Abstract
Viral capsid proteins (CPs) can regulate gene expression and encapsulate viral RNAs. Low-level expression of the brome mosaic virus (BMV) CP was found to stimulate viral RNA accumulation, while higher levels inhibited translation and BMV RNA replication. Regulation of translation acts through an RNA element named the B box, which is also critical for the replicase assembly. The BMV CP has also been shown to preferentially bind to an RNA element named SLC that contains the core promoter for genomic minus-strand RNA synthesis. To further elucidate CP interaction with RNA, Available online we used a reversible cross-linking–peptide fingerprinting assay to identify 27 May 2009 peptides in the capsid that contact the SLC, the B-box RNA, and the encapsidated RNA. Transient expression of three mutations made in residues within or close by the cross-linked peptides partially released the normal inhibition of viral RNA accumulation in agroinfiltrated Nicotiana benthamiana. Interestingly, two of the mutants, R142A and D148A, were found to retain the ability to down-regulate reporter RNA translation. These two mutants formed viral particles in inoculated leaves, but only R142Awas able to move systemically in the inoculated plant. The R142A CP was found to have higher affinities for SLC and the B box compared with those of wild-type CP and to alter contacts to the RNA in the virion. These results better define how the BMV CP can interact with RNA and regulate different viral processes.
Keywords: positive-strand RNA virus, brome mosaic virus, capsid protein, RNA replication, translation
Introduction
Viral capsid proteins (CPs) are important in many aspects of the viral infection cycle, including modulating gene expression, RNA replication, virus assembly and disassembly, and virus trafficking.1 For example, the CPs of alfalfa mosaic virus and bacteriophage MS2 could regulate RNA synthesis and translation by binding to the RNA motif in the viral RNAs.2–5
Brome mosaic virus (BMV) is a segmented positive-strand RNA virus. The BMV genome contains three capped messenger-sense genomic RNAs packaged in T=3 icosahedral particles.6 Genomic RNA1 and RNA2 are encapsulated into separate viral particles, while RNA3 is co-packaged with subgenomic RNA4.7 The genome lacks a polyA tail but contains a tRNA-like structure within the 3′-untranslated region (UTR) that includes the promoter that directs minus-strand RNA synthesis. Genomic RNA1 and RNA2 encode replication proteins 1a and 2a, respectively, which interact to form the viral replicase.8 Genomic RNA3 encodes the movement protein, required for cell-to-cell and long-distance trafficking, and the CP.
Using the Agrobacterium-mediated gene expression system where BMV RNAs and proteins can be expressed individually or in combination,9 we found that higher levels of CP were associated with a dramatic inhibition of BMV RNA accumulation. A model summarizing some of the effects of the CP on BMV gene expression is shown in Fig. 1a. The inhibitory effect of the CP occurs at least partially through the suppression of the translation of the replication proteins through the B-box motif in the 5′-UTRs of BMV RNA1 and RNA2.10 Interestingly, the B box is also required to interact with the 1a protein to form the membrane-associated BMV replicase, suggesting a link between the regulation of translation and BMV replicase formation. The CP can also bind to the SLC motif that contains the core promoter to direct minus-strand RNA synthesis,11 a motif that contains a stem–triloop where two of the loop nucleotides stack with the stem and a key adenine base protrudes out into solution called a clamped adenine motif.12,13
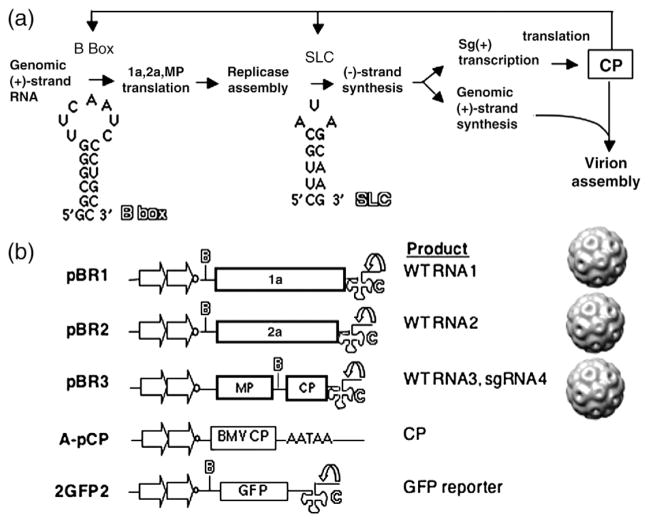
Fig. 1.
Model for the study and schematics of key constructs. (a) A model for the interaction between the BMV CP and various RNA elements in the BMV RNAs. The 5′-UTRs of BMV RNA1 and RNA2 contain a conserved motif named the B box, and the 3′-UTRs contain the SLC motif. (b) Constructs used in this study. Plasmids pBR1, pBR2, and pBR3 result in the transcription of the three BMV RNAs, which are individually encapsidated into separate virions. The plasmids are engineered to have tandem copies of the cauliflower mosaic virus 35S promoter (straight arrow) and a cis-acting ribozyme sequence (curved arrow) that will generate the authentic 3′ end of the BMV RNAs. The locations of the RNA motifs of the B box and SLC are identified by the letters B and C in outline fonts. Plasmid pCP was used for the transient expression of BMV CP from tandem copies of the 35S promoter and a translational enhancer 5′ of the BMV capsid coding sequence. The 3′-untranslated sequence contains a polyA processing signal (AATAA). Reporter RNA 2GFP2 contains the GFP coding sequence flanked by the 5′-UTR and 3′-UTR of BMV genomic RNA2.
Since RNA binding is a key to CP contribution to numerous regulatory events, we wanted to better define how the CP interacts with different RNAs involved in the regulation of translation, RNA replication, and RNA encapsidation using a reversible cross-linking, affinity purification, and peptide-fingerprinting assay by mass spectrometry (MS) (henceforth called RCAP).14 The mapping data led to the identification of three mutations in the BMV CP that differentially affect the inhibition of translation and the repression of viral RNA accumulation.
Results
Mapping BMV capsid interaction with RNAs
Previously, we demonstrated that overexpression of BMV subgenomically encoded CP could repress viral RNA accumulation using an Agrobacterium-mediated gene expression system.15 The repression of RNA replication is at least partially due to down-regulation of the translation of replication proteins through binding to the B-box RNA motif in the 5′-UTRs of RNA1 and RNA2. On the other hand, lower expression of CP was found to stimulate RNA accumulation, probably by binding to the SLC motif in the 3′-UTRs, which contain the core promoter for minus-strand RNA synthesis (Supplementary Fig. 1).
To further understand how CP–RNA interaction is involved in regulation of translation and replication, we sought to identify mutations in the CP that could impact the regulatory effects on BMV RNA accumulation. RCAP was used to map CP binding to the B-box RNA motif (19 nucleotides)10 and to SLD4 (25 nucleotides), a derivative of SLC (Fig. 1a and Refs. 12, 16, and 17). Briefly, formaldehyde was used to cross-link the CP to an RNA that has been covalently attached to an inert resin through a 5′ amino group (Fig. 2a). The cross-linked complex was then subjected to trypsin cleavage, followed by extensive washing and purification of the RNA resin to remove noncovalently associated peptides. The bound peptides were released from the RNA resin by reversing the cross-links and were identified using matrix-assisted laser desorption/ionization time-of-flight (MALDI-ToF) MS and matched to the expected trypsin fragmentation pattern of the CP. Versions of this method were employed to generate a map of how several RNA-binding proteins can contact RNA and were further modified to chemically synthesize RNA coupled to resin.14,19,20
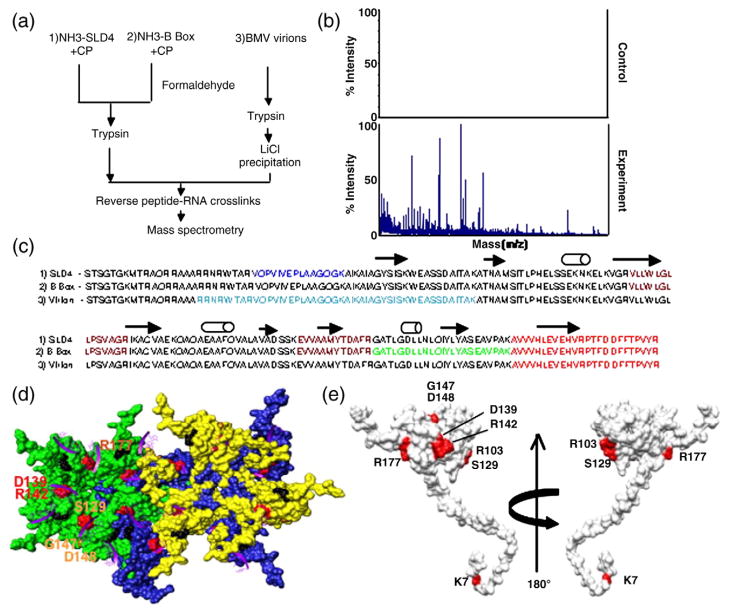
Fig. 2.
Mapping of the RNA-binding residues in the BMV CP. (a) Schematic of the RCAP protocol used to map the CP peptide–RNA interaction. Three RNA samples were analyzed as diagrammed. (b) Representative MALDI-ToF MS spectra showing the signals obtained from a control sample and a sample of the CP cross-linked to the SLD4 RNA. Both were treated identically except that the control was not subjected to formaldehyde. Signals that are three standard errors above the background were considered to be significant. (c) Sequence of the BMV CP from residues 2–189 with the peptides identified by the three RNAs in color. The peptide cross-linked to all three RNAs is shown in red. The peptide that was cross-linked to the B box and SLD4 is shown in maroon. The N-terminal methionine was not included because it was processed from the mature capsid.18 (d) Modeling of CP interaction with virion RNA. The region in the globular domain of the CP is emphasized since it was preferentially used to interact with SLD4 and the B box. The subunits that form the pentamer and hexamer are shown in green, blue, and yellow to allow better visualization of the subunits. The locations of several of the potential RNA-binding residues are identified by the names of the residues. The purple ribbons represent the fragments of RNAs seen in the CCMV crystal structure that are positioned onto the structure of the BMV capsid. The amino acids that could contact viral RNA are shown in the context of a hexamer of the BMV capsid. (e) A view of the residues in a monomer of the BMV CP that were subjected to mutational analysis.
The reactions led to robust signals reproducibly above background (Fig. 2b). Several of the peptides were nested due to missed cleavage at lysines and arginines by trypsin, as would happen when the RNA prevents cleavage. The nested peptides increased confidence in the assignments of the peptides. Both SLD4 and the B-box RNAs contacted several portions of the BMV CP (Fig. 2c). The 25 N-terminal residues of the CP, a known RNA-binding sequence,21,22 did not produce peptides. The remaining two-thirds of the capsid formed a more globular structure and a C-terminal stub (Fig. 2d and e). The two RNAs were associated with three identical peptides (from residues 90–103, residues 131–142, and residues 166–189), but SLD4 was associated with a unique peptide from residues 27–41 and the B-box motif was associated with a peptide from residues 143–165 (Fig. 2c and Supplementary Fig. 2).
We also examined how the CP interacted with the RNAs in the context of virions. BMV virions purified from CsCl density gradients were cross-linked and digested with trypsin, and LiCl was used to selectively precipitate the viral RNAs and cross-linked peptides. Interestingly, six of the seven ions in the mass spectra were from residues 20–64 (Supplementary Fig. 2). In addition, residues 166–189 from the globular domain of the CP were also found, indicating that the C-terminus of the CP can contact both the virion RNA and the shorter RNA motifs. These results suggest that the N-terminus of the CP plays a prominent role in the context of the virion but that the central and C-terminal portions of the CP play a role in contacting all three RNAs.
While the RCAP data provide general guidance about RNA–CP contacts, we sought to dissect the contacts that could affect the biological activities of the CP through mutational analysis. To identify residues to mutate, we used molecular modeling to identify possible RNA-binding residues in the globular domain of the CP (Fig. 2d). The RNAs within the BMV virion have not been resolved by structural analyses.6 However, a portion of the RNA was observed in the closely related cowpea chlorotic mottle virus (CCMV) virion and provided guidance for BMV.23 Oligomer structures of the BMV capsid [Protein Data Bank (PDB) ID 1js9] and the CCMV capsid (PDB ID 1cwp) were produced using a ViperDB oligomer generator24 and then superimposed (the RMSD was 0.952) and modeled using the UCSF Chimera program.25 Residues R103, S129, D139, R142, G147, and D148 within or adjacent to the cross-linked portions of the BMV CP were selected for substitutions with alanines. In addition, we tested residue K7 as a representative residue from the N-terminal tail.
Functional analysis of the effects of CP mutations on BMV RNA accumulation
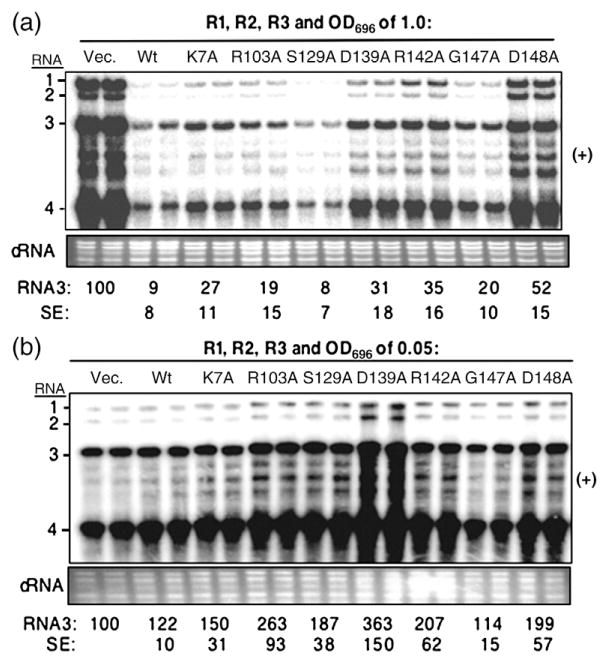
To determine whether the mutations in the CP affected its ability to repress BMV RNA accumulation, we infiltrated Agrobacterium culture [optical density at 595 nm (OD595) of 1.0], which carries the plasmid encoding the gene for the wild type (WT) or mutant CP, into Nicotiana benthamiana along with cultures expressing the three BMV genomic RNAs (OD595 of 0.1 each). RNA accumulation was detected by Northern blot analysis 2 days after infiltration. CP mutants D139A, R142A, and D148A were found to have increased BMV RNA accumulation compared with that of WT CP, suggesting that the mutations affected the repression of RNA synthesis. Relative to the RCAP mapping data, D139 and R142 were in the peptide that cross-linked with the SLD4 and B-box RNA motifs, but D148 was not (Fig. 2c and Supplementary Fig. 2).
To examine which mutations affected the ability of the WT CP stimulation of RNA synthesis when expressed at low levels, we infiltrated plants with cultures expressing the three BMV RNAs and either the WT or a mutant capsid (OD595 of 0.05). Except for mutant G147A, the other mutants increased BMV RNAs at levels comparable with or higher than those of plants that expressed the WT CP (Fig. 3b), indicating that most of the mutations, including D139A, R142A, and D148A, did not affect the ability of the CP to stimulate BMV RNA synthesis.
Fig. 3.
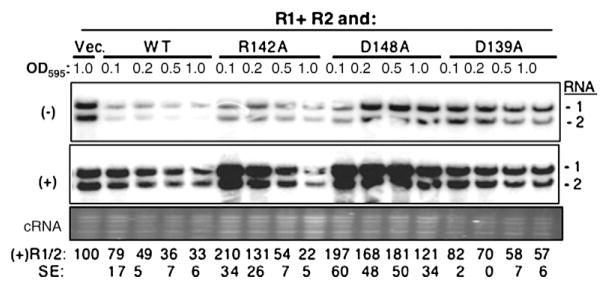
Effects of transient expression of mutant BMV CPs on viral RNA accumulation. (a) Over-expression of the WT and mutant CP on BMV RNA accumulation. A mixture of Agrobacterium expressing RNA1, RNA2, and RNA3 (each at an OD595 of 0.1) was co-infiltrated with the culture that can express either WT CP (A-pCP) or mutant derivatives (each at an OD595 of 1.0). Agrobacterium harboring the vector was used as a negative control. Viral RNA was detected by Northern blot assay. The ethidium-bromide-stained cellular RNA was used as a loading control. Quantification of RNA3 is shown at the bottom of the image as a percentage of the vector control. The standard error (SE) values represent one standard deviation from the mean of at least three independently assayed samples. (b) Effect of WT and mutant BMV CP on RNA accumulation when infiltrated at an OD595 of 0.05. The samples were processed as described above.
The above analysis prompted us to focus on mutants D139A, R142A, and D148A. Plants infiltrated to express only the WT or one of these three mutants showed comparable levels of CP in Western blots (Fig. 4a). Therefore, the effects of the mutations were unlikely to be through differential levels of expression and/or stability.
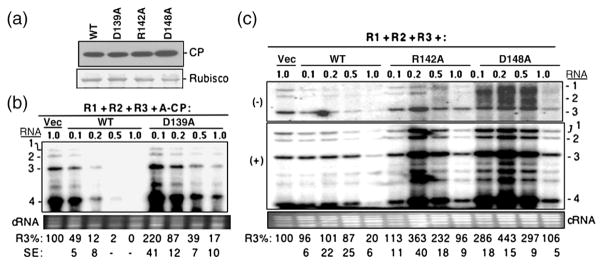
Fig. 4.
Characterization of the three BMV CP mutants that affected the ability to repress RNA accumulation. (a) Western blot of CP expression level in N. benthamiana plants infiltrated with Agrobacterium (OD595 of 1.0) harboring the empty vector or pCP that can express either WT or mutant BMV CP. The entire leaf was macerated and clarified by centrifugation at 13,000g, and then an aliquot was subjected to SDS-PAGE and transferred onto PVDF membrane. The primary antibody was rabbit anti-CP polyclonal antibody. A Coomassie blue-stained small subunit of Rubisco was used as the protein loading control. The mean and standard error values of at least three independently assayed samples are shown below the gel image. (b) Titration of Agrobacterium expressing CP mutant D139A on BMV RNA accumulation. N. benthamiana plants were infiltrated with a mixture of Agrobacterium expressing three genomic RNAs (each at an OD595 of 0.1) along with culture that can express either WT CP or the D139A mutant at the concentrations noted above the gel image. Viral RNA was detected by a Northern blot assay, and RNA3 was quantified at the bottom of the image as a percentage of the vector control. The standard error values represent one standard deviation from at least three independently assayed samples. Those dented with a dash were deemed to be below the limit of accurate detection by phosphorimage analysis. (c) Effects of titration of Agrobacterium expressing CP mutants R142A and D148A on RNA accumulation. The minus-strand (top panel) and plus-strand (middle panel) RNAs were detected by Northern blot with specific probe. The bottom panel contains ethidium-bromide-stained cellular RNA used as a loading control. Quantification of the plus-strand BMV RNAs is shown at the bottom of the gel image and is representative of two independent experiments with four independent samples.
The effects of the three mutants were analyzed over a range of inoculum concentrations. Both D139A and WT CP inhibited BMV RNA accumulation in a concentration-dependent manner, but the inhibition was more severe with the WT CP (Fig. 4b). At an OD595 of 0.2, D139A had BMV RNA accumulation at 87% of the vector control, while WT CP had RNA accumulation at 49% (Fig. 4b). This difference is consistent with the observations that D139A could not repress BMV RNA replication at the level seen with the WT CP. Mutants R142A and D148A had more pronounced defect in the repression of BMV RNA accumulation. At an OD595 of 0.2, BMV RNA3 accumulation increased by three- to fivefold (Fig. 4c). The stimulatory effect was observed even at an OD595 of 0.5. However, at the higher concentration, both R142A and D148A repressed BMV minus-and plus-strand RNA accumulations (Fig. 4c).
The presence of the A-pCP along with all three BMV RNAs raises the possibility that the RNA encoding the CP could have an effect on the stability of BMV RNA3, which then indirectly affects BMV accumulation. While that single amino acid substitutions affected BMV RNA accumulation argues strongly against this idea, we wanted to rule out the possibility that the mutations in the CP act through RNA silencing. BMV RNA1 and RNA2 introduced by Agrobacterium can replicate in N. benthamiana in the absence of RNA3.9 Furthermore, transient expression of the WT CP could regulate RNA1 and RNA2 accumulations in a concentration-dependent manner.9,10 Therefore, we examined whether the CP could affect BMV RNA1 and RNA2 accumulations. A range concentration of the three mutant CPs was tested along with BMV RNA1 and RNA2. At an OD595 of 0.2, the WT CP reduced RNA1 accumulation to 49%, while the presence of the three mutants resulted in BMV RNAs at levels in excess of the absence of CP (Fig. 5). At all ranges of inoculum tested, the three mutant CPs resulted in higher levels of BMV RNA1 and RNA2 accumulations. These results show that the three mutant CPs, R142A, D148A, and D139A, could regulate BMV RNA1 and RNA2 accumulations.
Fig. 5.
Effects of the three CP mutants on BMV RNA1 and RNA2 accumulations. N. benthamiana was agroinfiltrated to express RNA1, RNA2, and either WT or mutant CP from A-pCP or the empty expression vector (Vec.). The RNAs were identified by Northern blot analysis using probes to the 3′-UTRs of the BMV RNAs. Quantifications were determined by phosphorimage analysis and were normalized for the input RNA, as estimated by the staining of cellular RNAs (cRNA). The accumulation of the minus-strand RNAs (not shown) parallels that of the plus-strand RNAs.
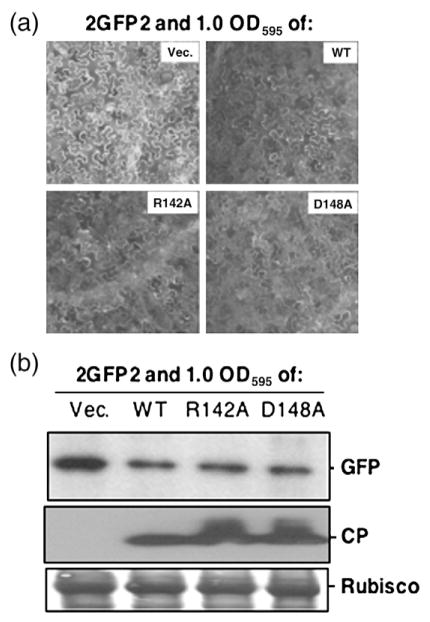
Will the mutant CPs retain the ability of the WT CP to regulate translation from BMV RNAs? A reporter RNA named 2GFP2 that contained the 5′-and 3′-UTRs of BMV RNA2 flanking the green fluorescent protein (GFP) coding sequence (Fig. 1b) was previously shown to be inhibited by the WT CP in a manner that required the B box in the 5′-UTRs of BMV RNA1 and RNA2.10 Mutant D139A was previously shown to decrease the ability to inhibit GFP production.10 Interestingly, mutant R142A and D148A infiltrated at an OD595 of 1.0 along with 2GFP2-decreased GFP expression (Fig. 6a). Western blots of lysates from inoculated leaves probed for the GFP confirmed that R142A and D148A repressed GFP expression in a manner similar to the WT CP (Fig. 6b).
Fig. 6.
Effects of CP mutants on reporter RNA translation. (a) Representative fluorescent microscopic examination of GFP expression from the 2GFP2 reporter in N. benthamiana leaves. Agrobacterium harboring 2GFP2 (at an OD595 of 0.2) was co-infiltrated with the culture that can express either WT CP or the mutants (each at an OD595 of 1.0). GFP images were taken at 40 hpi. (b) Western blot analysis of GFP and CP expression. The leaves were macerated, and approximately equal amounts of protein lysates were subjected to SDS-PAGE followed by Western blot with either rabbit anti-CP polyclonal antibody or mouse anti-GFP monoclonal antibody. The secondary antibodies used were a goat anti-rabbit or a goat anti-mouse. “Rubisco” denotes the small subunit of the Rubisco protein that served as a protein loading control.
In summary, mutants R142A and D148A increased BMV RNA accumulation without affecting the ability to regulate translation. The results suggest that the mutations alter but do not abolish CP binding to BMV RNA.
Analysis of the effects of the mutations on virus production
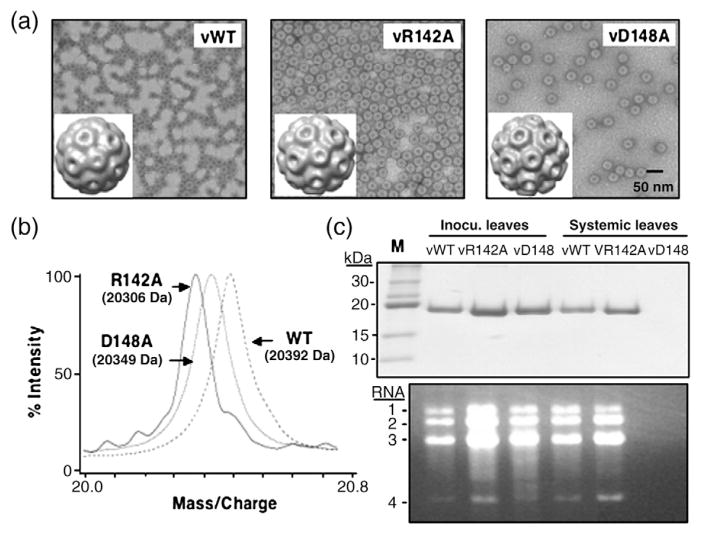
We wanted to determine whether the R142A and D148A mutations will affect BMV virion production and systemic spread. The two mutations were introduced separately into a plasmid that can express BMV RNA3. Ten days after infiltration into N. benthamiana leaves, entire plant leaves were used for virus purification and virions were detected with both mutants (Fig. 7a, top panel). However, the abundance of virions from mutant D148A, designated vD148A, was reduced in two independent experiments (Fig. 7a and other data not shown). When capsids from vR142A and vD148A were subjected to MS, they produced only one peak per sample that was at the molecular mass consistent with the retention of the respective amino acid substitutions (Fig. 7b). Therefore, both mutations are stable for the time frame of the analysis of our experiments.
Fig. 7.
Characterization of mutant viruses in N. benthamiana. (a) Electron micrographs of WT and mutant virions and three-dimensional reconstruction of viral particles. Viral particles were visualized at a magnification of 39,000× with a JEOL 1200EX electron microscope at 100 kV. The micrographs were digitalized using an Epson Projection 3200 scanner at 1200 dpi, corresponding to 5.5 Å/pixel. The EMAN software package run under Linux was used for single-particle image analysis and three-dimensional reconstruction.26 The bar represents 50 nm. (b) The masses of the CP subunit from the WT virion and the mutants vR142A and vD148A determined by MS. (c) SDS-PAGE to detect the viral CP from the local inoculated leaves and systemic infection leaves. N. benthamiana leaves were infiltrated with a mixture of Agrobacterium expressing BMV RNA1 and RNA2 along with culture expressing either WT RNA3 or mutant RNA3 (R142A or D148A). The virions were separately isolated from locally inoculated and systemically infected leaves, respectively, and subjected to SDS-PAGE analysis. The agarose gel was used to detect the viral RNA isolated from WT virus or the mutant virion. Viral RNA was extracted and loaded on 1.2% native agarose gel. RNA was visualized by ethidium bromide staining.
The reduced levels of vD148A prompted us to examine the virions separately in the infiltrated and noninfiltrated leaves. In the infiltrated leaves, both mutant virions accumulated to levels at least comparable with those of the WT. Approximately 6–7 days after the initial infiltration, WT BMVand the vR142A mutant moved systemically into noninfiltrated leaves, but vD148A did not. No vD148A virion was observed even 10 days after the initial inoculation (Fig. 7c, upper panel). In all cases where virions were found, they contained BMV RNAs (Fig. 7c, lower panel). In a denaturing gel, RNA4 was also abundantly represented in the preparations that had virions (G. Yi, unpublished data). These results show that the mutation in vD148A renders it unable to infect systemically even though there was no obvious effect on RNA packaging.
To examine whether the mutations caused an obvious defect in virion formation, we randomly picked more than 1000 single virus particles from micrographs of each of the three virions and performed single-particle reconstruction using the EMAN suite of computer programs.26 The reconstructions showed particles composed of pentamers and hexamers arranged in a manner similar to that for the WT BMV, and their diameters were not different from those in the WT (Fig. 7a, insets, and other data not shown). These results suggest that the defects in inhibiting RNA accumulation (in mutants R142A and D148A) and in systemic infection are unlikely to be due to effects on capsid subunit interactions.
Characterization of viruses with the R142A mutation
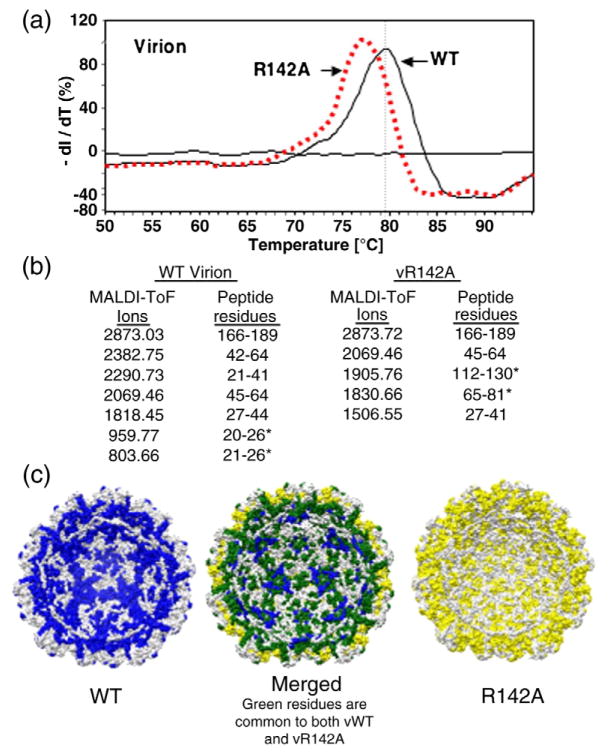
For the remainder of this analysis, we focus on the R142A mutation since we can prepare high-quality CPs from CsCl-purified virions. While single-virus-particle reconstruction with vR142A did not reveal any defects in virion structure, we wanted to assess whether interaction with the packaged RNA was affected. A differential scanning fluorimetry assay was used to assess the stability of the vR142A relative to WT virions. In this assay, the virion was subjected to a controlled increase in temperature in the presence of the dye SYPRO orange, which fluoresces in the presence of hydrophobic environment, such as denatured proteins. The temperature at the greatest transition in fluorescence is thus an apparent Tm for the sample. vR142A had a Tm value (77.3 °C) reproducibly lower than that of the WT BMV (79.5 °C) at pH 7.0 (Fig. 8a), implying that vR142A had thermal dynamic properties distinguishable from the WT BMV. In this assay, a difference of 1 °C can be interpreted to be significant.27
Fig. 8.
Biophysical characterization of the R142A virion. (a) Differential scanning fluorimetry to determine the Tm values of WT and R142A virions. The experiment was performed in an Eppendorf Master-cycler EP Realplex machine with SYPRO orange as the molecular probe. The 96-well plate containing all the samples was heated at a rate of 1.0 °C/min, from 25 to 95 °C, and the fluorescence intensity was measured with excitation/emission wavelengths of 470/550 nm. The Tm values were calculated by obtaining the maximum of the first derivative using KaleidaGraph. (b) MS of peptides that interact with viral RNA in WT and R142A virions. Data collection was performed on a Bruker Biflex III MALDI-ToF mass spectrometer. Samples were desalted using a ZipTip and spotted on the sample plate in α-cyano-4-hydroxycinnamic acid matrix (mass=189.17 Da). Positive ions from masses 400–4000 Da were analyzed in reflectron mode. The MS data are shown in Supplementary Fig. 2. (c) A molecular model of the RNA-contacting residues within the inner cavity of the R142A mutant virion (maize), the WT virion (blue), and the merged models that show the differences in the RNA contacts (spartan green).
We used the RCAP assay to gain further insight into the effect of the R142A substitution on interaction with the virion RNA. Several peptides were identical in the R142A and the WT virions, such as those containing residues 27–64 and residues 166–189 (Supplementary Fig. 3). Interestingly, vR142A lacked peptide 21–26 that was observed in the WT virion, while the R142A virion had two additional peptides, one that extended the block of residues from the N-terminal arginine-rich motif (residues 65–81) and another in the central portion of the CP (residues 112–130) (Fig. 8b). In the context of the models of the BMV virion, the locations of the RNA-cross-linked peptides in vR142A are obviously distinct from those in the WT (Fig. 8c). While the effects of the mutations could be complex, these results indicate that the R142A qualitatively affected CP recognition of RNA in the context of virions.
Interaction of the disassembled R142A with BMV RNA motifs
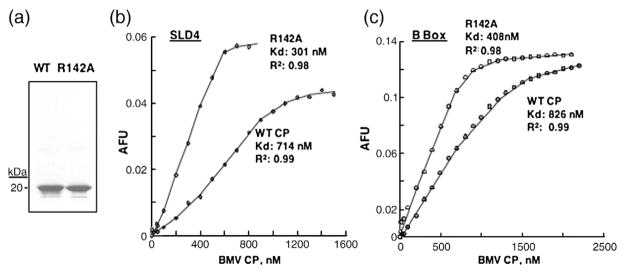
Fluorescence anisotropy was used to examine whether the R142A substitution affected the binding affinity for RNA elements that regulate BMV RNA accumulation. SLD4 and the B-box RNA motifs were chemically synthesized to contain 5′ fluorophores and titrated with increasing concentrations of the WT and R142A CP subunits dissociated from CsCl-purified virions28 (Fig. 9a). The WT CP binds to the SLD4 with a Kd of 714 nM, while R142A had a Kd of 301 nM (Fig. 9b). Thus, the R142A substitution increased RNA-binding affinity by approximately twofold. A similar twofold increase in the affinity of the R142A CP on binding the B-box RNA was observed, demonstrating that the substitution did not affect the specific recognition for the two RNA motifs (Fig. 9c).
Fig. 9.
Affinity of WT and the R142A CP for binding the B-box and SLC RNA motifs. (a) SDS-PAGE showing the quantity and purity of WT CP and R142A subunits purified from virions. The virion was purified from CsCl gradients and dissociated as described in Materials and Methods. (b) Affinity anisotropy experiment to determine the binding affinity of the CP to fluorescence-labeled SLD4. The RNA, at a final concentration of 0.2 μM, was in a 500-μL cuvette containing a magnetic stir bar, suspended in a buffer containing 50 mM Tris, pH 7.5, and 50 mM NaCl. WT CP or R142A subunits were titrated into the cuvettes, and at least 10 anisotropy values were taken after each aliquot of CP was added. All data were corrected for the background intensity of the buffer. Binding data were fitted into the Hill equation using KaleidaGraph software. (c) Anisotropy experiment to determine the affinity of WT CP or mutants for the B-box RNA.
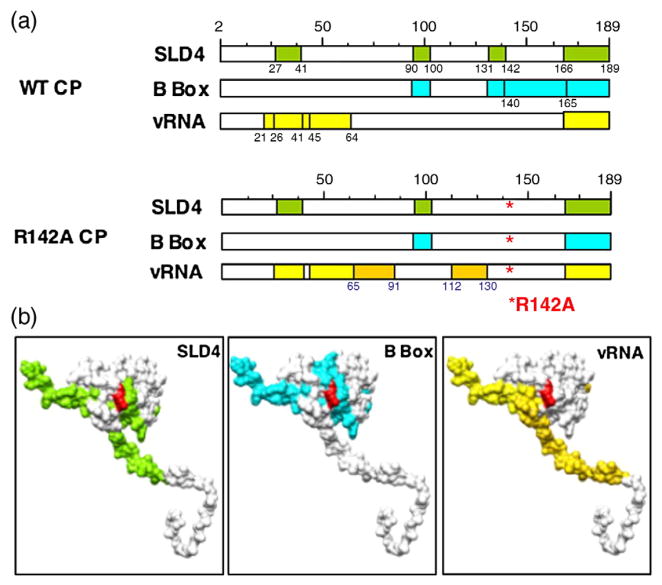
Lastly, we mapped the peptides in R142A CP that can be cross-linked to the RNA motif SLD4 or the B box using the RCAP assay. Similar to the WT CP recognition of SLD4 and the B box, peptides 90–103 and 166–189 (including the C-terminal stub) from R142A were cross-linked to both SLD4 and the B-box RNA (Fig. 10a and Supplementary Fig. 2). Only the peptide containing residues 27–41 differed with SLD4 or the B box as bait in the cross-linking reaction. As with the WT CP, peptide 27–41 was the B box, was missing in R142A. While it was cross-linked to SLD4 but not to the B box (Fig. 10a expected because the R142A substitution will pre-and Supplementary Fig. 2c and d). Peptide 131–142, vent trypsin cleavage at the C-terminus of R142, we which in the WT CP interacts with both SLD4 and did not find a peptide that could be generated by trypsin at the next cleavage site (residues 131–165). This may be due to a decreased sensitivity of the MALDI-ToF for ions of higher masses. Nonetheless, this and previous mapping data clearly show that the R142A CP binds to the two RNA motifs and to the BMV genome differently (Figs. 2, 8, and 10a). The locations of the peptides that contact the different small RNA motifs are summarized in Fig. 10b.
Fig. 10.
A summary of the peptides in the CP that contact the B-box motif, SLD4, or the virion RNA and a comparison with the peptides from the WT CP that were cross-linked to the same RNAs. (a) Schematics of the locations of the peptides that bind to the three RNAs used in the mapping studies. The peptides that differed between the WT and R142A capsid are highlighted in a darker shade. (b) Models of the BMV CP with the peptides cross-linked to each RNA highlighted in the same color scheme as that in (a). The location of the R142 residue is shown in red.
Discussion
The BMV CP has at least three activities that are mediated through RNA binding: (1) encapsidation of BMV vRNAs, (2) repression of RNA accumulation and translation associated with higher levels of BMV CP, and (3) stimulation of BMV RNA accumulation at lower CP levels. These activities likely function in an ordered sequence to facilitate successful BMV infection. To better understand the activities of the multifunctional BMV CP, we mapped its RNA binding using an RCAP assay and then identified three mutations that affect one or more of the CP-associated activities in BMV-infected plants.
The C-terminus and the globular domain of the CP play an important role in systemic movement and encapsidation.29 The N-terminal arginine-rich motif and adjacent regions of the CP can affect selective packaging of the BMV RNAs and direct the morphology of BMV virion assembly.21,22,30 We have found that residues 166–189 at the C-terminus of the CP were cross-linked to all three RNAs we analyzed. This sequence contains only three basic residues (two histidines and an arginine at the very C-terminus) but does contain a stretch with three phenylalanines that lie along the base of an ~20-Å channel formed by the C-terminal stub. It may be informative to examine this sequence in more detail in a future study.
Binding to the B box and SLC that directs replicase assembly and minus-strand RNA synthesis occurs primarily in the globular portions of the CP and the C-terminal stub. The same portions of the CP and residues 27–41 adjacent to the arginine-rich N-terminus were cross-linked to SLD4, which directs minus-strand RNA replication.13 Interestingly, the N-terminal arginine-rich residues of the BMV CP are prominently represented only when examined in the context of the virion. This motif extends into the cavity of the virion,6 and our observations suggest that the arginine-rich motif may play a more prominent role in interacting with the encapsidated RNA.
Our mapping results did not identify residues 2–20 in any of the spectra, but this could be due to the peptides being smaller than 500 Da, which is difficult to distinguish from ions associated with the buffer and matrix in our analysis. Nonetheless, the results suggest that the CP binding to the B box and SLC will be distinct from contacts with the virion-associated RNA. Another factor that could influence RNA recognition is the oligomerization state of the CP, a function dependent on the concentration of the CP. We were unable to distinguish the effects of CP oligomerization. However, we did notice that binding to the small RNA motifs did not appear to be cooperative.
The mapping data provided guidance to mutational analysis of RNA binding by the BMV CP. With only seven mutations in the CP, we were surprised to find that three in the globular domain of the CP (D139A, R142A, and D148A) were decreased in the shutoff of BMV RNA accumulation. The three mutants did differ in other CP-associated activities. D139A was shown to decrease the ability to repress accumulation of BMV RNAs (Figs. 4b and 5) and the translation from the reporter construct 2GFP2.10 R142A and D148A retained the ability to inhibit translation but increased RNA synthesis when expressed at concentrations that are expected to be inhibitory to RNA accumulation. Although R142A mutant can replicate, encapsidate, and move systematically, the virion appears to be less stable compared with the WT BMV as determined by differential scanning fluorimetry (Fig. 7b). The D148A virion has lost the ability to move systematically, although it can replicate and form the virion. This result suggests that the C-terminus of BMV CP might control viral long-distance movement, as has been reported by Okinaka et al.31 Since CP could bind with the host cellular protein,32 it raises the possibility that D148A mutant could affect the ability to interact with the host factor(s), which is critical for viral trafficking. Indeed, it was recently reported that cellular proteins’ Cajal body and nucleolus are required for groundnut rosette virus to move viral RNA long distances through the phloem.33,34
The effect of the three mutants on RNA accumulation was seen with both minus- and plus-strand RNAs (Fig. 4c), suggesting that replication activity has been increased with these two mutations. At this point, our results suggest that R142A contributes to RNA-binding affinity without affecting the specific recognition of a regulatory RNA motif. However, a number of factors are compatible with the BMV CP having a role in RNA replication. First, mutations in the CP, such as R142A and D148A, can increase BMV RNA accumulation. Second, the CP can selectively bind the promoter for BMV minus-strand RNA synthesis.11 Third, the R142A substitution that increased RNA accumulation increased the affinity for binding to RNA motifs.
The BMV RNA replication field has generally thought that the replicase does not require the CP. This is in a large part due to BMV RNA1 and RNA2 being able to replicate in the absence of BMV RNA3.9,35,36 However, RNA accumulation in the absence of RNA3 is reduced by at least a log and the CP expressed in trans can rescue RNA accumulation to levels comparable with those seen with RNA3.9 Results herein suggest that the CP should be reevaluated as an important component of BMV RNA replication in plants, as is the case with alfalfa mosaic virus replication.2,37
The multiple roles of the BMV CP in the infection process may take place as follows: After the three genomic RNAs enter the cell’s cytoplasm, the viral RNAs can serve as templates for the translation of replication proteins 1a and 2a. Protein 1a induces invaginations at the ER membrane38 and recruits RNA template as well as binds the 2a polymerase and/or host factor(s).39–41 The progeny RNAs will template the translation of additional BMV proteins, including the CP. At low CP concentrations, it binds to specific RNA motifs to modulate replication.11 At higher CP concentrations, binding to specific RNA elements could prevent translation of BMV proteins and/or replicase formation. It is also possible that the CP could bind elements in the three BMV RNAs with different affinities or different oligomerization states. There is already a suggestion that the one nucleotide difference in the B boxes of RNA2 and RNA3 affected RNA binding by the CP.10 Lastly, we note that, despite being a positive-sense RNA virus with a single-stranded RNA genome, the encapsidated viral RNA may contain a significant amount of RNAs that are base paired.42 It is possible that the CP can serve an important role in preventing the recognition of the double-stranded RNA by the host innate immunity receptors. Clearly, additional roles for the multifunctional CP remain to be elucidated.
Materials and Methods
Materials
The Agrobacterium-mediated gene delivery system to express the three BMV genomic RNAs and the BMV-encoded CP was developed by Gopinath et al.9 The CP mutants for transient expression and RNA3 mutants harboring R142A and D148Awere made by PCR-mediated site-directed mutagenesis with appropriate primers (sequences available upon request). The sequences of the cDNAs were confirmed to have no other mutations. 2GFP2 was previously described by Yi and Kao.43 All RNA oligonucleotides were synthesized by IDT (Coralville, IA).
Agroinfiltration
All the plasmids were introduced into Agrobacterium tumefaciens C58C1 by electroporation. The cultures were grown and infiltrated into N. benthamiana as described by Gopinath et al.9 Agrobacterium cultures harboring the 2GFP2 reporter construct and CP could vary in each experiment, and the amounts infiltrated are stated in the figures or figure legends. For each condition tested, at least two independently infiltrated samples were analyzed, and each result shown was reproduced in at least one other independent experiment that also contained two infiltrated plants.
RNA extraction and Northern blots
Total RNA extraction and Northern blots were performed as described by Yi and Kao.43 Total RNA was isolated from 50 mg of leaf tissue by macerating the tissue in the presence of a lysis buffer (0.1 M glycine, 40 mM ethylenediaminetetraacetic acid, 100 mM NaCl, 2% SDS, and 0.05% bentonite, pH 9.0) and then extracting with an equal volume of phenol and chloroform and precipitating with an equal volume of isopropanol. Glyoxylated RNA (usually 7.5 μg of total RNA denatured at 60 °C for 30 min) was loaded into a 1.2% agarose gel. After transferring the RNA to nylon membrane, Northern blots were performed with α-32P-labeled strand-specific riboprobes. Briefly, to detect minus-strand RNAs, the EcoRI-linearized plasmid pB3HE1, which contains 200 nucleotides of the tRNA-like sequence of BMV 3′-UTR, was used as template for in vitro transcription by a T7 transcription kit (EPICENTRE). The HindIII-linearized plasmid was used as template for in vitro transcription with SP6 polymerase to detect plus-strand RNA.
Protein analysis and Western blot
Leaf tissue was macerated in a buffer containing 50 mM Tris–acetate, pH 7.4, 10 mM MgCl2, 5 mM dithiothreitol (DTT), 1 mM PMSF, and 10% glycerol. The lysate was centrifuged at 18,000g for 30 min at 4 °C to remove the insoluble materials. The supernatant was quantified by spectrometry (nanodrop), and about 10 μg of total protein was subjected to SDS-PAGE and Western blot analysis with monoclonal anti-GFP antibody (Invitrogen) or rabbit anti-CP antibody (ATCC).
Purification of BMV particles and dissociated CPs
BMV virions were purified from N. benthamiana leaves infiltrated with Agrobacterium expressing genomic RNA1 and RNA2 together with either WT or mutant RNA3. Ten days post-inoculation (dpi), the leaves were homogenized in a 1:3 ratio of SAMA extraction buffer (250 mM sodium acetate and 10 mM MgCl2, pH 4.5) and the supernatant was vortexed with 10% chloroform for 10 min. After centrifugation at 18,000g for 10 min, the supernatant was layered on a 10% sucrose cushion prepared in SAMA buffer (50 mM sodium acetate and 10 mM MgCl2, pH 4.5) and centrifuged for 3 h at 28,000g. The viral pellets were dissolved in a 37.5% CsCl solution and subjected to centrifugation at 45,000g in a Beckman Ti 70.1 rotor for 24 h. The band of virions was pulled off by use of a 3-mL syringe fitted with a 23-gauge needle. The virions were dialyzed with a minimum of three changes of SAMA buffer and stored at −80 °C until use.
Dissociated CPs were prepared from CsCl-purified virions by dialysis in a buffer containing 500 mM CaCl2, 1 mM DTT, and 50 mM Tris–HCl, pH 7.5, for 24 h at 4 °C. Following centrifugation at 12,000g for 1 h to remove the precipitated RNAs, the supernatant containing CPs was dialyzed in the buffer containing 300 mM NaCl, 50 mM Tris–HCl, pH 7.5, and 1 mM DTT for 24 h. The concentration of dissociated CP was determined by spectrophotometry and checked for purity by SDS-PAGE.
Fluorescence spectroscopy
All fluorescence measurements were made using an LS55 spectrometer and cuvettes (Perkin-Elmer) with an optical path length of 0.4 cm at room temperature (22 to 23 °C). The fluorophore-labeled RNA was at 0.2 μM in a buffer containing 50 mM Tris–Cl, pH 7.5, and 50 mM NaCl. BMV CP was added in aliquots, with the final added volume not exceeding 10% of the total sample volume. The samples were excited by light set at a wavelength of 495 nm, and the emission was scanned with a wavelength from 510 to 560 nm. The emission scan was repeated 10 times, and all results were averaged. All data were corrected for the background intensity of the buffer and for dilution.
Differential scanning fluorimetry
Differential scanning fluorimetry was performed in an Eppendorf Mastercycler EP Realplex machine. Each sample was prepared in a total volume of 50 μL containing 2.5 μM solutions of WT CP and mutant R142A final concentration, and SYPRO orange (Molecular Probes) was at 2.5× final concentration in buffer T (100 mM Tris, pH 7.0, 50 mM KCl, and 5 mM MgCl2). Each sample was tested in triplicate. The 96-well plate containing all the samples was heated at a rate of 1.0 °C/min, from 25 to 95 °C, and the fluorescence intensity was measured with excitation/emission wavelengths of 470/550 nm. The Tm values were calculated using KaleidaGraph (Synergy Software, Reading, PA).
Electron microscopy and three-dimensional reconstruction of BMV particles
BMV virus particles (in 10-μL aliquots) were adsorbed to a glow-discharged carbon-coated copper grid, negatively stained with 1% uranyl acetate for 20 s, and visualized at a magnification of 39,000× with a JEOL 1200EX electron microscope at 100 kV. Electron micrographs were digitalized using an Epson Projection 3200 scanner (Long Beach, CA) at 1200 dpi, corresponding to 5.5 Å/pixel. The EMAN software package run under Linux was used for single-particle image analysis and three-dimensional reconstruction.26 For all preparations, over 1000 particles were selected from the electron micrographs using EMAN’s Boxer routine. The images were then filtered to remove high- and low-frequency noises, translationally and rotationally aligned, classified, and averaged without applying symmetry. For three-dimensional reconstruction, an initial model was produced from the centered particles with EMAN’s Starticos program and subjected to eight cycles of refinement by projection matching. The final three-dimensional reconstruction was scaled to 5.5 Å/pixel with EMAN’s Proc3d program and visualized with the University of California, Santa Cruz, Chimera software.25
Reversible cross-linking–peptide fingerprinting analysis (RCAP)
NHS-agarose resin (Sigma-Aldrich) was coupled to 5′-amine-modified oligonucleotides in 0.1 M sodium tetra-borate, pH 8.5, quenched with 1 M glycine, and washed extensively before use with a buffer of 20 mM Hepes, pH 7.5, 4 mM MnCl2, and 10 mM DTT. As a control, NHS-agarose resin was also incubated in the absence of any oligonucleotide substrate. CP (2 μM) was then added and incubated at room temperature for 5 min followed by the addition of formaldehyde to a final concentration of 0.1% in a 25-μL reaction. The cross-linking reaction was stopped by the addition of glycine at a final concentration of 0.2 M. After 5 min, sequencing-grade trypsin (Trypsin Gold, Promega, Madison, WI) was added at a protease/substrate ratio of 1:50 (w/w) in 100 mM NH4HCO3, pH 7.8, and incubated overnight at 37 °C. Unbound peptides were washed with 20 mM Hepes, pH 7.5, 1 M NaCl, 1 mM ethylenediaminetetraacetic acid, and 1 mM DTT. Nucleotide–peptide conjugates were then reversed by incubating the samples at 70 °C for 1 h. The samples were centrifuged at 3000g for 5 min, and the supernatants containing the peptides were desalted using a ZipTip (Millipore, Bedford, MA). The bound peptides were eluted in 2.5 μL of 70% acetonitrile and 0.1% trifluoroacetic acid and analyzed by MALDI-ToF MS.
Supplementary Material
Acknowledgments
Funding for this work was provided by the National Science Foundation Genome Systems (grant MCB0641362) and the National Institutes of Health (GM081929 for bioimaging). We thank the Indiana University Cereal Killers for their helpful discussions and support during this work and the Indiana University Mass Spectrometry Facility for allowing use of spectrometers. We thank Dr. Dong Qi for the helpful discussion.
Abbreviations used
- BMV
brome mosaic virus
- CP
capsid protein
- CCMV
cowpea chlorotic mottle virus
- MALDI-ToF
matrix-assisted laser desorption/ionization time of flight
- OD
optical density
- GFP
green fluorescent protein
- RCAP
a method coupling reversible cross-linking, affinity purification, and peptide fingerprinting
- dpi
days post-inoculation
- WT
wild type
- MS
mass spectrometry
- UTR
untranslated region
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2009.05.065
References
- 1.Callaway A, Giesman-Cookmeyer D, Gillock ET, Sit TL, Lommel SA. The multifunctional capsid proteins of plant RNA viruses. Annu Rev Phytopathol. 2001;39:419–460. doi: 10.1146/annurev.phyto.39.1.419. [DOI] [PubMed] [Google Scholar]
- 2.Guogas LM, Laforest SM, Gehrke L. Coat protein activation of alfalfa mosaic virus replication is concentration dependent. J Virol. 2005;79:5752–5761. doi: 10.1128/JVI.79.9.5752-5761.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neeleman L, Olsthoorn RC, Linthorst HJ, Bol JF. Translation of a nonpolyadenylated viral RNA is enhanced by binding of viral coat protein or polyadenylation of the RNA. Proc Natl Acad Sci USA. 2001;98:14286–14291. doi: 10.1073/pnas.251542798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bol JF. Replication of alfamo- and ilarviruses: role of the coat protein. Annu Rev Phytopathol. 2005;43:39–62. doi: 10.1146/annurev.phyto.43.101804.120505. [DOI] [PubMed] [Google Scholar]
- 5.Witherell GW, Gott JM, Uhlenbeck OC. Specific interaction between RNA phage coat proteins and RNA. Prog Nucleic Acid Res Mol Biol. 1991;40:185–220. doi: 10.1016/s0079-6603(08)60842-9. [DOI] [PubMed] [Google Scholar]
- 6.Lucas RW, Larson SB, McPherson A. The crystallographic structure of brome mosaic virus. J Mol Biol. 2002;317:95–108. doi: 10.1006/jmbi.2001.5389. [DOI] [PubMed] [Google Scholar]
- 7.Rao AL. Genome packaging by spherical plant RNA viruses. Annu Rev Phytopathol. 2006;44:61–87. doi: 10.1146/annurev.phyto.44.070505.143334. [DOI] [PubMed] [Google Scholar]
- 8.Noueiry AO, Ahlquist P. Brome mosaic virus RNA replication: revealing the role of the host in RNA virus replication. Annu Rev Phytopathol. 2003;41:77–98. doi: 10.1146/annurev.phyto.41.052002.095717. [DOI] [PubMed] [Google Scholar]
- 9.Gopinath K, Dragnea B, Kao CC. Interaction between brome mosaic virus proteins and RNAs: effects on RNA replication, protein expression and RNA stability. J Virol. 2005;79:14222–14234. doi: 10.1128/JVI.79.22.14222-14234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi G, Letteney E, Kim CH, Kao CC. Brome mosaic virus capsid protein regulates accumulation of viral replication proteins by binding to the replicase assembly RNA element. RNA. 2009;15:615–626. doi: 10.1261/rna.1375509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu J, Gopinath K, Murali A, Yi G, Hayward SD, Zhu H, Kao C. RNA-binding proteins that inhibit RNA virus infection. Proc Natl Acad Sci USA. 2007;104:3129–3134. doi: 10.1073/pnas.0611617104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim CH, Kao C. A mutant viral RNA promoter with an altered conformation retains efficient recognition by a viral RNA replicase through a solution-exposed adenine. RNA. 2001;7:1476–1485. [PMC free article] [PubMed] [Google Scholar]
- 13.Kim CH, Kao C, Tinoco I. RNA motifs that determine specificity between a viral replicase and its promoter. Nat Struct Biol. 2000;7:415–423. doi: 10.1038/75202. [DOI] [PubMed] [Google Scholar]
- 14.Kim YC, Russell WK, Ranjith-Kuman CT, Thomson M, Russell DH, Kao CC. Functional analysis of RNA binding by the hepatitis C virus RNA-dependent RNA polymerase. J Biol Chem. 2005;280:38011–38019. doi: 10.1074/jbc.M508145200. [DOI] [PubMed] [Google Scholar]
- 15.Gelvin SB. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev. 2003;67:16–37. doi: 10.1128/MMBR.67.1.16-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman MR, Kao CC. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J Mol Biol. 1999;286:709–720. doi: 10.1006/jmbi.1998.2503. [DOI] [PubMed] [Google Scholar]
- 17.Ranjith-Kumar CT, Zhang X, Kao CC. Enhancer-like activity of a brome mosaic virus RNA promoter. J Virol. 2003;77:1830–1839. doi: 10.1128/JVI.77.3.1830-1839.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.She YM, Haber S, Seifers DL, Loboda A, Chernushevich I, Perreault H, et al. Determination of the complete amino acid sequence for the coat protein of brome mosaic virus by time-of-flight mass spectrometry: evidence for mutations associated with change of propagation host. J Biol Chem. 2001;276:20039–20047. doi: 10.1074/jbc.M100189200. [DOI] [PubMed] [Google Scholar]
- 19.Bhardwaj K, Palaninathan S, Alcantara JM, Yi LL, Guarino L, Sacchettini JC, Kao CC. Structural and functional analysis of the severe acute respiratory syndrome coronavirus endo-ribonuclease Nsp15. J Biol Chem. 2008;283:3655–3664. doi: 10.1074/jbc.M708375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ranjith-Kumar CT, Duffy KE, Jordan JL, Eaton-Bassiri A, Vaughan R, Hoose S, et al. Single-stranded oligonucleotides can inhibit cytokine production induced by human toll-like receptor 3. Mol Cell Biol. 2008;28:4507–4519. doi: 10.1128/MCB.00308-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao AL, Grantham GL. Biological significance of the seven amino-terminal basic residues of brome mosaic virus coat protein. Virology. 1995;211:42–52. doi: 10.1006/viro.1995.1377. [DOI] [PubMed] [Google Scholar]
- 22.Rao AL, Grantham GL. Molecular studies on bromovirus capsid protein: II. Functional analysis of the amino-terminal arginine-rich motif and its role in encapsidation, movement, and pathology. Virology. 1996;226:294–305. doi: 10.1006/viro.1996.0657. [DOI] [PubMed] [Google Scholar]
- 23.Speir JA, Munshi S, Wang G, Baker TS, Johnson SE. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure. 1995;15:63–78. doi: 10.1016/s0969-2126(01)00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shepherd CM, Borelli IA, Lander G, Natarajan P, Siddavanahalli V, Bajaj C, et al. VIPERdb: a relational database for structural virology. Nucleic Acids Res. 2006;34:D386–D389. doi: 10.1093/nar/gkj032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 26.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 27.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 28.Dragnea B, Chen C, Kwak ES, Stein B, Kao CC. Gold nanoparticles as spectroscopic enhancers for in vitro studies on single viruses. J Am Chem Soc. 2003;125:6374–6375. doi: 10.1021/ja0343609. [DOI] [PubMed] [Google Scholar]
- 29.Takeda A, Kaido M, Okuno T, Mise K. The C terminus of the movement protein of brome mosaic virus controls the requirement for coat protein in cell-to-cell movement and plays a role in long-distance movement. J Gen Virol. 2004;85:1751–1761. doi: 10.1099/vir.0.79976-0. [DOI] [PubMed] [Google Scholar]
- 30.Calhoun SL, Speir JA, Rao AL. In vivo particle polymorphism results from deletion of a N-terminal peptide molecular switch in brome mosaic virus capsid protein. Virology. 2007;364:407–421. doi: 10.1016/j.virol.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okinaka Y, Mise K, Suzuki E, Okuno T, Furusawa I. The C terminus of brome mosaic virus coat protein controls viral cell-to-cell and long-distance movement. J Virol. 2001;75:5385–5390. doi: 10.1128/JVI.75.11.5385-5390.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okinaka Y, Mise K, Okuno T, Furusawa I. Characterization of a novel barley protein, HCP1, that interacts with the brome mosaic virus coat protein. Mol Plant–Microbe Interact. 2003;16:352–359. doi: 10.1094/MPMI.2003.16.4.352. [DOI] [PubMed] [Google Scholar]
- 33.Kim SH, Ryabov EV, Kalinina NO, Rakitina DV, Gillespie T, MacFarlane S, et al. Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J. 2007;26:2169–2179. doi: 10.1038/sj.emboj.7601674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SH, Macfarlane S, Kalinina NO, Rakitina DV, Ryabov EV, Gillespie T, et al. Interaction of a plant virus-encoded protein with the major nucleolar protein fibrillarin is required for systemic virus infection. Proc Natl Acad Sci USA. 2007;104:11115–11120. doi: 10.1073/pnas.0704632104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.French R, Ahlquist P. Intercistronic as well as terminal sequence are required for efficient amplification of brome mosaic virus RNA3. J Virol. 1987;61:1457–1465. doi: 10.1128/jvi.61.5.1457-1465.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pogue GP, Hall TC. The requirement for a 5′ stem–loop structure in brome mosaic virus replication supports a new model for viral positive-strand RNA initiation. J Virol. 1992;66:674–684. doi: 10.1128/jvi.66.2.674-684.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reichert VL, Choi M, Petrillo JE, Gehrke L. Alfalfa mosaic virus coat protein bridges RNA and RNA-dependent RNA polymerase in vitro. Virology. 2007;364:214–226. doi: 10.1016/j.virol.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahlquist P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat Rev Microbiol. 2006;4:371–382. doi: 10.1038/nrmicro1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen J, Ahlquist P. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J Virol. 2000;74:4310–4318. doi: 10.1128/jvi.74.9.4310-4318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Noueiry AO, Ahlquist P. Brome mosaic virus protein 1a recruits viral RNA2 to RNA replication through a 5′ proximal RNA2 signal. J Virol. 2001;75:3207–3219. doi: 10.1128/JVI.75.7.3207-3219.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kao CC, Ahlquist P. Identification of domains required for direct interaction of the helicase-like and polymerase-like RNA replication proteins of brome mosaic virus. J Virol. 1992;66:7293–7302. doi: 10.1128/jvi.66.12.7293-7302.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson KN, Tang L, Johnson JE, Ball LA. Heterologous RNA encapsidated in Pariacoto virus-like particles forms a dodecahedral cage similar to genomic RNA in wild-type virions. J Virol. 2004;78:11371–11378. doi: 10.1128/JVI.78.20.11371-11378.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yi G, Kao C. cis- and trans-acting functions of brome mosaic virus protein 1a in genomic RNA1 replication. J Virol. 2008;82:3045–3053. doi: 10.1128/JVI.02390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.