727–3736. doi: 10.1182/blood-2003-02-0412. [DOI] [PubMed] [Google Scholar]
- 8.Muller-Tidow C, Steffen B, Cauvet T, Tickenbrock L, Ji P, Diederichs S, Sargin B, Kohler G, Stelljes M, Puccetti E, Ruthardt M, deVos S, Hiebert SW, Koeffler HP, Berdel WE, Serve H. Translocation products in acute myeloid leukemia activate the Wnt signaling pathway in hematopoietic cells. Mol Cell Biol. 2004;24:2890–2904. doi: 10.1128/MCB.24.7.2890-2904.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lacorazza HD, Miyazaki Y, Di Cristofano A, Deblasio A, Hedvat C, Zhang J, Cordon-Cardo C, Mao S, Pandolfi PP, Nimer SD. The ETS protein MEF plays a critical role in perforin gene expression and the development of natural killer and NK-T cells. Immunity. 2002;17:437–449. doi: 10.1016/s1074-7613(02)00422-3. [DOI] [PubMed] [Google Scholar]
- 10.Lacorazza HD, Yamada T, Liu Y, Miyata Y, Sivina M, Nunes J, Nimer SD. The transcription factor MEF/ELF4 regulates the quiescence of primitive hematopoietic cells. Cancer Cell. 2006;9:175–187. doi: 10.1016/j.ccr.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 11.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7:1074–1082. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- 12.Wei D, Kanai M, Huang S, Xie K. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis. 2006;27:23–31. doi: 10.1093/carcin/bgi243. [DOI] [PubMed] [Google Scholar]
- 13.Zhao W, Hisamuddin IM, Nandan MO, Babbin BA, Lamb NE, Yang VW. Identification of Kruppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene. 2004;23:395–402. doi: 10.1038/sj.onc.1207067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yasunaga J, Taniguchi Y, Nosaka K, Yoshida M, Satou Y, Sakai T, Mitsuya H, Matsuoka M. Identification of aberrantly methylated genes in association with adult T-cell leukemia. Cancer Res. 2004;64:6002–6009. doi: 10.1158/0008-5472.CAN-04-1422. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 16.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 18.Buckley AF, Kuo CT, Leiden JM. Transcription factor LKLF is sufficient to program T cell quiescence via a c-Myc--dependent pathway. Nat Immunol. 2001;2:698–704. doi: 10.1038/90633. [DOI] [PubMed] [Google Scholar]
- 19.Tzachanis D, Freeman GJ, Hirano N, van Puijenbroek AA, Delfs MW, Berezovskaya A, Nadler LM, Boussiotis VA. Tob is a negative regulator of activation that is expressed in anergic and quiescent T cells. Nat Immunol. 2001;2:1174–1182. doi: 10.1038/ni730. [DOI] [PubMed] [Google Scholar]
- 20.Medema RH, Kops GJ, Bos JL, Burgering BM. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404:782–787. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- 21.Bista P, Mele DA, Baez DV, Huber BT. Lymphocyte quiescence factor Dpp2 is transcriptionally activated by KLF2 and TOB1. Mol Immunol. 2008;45:3618–3623. doi: 10.1016/j.molimm.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baksh S, Widlund HR, Frazer-Abel AA, Du J, Fosmire S, Fisher DE, DeCaprio JA, Modiano JF, Burakoff SJ. NFATc2-mediated repression of cyclin-dependent kinase 4 expression. Mol Cell. 2002;10:1071–1081. doi: 10.1016/s1097-2765(02)00701-3. [DOI] [PubMed] [Google Scholar]
- 23.Hou S, Hyland L, Ryan KW, Portner A, Doherty PC. Virus-specific CD8+ T-cell memory determined by clonal burst size. Nature. 1994;369:652–654. doi: 10.1038/369652a0. [DOI] [PubMed] [Google Scholar]
- 24.Wherry EJ, Teichgraber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, Ahmed R. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol. 2003;4:225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- 25.Kaech SM, Wherry EJ. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity. 2007;27:393–405. doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marsden VS, Kappler JW, Marrack PC. Homeostasis of the memory T cell pool. Int Arch Allergy Immunol. 2006;139:63–74. doi: 10.1159/000090000. [DOI] [PubMed] [Google Scholar]
- 27.Cannarile MA, Lind NA, Rivera R, Sheridan AD, Camfield KA, Wu BB, Cheung KP, Ding Z, Goldrath AW. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nat Immunol. 2006;7:1317–1325. doi: 10.1038/ni1403. [DOI] [PubMed] [Google Scholar]
- 28.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang M, Byrne S, Liu N, Wang Y, Oxenius A, Ashton-Rickardt PG. Differential survival of cytotoxic T cells and memory cell precursors. J Immunol. 2007;178:3483–3491. doi: 10.4049/jimmunol.178.6.3483. [DOI] [PubMed] [Google Scholar]
- 30.Marrack P, Bender J, Hildeman D, Jordan M, Mitchell T, Murakami M, Sakamoto A, Schaefer BC, Swanson B, Kappler J. Homeostasis of alpha beta TCR+ T cells. Nat Immunol. 2000;1:107–111. doi: 10.1038/77778. [DOI] [PubMed] [Google Scholar]
- 31.Goldrath AW, Luckey CJ, Park R, Benoist C, Mathis D. The molecular program induced in T cells undergoing homeostatic proliferation. Proc Natl Acad Sci U S A. 2004;101:16885–16890. doi: 10.1073/pnas.0407417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masopust D, Vezys V, Marzo AL, Lefrancois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291:2413–2417. doi: 10.1126/science.1058867. [DOI] [PubMed] [Google Scholar]
- 33.Warnock RA, Askari S, Butcher EC, von Andrian UH. Molecular mechanisms of lymphocyte homing to peripheral lymph nodes. J Exp Med. 1998;187:205–216. doi: 10.1084/jem.187.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bai A, Hu H, Yeung M, Chen J. Kruppel-like factor 2 controls T cell trafficking by activating Lselectin (CD62L) and sphingosine-1-phosphate receptor 1 transcription. J Immunol. 2007;178:7632–7639. doi: 10.4049/jimmunol.178.12.7632. [DOI] [PubMed] [Google Scholar]
- 35.Carlson CM, Endrizzi BT, Wu J, Ding X, Weinreich MA, Walsh ER, Wani MA, Lingrel JB, Hogquist KA, Jameson SC. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 36.Sebzda E, Zou Z, Lee JS, Wang T, Kahn ML. Transcription factor KLF2 regulates the migration of naive T cells by restricting chemokine receptor expression patterns. Nat Immunol. 2008;9:292–300. doi: 10.1038/ni1565. [DOI] [PubMed] [Google Scholar]
- 37.Sprent J, Cho JH, Boyman O, Surh CD. T cell homeostasis. Immunol Cell Biol. 2008 doi: 10.1038/icb.2008.12. [DOI] [PubMed] [Google Scholar]
- 38.Surh CD, Sprent J. Homeostatic T cell proliferation: how far can T cells be activated to self-ligands? J Exp Med. 2000;192:F9–F14. doi: 10.1084/jem.192.4.f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kerdiles YM, Beisner DR, Tinoco R, Dejean AS, Castrillon DH, DePinho RA, Hedrick SM. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol. 2009;10:176–184. doi: 10.1038/ni.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lacombe MH, Hardy MP, Rooney J, Labrecque N. IL-7 receptor expression levels do not identify CD8+ memory T lymphocyte precursors following peptide immunization. J Immunol. 2005;175:4400–4407. doi: 10.4049/jimmunol.175.7.4400. [DOI] [PubMed] [Google Scholar]
- 41.Badovinac VP, Messingham KA, Jabbari A, Haring JS, Harty JT. Accelerated CD8+ T-cell memory and prime-boost response after dendritic-cell vaccination. Nat Med. 2005;11:748–756. doi: 10.1038/nm1257. [DOI] [PubMed] [Google Scholar]
- 42.Rowland BD, Peeper DS. KLF4, p21 and context-dependent opposing forces in cancer. Nat Rev Cancer. 2006;6:11–23. doi: 10.1038/nrc1780. [DOI] [PubMed] [Google Scholar]
- 43.Mahatan CS, Kaestner KH, Geiman DE, Yang VW. Characterization of the structure and regulation of the murine gene encoding gut-enriched Kruppel-like factor (Kruppel-like factor 4) Nucleic Acids Res. 1999;27:4562–4569. doi: 10.1093/nar/27.23.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao JJ, Liu Y, Lacorazza HD, Soslow RA, Scandura JM, Nimer SD, Hedvat CV. Tumor promoting properties of the ETS protein MEF in ovarian cancer. Oncogene. 2007;26:4032–4037. doi: 10.1038/sj.onc.1210170. [DOI] [PubMed] [Google Scholar]
- 45.Sandberg ML, Sutton SE, Pletcher MT, Wiltshire T, Tarantino LM, Hogenesch JB, Cooke MP. c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Dev Cell. 2005;8:153–166. doi: 10.1016/j.devcel.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 46.Veiga-Fernandes H, Rocha B. High expression of active CDK6 in the cytoplasm of CD8 memory cells favors rapid division. Nat Immunol. 2004;5:31–37. doi: 10.1038/ni1015. [DOI] [PubMed] [Google Scholar]
- 47.Yusuf I, Kharas MG, Chen J, Peralta RQ, Maruniak A, Sareen P, Yang VW, Kaestner KH, Fruman DA. KLF4 is a FOXO target gene that suppresses B cell proliferation. Int Immunol. 2008;20:671–681. doi: 10.1093/intimm/dxn024. [DOI] [PubMed] [Google Scholar]
- 48.Hogquist KA, Weinreich MA, Jameson SC. T-cell migration: T-cell migration Kruppeled T cells move again. Immunol Cell Biol. 2008 doi: 10.1038/icb.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vezys V, Yates A, Casey KA, Lanier G, Ahmed R, Antia R, Masopust D. Memory CD8 T-cell compartment grows in size with immunological experience. Nature. 2009;457:196–199. doi: 10.1038/nature07486. [DOI] [PubMed] [Google Scholar]
- 50.Ginaldi L, De Martinis M, D'Ostilio A, Marini L, Loreto F, Modesti M, Quaglino D. Changes in the expression of surface receptors on lymphocyte subsets in the elderly: quantitative flow cytometric analysis. Am J Hematol. 2001;67:63–72. doi: 10.1002/ajh.1082. [DOI] [PubMed] [Google Scholar]
- 51.Katz JP, Perreault N, Goldstein BG, Lee CS, Labosky PA, Yang VW, Kaestner KH. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krieg C, Boyman O, Fu YX, Kaye J. B and T lymphocyte attenuator regulates CD8+ T cell-intrinsic homeostasis and memory cell generation. Nat Immunol. 2007;8:162–171. doi: 10.1038/ni1418. [DOI] [PubMed] [Google Scholar]
- 53.Galan-Caridad JM, Harel S, Arenzana TL, Hou ZE, Doetsch FK, Mirny LA, Reizis B. Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell. 2007;129:345–357. doi: 10.1016/j.cell.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.