SUMMARY
The Hedgehog (Hh) signaling pathway initiates an evolutionarily conserved developmental program required for the proper patterning of many tissues. Costal2 (Cos2) is a requisite component of the Hh pathway, whose mechanistic role is not well understood. Cos2 was initially predicted, based on its primary sequence, to function as a microtubule-associated (MT) molecular motor. However, despite being identified over a decade ago, evidence showing that Cos2 function might require kinesin-like properties has for the most part been lacking. Thus the prevailing dogma in the field is that Cos2 functions solely as a scaffolding protein during Hh signal transduction. Here, we provide the first evidence that Cos2 motility is required for its biological function, and that this motility may be Hh regulated. We show that Cos2 motility requires an active motor domain, ATP and microtubules. Additionally, Cos2 recruits and transports other components of the Hh signaling pathway, including the transcription factor Cubitus interruptus (Ci), throughout the cell. Drosophila expressing cos2 mutations that encode proteins that lack motility are attenuated in their ability to regulate Ci activity and exhibit phenotypes consistent with attenuated Cos2 function. Combined, these results demonstrate that Cos2 motility plays an important role in its function, regulating the amounts and activity of Ci that ultimately interpret the level of Hh to which cells are exposed.
Keywords: hedgehog, Costal2, microtubules, kinesin, development
RESULTS AND DISCUSSION
Cos2 associates with the transcription factor Ci [1, 2], the transmembrane protein Smoothened (Smo) [3–6], and a number of protein kinases [7–10] that regulate the Hh pathway, consistent with it acting as a scaffolding protein [7, 10–12]. We have previously shown, by subcellular fractionation of Drosophila cell extracts, that the majority of Cos2 migrates with a vesicular membrane enriched fraction [13]. Conversely, Cos2 enriches in a cytoplasm enriched fraction when isolated from cells exposed to Hh, indicating that its membrane association is negatively regulated by Hh. Although Hh appears to regulate the affinity of Cos2 for membrane vesicles [13] and microtubules (MTs) [7], the relationship between Cos2 function and its association with these subcellular structures is largely unknown.
To explore the link between Cos2 binding to both MTs and vesicular membranes, we first expressed a GFP-(Green Fluorescent Protein)-tagged wild-type (wt) cos2 construct (cos2-GFP), similar to one capable of rescuing cos2 mutant Drosophila [14], and determined its subcellular location in live S2 cells (Fig. 1a). S2 cells are a Hh responsive Drosophila cultured cell line that is able to perform most responses to the Hh signal, but lacks ci expression [15, 16]. Similar to a truncated Cos2-GFP fusion protein previously described [13], full length Cos2-GFP enriched in various sized puncta throughout the cytoplasm (Fig. 1a). The number and relative size of these puncta appeared somewhat variable within the population of transfected cells (for examples see Fig. 1a, 1b, 1d, and Fig. 2a), as did the amount of Cos2 that localized in a more diffuse manner (Table 1). Hh attenuated the localization of Cos2 to these distinct puncta, as Cos2-GFP localization appeared relatively more diffuse in S2 cells exposed to Hh (Fig. 1a’), but had little effect on the levels of Cos2-GFP found in cells (see Fig. S1g). Similar results were obtained when smo fused to GFP (smo-GFP) was co-expressed with cos2-RFP (Cos2 tagged with Red Fluorescent Protein) (Fig. 1b’ compared to Fig 1b and Table 1), as high levels of Smo are known to activate Hh signaling [17]. The number of cells exhibiting diffuse Cos2-RFP localization, versus the number of cells with Cos2-RFP in puncta, increased as the amount of smo-GFP transfected was increased. Furthermore, this increase in diffuse Cos2-RFP localization only occurred in the population of cells co-expressing smo-GFP (data not shown).
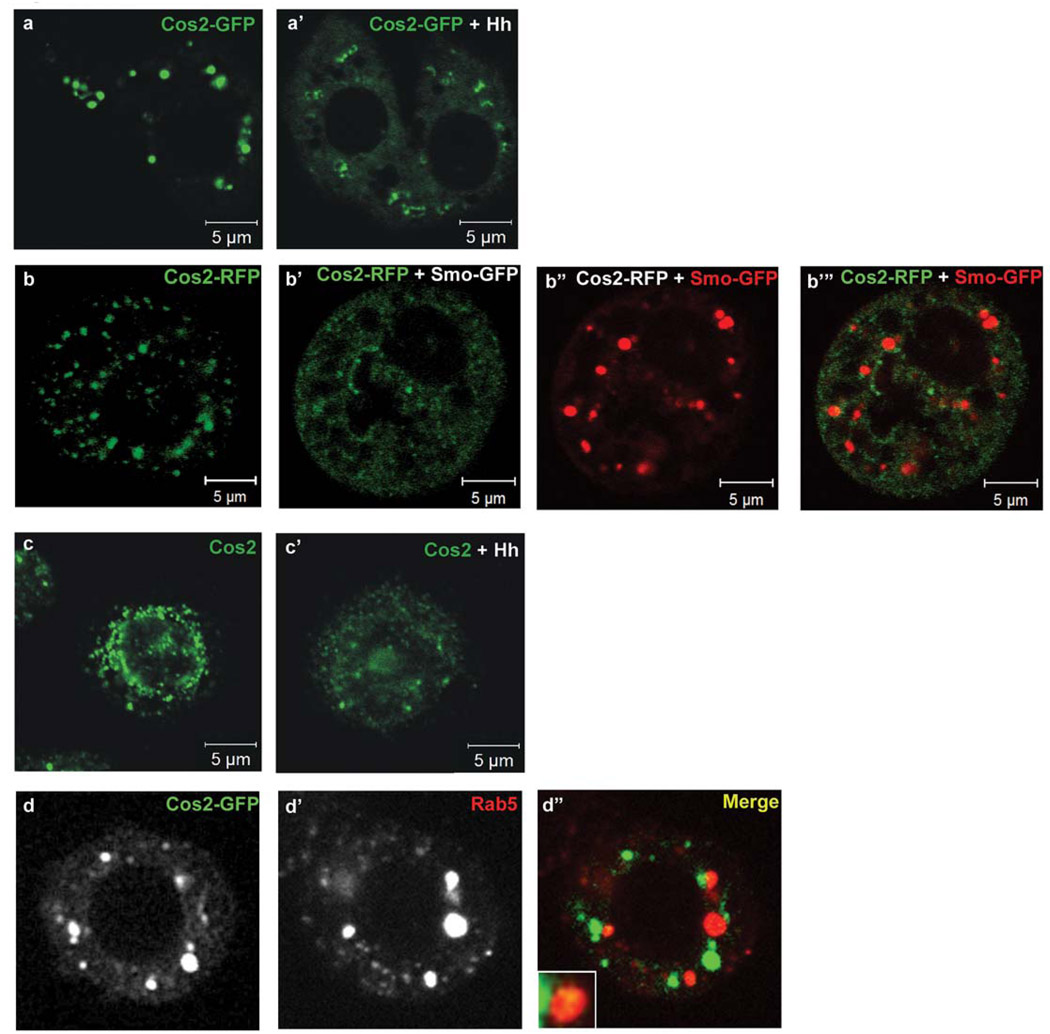
Figure 1. Hh regulates the subcellular localization of Cos2.
(a–a’) Live images of S2 cells expressing cos2-GFP with (+) or without hh. The bulk of Cos2-GFP is found in puncta in the absence of Hh. Hh exposure results in a more diffuse localization of Cos2-GFP. (b–b’”) Live images of S2 cells expressing (b) cos2-RFP only, pseudo-colored green, or (b’–b’”) co-expressing smo-GFP, pseudo-colored red. As with Cos2-GFP, Cos2-RFP is found primarily in puncta. However, co-expression with smo-GFP results in a shift to a more diffuse localization of Cos2-RFP. (c–c’) Indirect immunofluorescent staining of endogenous Cos2 in S2 cells, with (+) or without hh. The majority of Cos2 appears punctate throughout the cytoplasm of the cells. Exposure to Hh leads to an apparent net reduction in fluorescence intensity, decreased localization to puncta and increased diffuse staining of Cos2. (d–d”) Cos2-GFP puncta partially co-localize with Rab5. Approximately 30% of the total Cos2-GFP fluorescence co-localizes with Rab5, a marker of early endosomal vesicles, delineating one pool of membrane vesicle associated Cos2. Inset in lower left-hand corner shows an enlarged area of Cos2 and Rab5 co-localization.
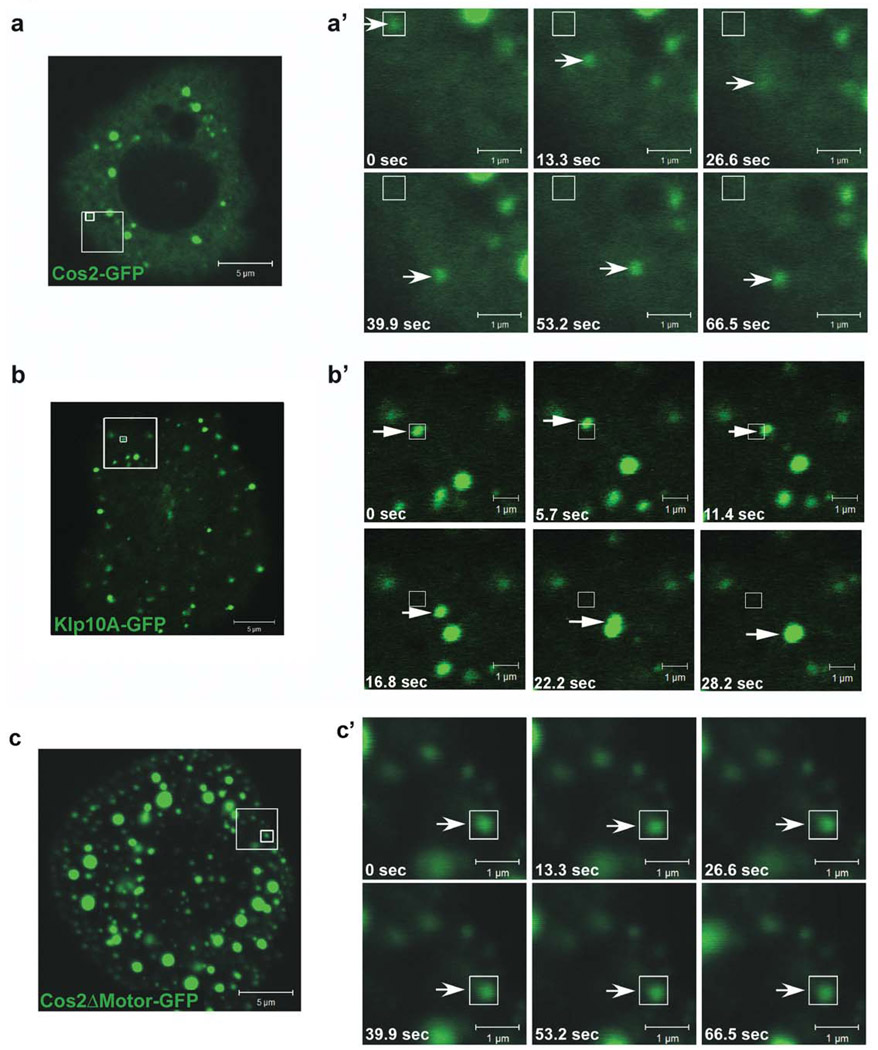
Figure 2. Cos2 puncta are motile.
S2 cells expressing cos2-GFP (a), klp10A-GFP (b), a known kinesin family member, or cos2ΔMotor-GFP (c), a GFP-fused truncated form of Cos2 that lacks its putative motor domain. The large box in each panel indicates the enlarged area depicted in the corresponding time series in (a’), (b’) and (c’). The smaller box indicates the origin of a single puncta that is tracked, over time, with arrows in (a’), (b’) and (c’). (a’, b’) Time-lapse images showing a Cos2-GFP puncta (a’) and a Klp10A-GFP puncta (b’) both moving away from their original locations (small boxes). (c’) Time-lapse images showing a Cos2ΔMotor-GFP puncta that remains essentially immobile over the same time period as in a’.
Table 1. Quantification of the number of cells with diffuse Cos2 localization in response to various treatments.
Cos2 exhibits a more diffuse localization in response to different conditions, and the percentages shown represent the penetrance of this effect within populations of cells. The results shown here are the quantitation of data shown in Figs. 1b, 1c, Figs. S1a, and S1c. “n” represents total number of cells counted from random fields of cells. Cos2-RFP diffuseness in the presence of Smo (+ Smo) was quantified only from cells visibly expressing both cos2-RFP and smo-GFP. Cells in the various experiments exhibited a 25–30% overall transfection efficiency.
| Conditions | n | Cells with diffuse Cos2 localization (%) | |
|---|---|---|---|
| Endogenous Cos2 | −Hh | 38 | 10.5 |
| +Hh | 76 | 30.3 | |
| + Fu dsRNA | 90 | 10 | |
| + Fu dsRNA; + Hh | 113 | 10.6 | |
| Cos2- RFP | − Smo | 49 | 14.2 |
| + Smo | 48 | 77.9 |
To verify that the localization of Cos2-GFP was similar to that of native Cos2 we examined the localization of endogenous Cos2 in S2 cells transfected with a hh expression vector or empty vector control (Fig. 1c and Fig. S1a). Endogenous Cos2 also localized to different sized punctate structures. Furthermore, this punctate localization of Cos2 is significantly altered in cells exposed to Hh, exhibiting a more diffuse localization than Cos2 in cells not exposed to Hh (Fig. 1c’ compared to Fig. 1c, Fig. S1a, and Table 1). These puncta indeed represent localization of endogenous Cos2, as they are largely absent in S2 cells treated with Cos2-specific dsRNA (Fig. S1d). Taken together, our observations are consistent with Cos2-GFP localizing in a manner similar to endogenous Cos2, both in the presence and absence of Hh.
We have previously shown that Cos2 co-migrates with vesicular markers, suggesting that some fraction of the Cos2 puncta we observe is localized to discrete membrane vesicles [13]. To begin to identify what types of vesicles Cos2 localizes to, we expressed cos2-GFP and probed for a series of endogenous vesicular markers (Fig. 1d, Fig. S2 and Table S1). We obtained significant overlap of Cos2-GFP puncta with the early endosome specific marker Rab5 (∼30%) (Fig. 1d and Fig. S2a), but also with the recycling endosome marker Rab11 (Fig. S2b), the early and late endosome marker hepatocyte growth factor regulated tyrosine kinase substrate (Hrs) (Fig. S2c), and the late endosome marker lysobisphosphatidic acid (LBPA) (Fig. S2d). Thus, a subset of the Cos2 puncta observed here likely represent endosomal vesicles.
To identify what factors might be regulating Cos2 localization to these puncta, we knocked down the expression of Smo by RNAi and examined the ability of Cos2 to translocate from a membrane enriched fraction to a cytoplasm enriched fraction (Fig. S1b). Consistent with Smo being required for all Hh signaling [18–20], its knockdown attenuated the ability of Cos2 to translocate from the membrane enriched fraction to a cytoplasm enriched fraction in response to Hh. Similar results were obtained when the level of the Hh regulated protein kinase Fused (Fu) was knocked down by RNAi (Figs. S1b, S1c, and Table 1), suggesting that Hh may regulate Cos2 relocalization in a manner dependent on Fu function. Hh normally induces the Fu-dependent phosphorylation of Cos2 [7, 21]. However, over-expression of both fu and cos2 leads to the phosphorylation of Cos2 at its physiologically relevant site [21], suggesting that Fu may be constitutively active when over-expressed with Cos2 (see also Fig. S1f). Consistent with this latter suggestion, co-expression of fu with cos2-GFP also affected its subcellular distribution in a dose-dependent manner, with much of Cos2-GFP appearing more diffuse in the presence of high levels of exogenous Fu (Fig. S1e and data not shown). These results suggested a positive role for Smo and Fu in Hh dependent Cos2 relocalization.
Many of the Cos2-GFP puncta appeared to move rapidly throughout the cytoplasm of transfected S2 cells (Fig. 2a and Movie 1A and Movie 1B). The Cos2 enriched puncta exhibited a velocity inversely related to their size, with smaller puncta moving faster than larger puncta (Fig. S3c, Movie 1A and Movie 1B). For ease of comparison we divided the different sized puncta into three groups, designated large, medium or small (Fig. S3a), and compared their velocities (Fig. S3c and Table S2). The average velocity of the small wt Cos2-GFP puncta was 62 nm/sec, which was within the range reported for other Kinesin-like proteins (KLPs) [22–25]. However, this average Cos2-GFP velocity was less than the average velocity of the KLP we examined as a control (Klp10A) [26, 27] (Fig. 2b and Table 2), consistent with Cos2 being at the slower end of the spectrum of KLPs. Many of the Cos2-GFP puncta were also quite dynamic in nature, with small puncta appearing to fuse with and even bud off from the larger puncta (Fig. S4a and Movie 14A). Photobleaching experiments verified the dynamic nature of Cos2-GFP populating these puncta (Fig. S4b and Movie 14B), which quickly recovered from the photobleaching procedure.
Table 2. Comparison of mean velocities of small Cos2 puncta.
Velocities were measured as described in Methods section. Small Cos2 puncta (i.e. puncta that fall within the 0.10–0.40 µm size range) had a mean velocity of 61.7 nm/sec. Mutations or conditions predicted to impede or prevent movement, such as Cos2ΔMotor, Cos2-S182N, and ATP depletion, all resulted in a significant decrease in Cos2 velocity. The velocity of Cos2 puncta co-localized with Fu-tail or Ci-CORD were not significantly different from that of Cos2 alone, whereas the velocity of Cos2 co-localized with Smo puncta was significantly faster than Cos2 alone. Standard error of the mean (SE) is shown in the rightmost column and ** indicates that values shown are significantly different when compared to Cos2 (p ≤ 0.02).
| GFP-tagged Constructs | Velocity (nm/sec) | SE |
|---|---|---|
| Cos2 | 61.7 | 6.8 |
| Cos2-ΔMotor | 12.2** | 6.6 |
| Cos2-S182N | 2.9** | 1.4 |
| ATP-depleted Cos2 | 12.8** | 6.1 |
| ATP-depletion mock treatment | 73.6 | 23.5 |
| Cos2 colocalized with Fu-Tail | 87.5 | 14.1 |
| Cos2 colocalized with Ci-CORD | 77.7 | 9.5 |
| Cos2 colocalized with Smo | 167.3** | 33.8 |
| Klp10A | 253.5** | 75.1 |
To confirm that Cos2-GFP movement was an active process dependent on its kinesin-like activity and not due to basal cytoplasmic streaming, we compared the movement of wt Cos2-GFP to Cos2ΔMotor-GFP (Fig. 2c also Movie 1A compared to Movie 3). Cos2ΔMotor is a mutant Cos2 fusion protein that lacks its putative motor domain and would be predicted to be immotile. A similar motorless GFP fusion protein has been shown to function as a dominant negative inhibitor of wt Cos2 in vivo [14], presumably through association with endogenous wt Cos2. Although some minor movement of Cos2ΔMotor-GFP puncta was observed, a significant portion of the Cos2-GFP puncta translocated over a greater distance during the same time frame (compare Fig. 2a’ and 2c’, as well as Table 2 and Table S2). The immobile Cos2ΔMotor-GFP puncta were more numerous and larger than those observed with wt Cos2-GFP (Fig. 2c), and the average velocity of small Cos2ΔMotor-GFP puncta was significantly slower than that of similarly sized Cos2-GFP puncta (Table 2 and Fig. S3c). Residual movement of these Cos2ΔMotor-GFP puncta was slightly greater than that observed in ATP depleted S2 cells. As the cargo of KLPs typically associate with both an anterograde motor and a retrograde motor, the movement of this small subset of puncta may be due to transport by another motor protein [28]. Many smaller immobile Cos2ΔMotor-GFP puncta also appeared to enrich near the plasma membrane, consistent with the hypothesis that Cos2 normally may traffic to and from the plasma membrane. Thus, these results begin to correlate the motility of Cos2 with its function, as a mutant of Cos2 which lacks motility has attenuated function in vivo [14].
We noted that many of the Cos2-GFP puncta observed in cells appeared to move in discrete linear stretches, as if moving along a MT network. KLP motor activity requires, at its most basic, ATP and intact MTs [29, 30]. Therefore, we examined the ability of Cos2 to move in the absence of either ATP or an intact MT network (Fig. 3). S2 cells that were depleted of ATP [31] still contained Cos2 puncta, but these puncta were almost completely immobile (Fig. 3a and a’, Table 2 and Movie 4). However, upon restoration of ATP levels, Cos2-GFP puncta motility was quickly re-established (Fig. 3b and b’, also Movie 5). Note also that Cos2-GFP puncta in vehicle treated S2 cells remained motile (Table 2 and Fig. S3b). To then test whether these Cos2 puncta co-localize with MTs, we expressed cos2-RFP in cells stably expressing GFP-α-tubulin. Whereas the bulk of RFP alone was predominantly nuclear (data not shown), the majority of punctate Cos2-RFP appeared to co-localize with the MT network (Fig. 3c and Fig. S5b). We next used nocodazole to disrupt the MT network of individual S2 cells expressing cos2-GFP and examined the localization of Cos2-GFP to discrete MT associated puncta using live imaging (Fig. 3d and Fig. S5a). The loss of punctate Cos2-GFP in a nocodazole treated cell correlated with the destabilization of the MT network, with loss of peripheral MTs and peripheral puncta preceding loss of more central MTs and puncta. These results show that Cos2 co-localizes with MTs and that Cos2’s enrichment in punctate structures depends on an intact MT network. Since Hh is known to disrupt the physical association between Cos2 and MTs [7, 32], Hh might control Cos2’s ability to form motile puncta by regulating its MT association. We note however, that despite its lack of a motor domain, Cos2ΔMotor-GFP is still able to form puncta (Fig. 2c). We hypothesized that either Cos2ΔMotor-GFP is able to bind MTs through a domain distinct from its motor domain, which normally harbors a MT binding motif [33], or that Cos2ΔMotor-GFP’s ability to form puncta is due to dimerization with endogenous wt Cos2. Consistent with this latter suggestion, immunoprecipitation of Cos2ΔMotor-GFP was able to co-precipitate full-length wt Cos2 (Fig. S3d).
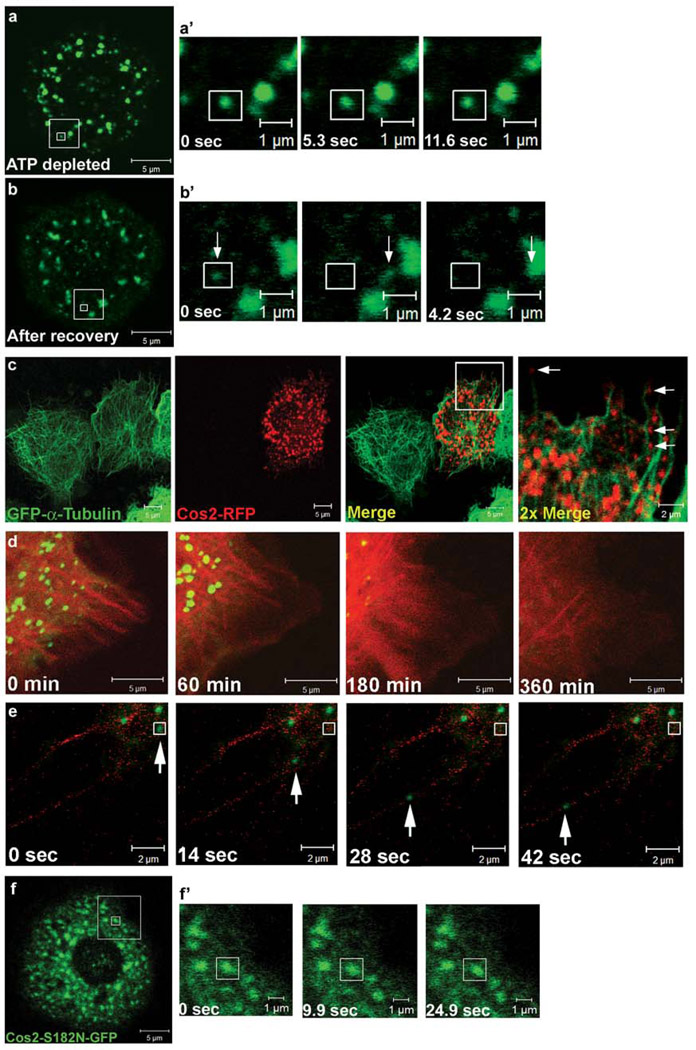
Figure 3. Cos2 motility requires microtubules and ATPase activity.
The large box in panels (a) and (b) indicates the enlarged area depicted in the corresponding time series (a’ and b’). The smaller boxes indicate the origin of a single puncta that is tracked, over time. (a– a’) S2 cells expressing cos2-GFP were ATP depleted by treatment with 5 mM sodium azide and 1 mM 2-deoxyglucose. Time-lapse images were taken 30 minutes after ATP depletion. The vast majority of Cos2-GFP puncta were immobile, relative to the vehicle treated S2 cells, which exhibited similar motility to untreated wt Cos2-GFP (Figure S3). (b– b’) Cos2 movement is recovered after ATP levels are restored, verifying the reversibility of the ATP depletion. The same cell as in (a) was reimaged after the ATP depletion solution was removed and the cell was allowed to recover for 30 minutes in fresh media. The movement of a single puncta (indicated by arrows) is tracked over a short time course. (c) Cos2 appears to localize along MT tracks. cos2-RFP was expressed in a S2 cell line stably expressing GFP-α-tubulin and imaged in live cells. The large box in the merge panel indicates the enlarged area depicted in the 2x merge panel. Individual Cos2-RFP puncta aligning with MTs are marked by arrows in the rightmost panel. (d) Live images of S2 cells co-transfected with plasmids expressing cos2-GFP and mCherry-tubulin, then treated with nocodazole. Images show a single cell over the course of 6 hours and show the correlation between dissociation of Cos2-GFP puncta and MT destabilization induced by nocodazole treatment. A more extensive time course of this nocodazole treated cell, showing Cos2-GFP and mCherry-tubulin in separate channels, is shown in Fig. S5a. (e) Live images of S2 cells expressing cos2-GFP and mCherry-tubulin, treated with 5 µM cytochalasin for 18 hours. Cytochalasin disrupts the actin cytoskeleton, allowing the formation of long MT-enriched projections, along which Cos2-GFP puncta appear to co-localize and move. The puncta indicated by the arrow traveled approximately 8 µm along the visible MT projection. (f–f’) Live images of S2 cells expressing cos2-S182N-GFP. (f) Cos2-S182N-GFP shows a highly punctate staining pattern in a subset of cells, similar to wt Cos2-GFP. The large box in panel (f) indicates the enlarged area depicted in the corresponding time series (f’) and the smaller box indicates the origin of a single puncta that is tracked, over time. (f’) Cos2-S182N-GFP puncta tracked over time do not exhibit significant motility (see also Fig. S3b).
To more directly establish that Cos2 puncta appear to move along MTs we initially analyzed Cos2-RFP movement in cells expressing GFP-α-tubulin, and observed many smaller Cos2-RFP puncta that appeared to track along MTs (Fig. S5b and Movie 6) [34]. To visualize longer and more defined MTs, we induced S2 cells expressing cos2-GFP and mCherry-tubulin to form long, thin, MT-enriched cellular extensions by treating them with the actin disrupting agent cytochalasin-D (Fig. 3e) [35]. Under these conditions, Cos2-GFP puncta also appear to move along these longer MT extensions (Movie 7A and Movie 7B). Approximately 80% of the Cos2-GFP puncta that localized to these MT-enriched cellular extensions exhibited significant mobility (Table 3). The Cos2-GFP puncta observed in this experiment moved at a slightly faster velocity than described earlier. The increased velocity may only be due to an increased ability to observe and measure uninterrupted stretches of MTs or may be due to the loss of some impediment to movement upon disruption of the actin cytoskeleton.
Table 3. Quantitation of Cos2-GFP puncta motility along microtubules.
S2 cells expressing cos2-GFP and mCherry-tubulin were treated with 5 µM cytochalasin for 18 hours. Cytochalasin disrupts the actin cytoskeleton, allowing formation of long MT projections, along which Cos2-GFP puncta appear to co-localize and move. MT projections are defined as a projection measuring more than 5µm in length and has a MT track that is labeled by mCherry-tubulin. Motile puncta were defined by previously described criteria outlined in the Methods section. Eighty-one percent of all puncta visible on MT projections moved in a manner consistent with them tracking along MTs.
| Cell Image | Microtubule projections per cell | Motile puncta per microtubule projections | Total puncta observed | Total puncta on microtubules exhibiting motility (%) |
|---|---|---|---|---|
| 1 | 10 | 11 | 21 | 52.4 |
| 2 | 12 | 3 | 3 | 100 |
| 3 | 7 | 27 | 29 | 93.1 |
| 4 | 2 | 5 | 5 | 100 |
| 5 | 5 | 18 | 21 | 85.7 |
| Totals: | 36 | 64 | 79 | 81 |
Molecular motors utilize ATP hydrolysis to generate force, which is then translated into movement [36]. A putative Cos2 ATPase-deficient mutant, Cos2-S182N, has been shown to function as a dominant negative inhibitor of endogenous Cos2 in vivo [14], suggesting that Cos2 requires efficient ATPase activity for function. Mutation of the S182 residue in the putative P-loop domain of Cos2 [32] is hypothesized to prevent ATP hydrolysis and render it immotile in MT-bound rigor complexes [14, 37]. We expressed GFP tagged cos2-S182N in cells to determine the contribution of ATPase activity to Cos2’s ability to move along MTs. Consistent with the prediction that this mutation should lack motor activity, we found that while Cos2-S182N-GFP was able to enrich in puncta, these puncta did not exhibit any significant motility (Fig. 3f, Table 2, and Movie 8). Thus, in the absence of a functional ATPase domain, the mobility of Cos2-GFP enriched puncta is significantly attenuated, exhibiting motility comparable to that observed in ATP depleted S2 cells (Fig. S3b and Table 2). Although Cos2-S182N-GFP was observed in immobile puncta in all cells, in general the localization of Cos2-S182N-GFP appeared more diffuse than that observed with wt Cos2-GFP (data not shown). This increased degree of diffuse localization of Cos2 is consistent with a decreased affinity for MTs. Interestingly, mutations in conserved nucleotide binding motifs in other KLPs can also affect their MT affinity [38, 39]. These results suggest that Cos2 requires a functional ATPase domain for efficient MT dependent movement, as would be expected for a classical KLP. Taken together with the phenotype of this cos2 mutant in Drosophila [14], these results, along with those observed with Cos2ΔMotor-GFP, suggest that Cos2 ATP-dependent motility is crucial for its activity.
Our results thus far suggest that Hh regulates some aspect of Cos2 function that involves MT-dependent vesicular trafficking. Such a role for Cos2 would certainly be consistent with its primary sequence similarity to KLPs. However, Cos2 is currently proposed to function as a molecular scaffold, binding to and enriching the local concentration of other members of the Hh signaling pathway [1–7, 9–12]. If Cos2 is able to move along the MT network, it is likely to transport these various binding partners throughout the cell. To test this hypothesis, we co-expressed wt cos2 with either the carboxyl-terminal domain of Fu tagged with GFP (GFP-fu-tail), or a region of Ci tagged with GFP (GFP-ci-CORD), as both of these regions of Fu and Ci associate with Cos2 [1, 10, 40] (Figs. 4b, 4d and Movie 9 and Movie 11). When expressed on their own, GFP-Fu-tail localized diffusely throughout the cytoplasm and nucleus, while GFP-Ci-CORD is predominantly nuclear [41] (Figs. 4a and 4c). However, when either GFP-fu-tail, or GFP-ci-CORD, was co-expressed with wt untagged cos2, they relocalized to discrete motile puncta that exhibited a similar velocity to that observed for Cos2-GFP alone (data not shown). Moreover, a high degree of co-localization and co-movement of Cos2-RFP with either Fu-tail-GFP or Ci-CORD-GFP was also observed (Fig. S6a, S6b, Table 2, Movie 10 and Movie 12). The velocity of these co-labeled puncta was similar to that of Cos2-GFP puncta alone (Fig. S3b and Table 2). These results suggest that Cos2 has the ability to tether two other Hh signaling components, Fu and Ci, to discrete puncta and transport them through the cytoplasm. Consistent with this suggestion, Fu, Cos2 and Ci appear to co-localize in various sized puncta when all three are co-expressed in S2 cells (Fig. S6d).
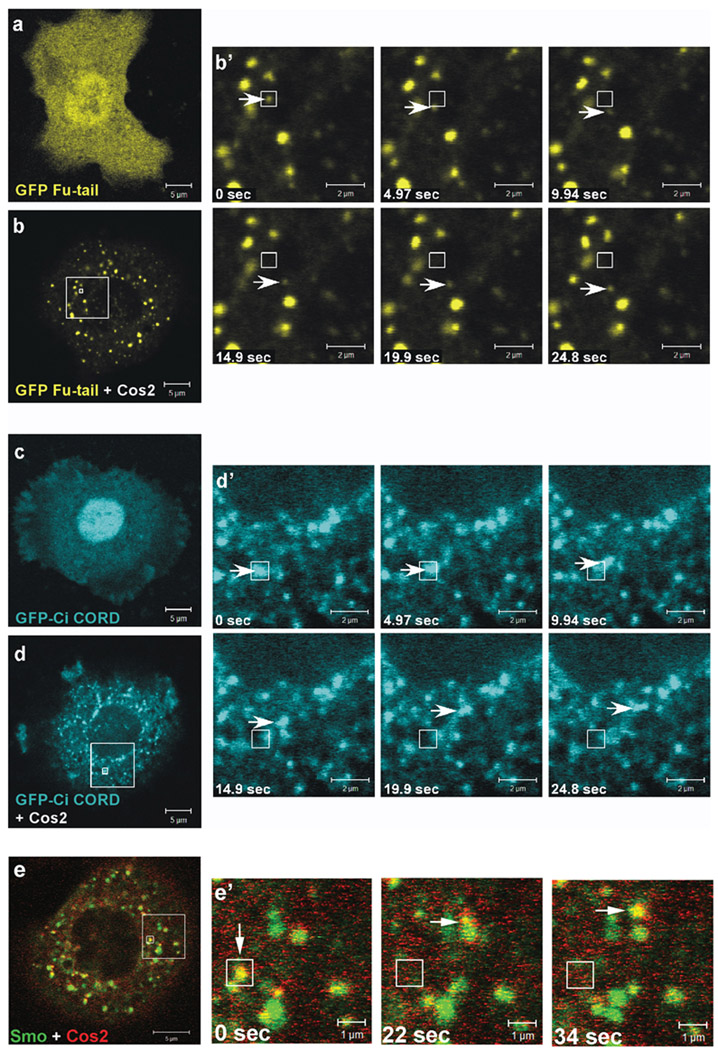
Figure 4. Cos2 recruits and moves other components of the Hh signaling pathway.
GFP-fu-tail (a, b) and GFP-ci-CORD (c, d) were expressed in S2 cells in the presence or absence of exogenous wt cos2. (a and b) Live images of S2 cells expressing GFP-fu-tail. GFP-Fu-tail localizes in a diffuse manner when expressed alone (a), but co-expression with wt cos2 dramatically relocalizes GFP-Fu-tail into puncta (b), which move with a similar velocity (b’) as Cos2 puncta. (c and d) Live images of S2 cells expressing GFP-ci-CORD. As previously observed in Cl8 cells [11], GFP-Ci-CORD is mostly nuclear (c). The expression of exogenous cos2 leads to a marked relocalization of Ci-CORD to cytoplasmic puncta (d), which were also shown to be motile (d’). (e) Cos2-RFP co-localizes with Smo-GFP in a subset of motile puncta. Live images of S2 cells expressing cos2-RFP and smo-GFP show that a subset of puncta co-localize and are motile (e’). Corresponding single fluorescence images of Smo-GFP and Cos2-RFP are shown in Fig. S6c. The large box in panels (b), (d), and (e) indicates the enlarged area depicted in the corresponding time series (b’, d’, and e’). The smaller boxes indicate the origin of a single particle (arrow) that is tracked, over time.
Given the apparent co-localized movement of Cos2 and Ci, and that both Cos2ΔMotor and Cos2-S182N appear to affect Ci stability (data not shown and [14]), Cos2 mobility may be necessary, directly or indirectly, to regulate Ci activity. Conversely, Fu is required to antagonize many aspects of Cos2’s function [42, 43], regulating its phosphorylation [21] and controlling its subcellular localization (see above). To test the hypothesis that Ci activity is dependent on Fu kinase attenuation of Cos2 function, we expressed Ci in S2 cells at a level that results in robust transcriptional activity (Fig. S1f). Addition of exogenous Cos2 leads to a reduction in Ci activity, consistent with its predicted role as a negative regulator of Ci in the absence of Hh [11, 32, 44]. Co-expression of wt fu relieves Cos2’s inhibitory repression of Ci, and this effect requires a functional kinase domain. These results are consistent with Hh regulating Ci activity, at least in part, via Fu-induced Cos2 relocalization.
As a fraction of Cos2 also associates with Smo [3], we examined the ability of this Smo/Cos2 complex to form and move together throughout the cell. As shown above (Fig. 1b), high levels of smo result in a more diffuse distribution of Cos2-GFP, consistent with high levels of Smo constitutively activating Hh signaling [17]. However, when cos2-RFP was co-expressed with lower levels of smo-GFP in S2 cells, we observed a significant degree of overlap between the two proteins (Fig. 4e). We also observed that many of the puncta enriched for both Cos2-RFP and Smo-GFP appeared to move together throughout the cell (Fig. 4e’ and Movie 13). The Smo-GFP/Cos2-RFP puncta had a velocity approximately three times that of Cos2-GFP alone or that of puncta containing Cos2-GFP with Ci-CORD or Fu-tail (Table 2). The reasons for this difference in apparent velocity are currently unknown, but are consistent with the puncta containing the Smo/Cos2 complex being distinct from the major pool of Cos2 puncta [45].
Our results demonstrate that Cos2 displays many of the hallmarks of a KLP, exhibiting MT-dependent motility that is dependent on a functional motor domain and on ATP. We show that Hh regulates the motility of Cos2, through directly regulating Cos2 movement, and/or through regulating Cos2’s affinity with MTs, which may require Fu activity. KLPs are known to transport specific cargos, which are distinct for the various family members. Consistent with Cos2 functioning as a KLP, we show that Cos2 is able to recruit many of its binding partners into motile puncta. Our results suggests that one important cargo for Cos2 is the transcription factor Ci, which ultimately determines all Hh readouts [46]. Interestingly, precedence for the idea that KLP motility may regulate the activity of a transcription factor was recently provided in a study linking TGF-β signaling to kinesin-1 function [49]. The activation and nuclear accumulation of the transcription factor SMAD2 was shown to be dependent upon kinesin-1 trafficking, with the loss of kinesin-1 dependent SMAD2 motility ultimately resulting in attenuated TGF-β signaling [49]. Similarly, Hh appears to regulate the Cos2 dependent movement of many of its signaling components, including Ci. Although we suggest that Cos2 motility is an intrinsic property of the protein, we cannot currently rule out the possibility that the motility we observe is indirect. In this latter scenario, the observed Cos2 motility would be driven by an as yet undiscovered molecular motor, whose function would be dependent on the motor domain of Cos2, as Cos2ΔMotor and Cos2-S182N exhibit little motility. To date, no such putative Cos2 binding protein has been described, despite numerous attempts to identify additional Cos2 binding proteins [7, 47, 48].
It was recently proposed that another KLP, KIF3A, regulates the proteolytic processing of the mammalian Ci orthologs GLI2 and GLI3. This regulation of GLI2 and GLI3 processing was suggested to result from the deregulation of the MT-dependent intraflagellar transport system found in the primary cilia of vertebrate cells [50–53]. As only a subset of Drosophila neuronal cells have primary cilia, the regulation of Ci most likely does not require localization to primary cilia [54]. In Drosophila cells, a requirement for Ci enrichment in a specific subcellular location, which for GLI2 and GLI3 would occur in primary cilia, might occur in a subset of the Cos2 puncta described here. Such a model may provide the basis behind the apparent lack of evolutionary conservation of Cos2 in mammals [55], with regulation of GLIs occurring in a defined subcellular organelle, the primary cilium, diminishing the requirement of a Cos2 ortholog in mammalian Hh signaling.
EXPERIMENTAL PROCEDURES
Molecular biology and cell culture
The following pActin 5.1, pUAS, or pMT/V5HisC based vectors have been previously described: pAc 5.1 (Vector ctrl.), GFP-Cos2ΔMotor, Hh, HhN, 3xHACos2, GAL4 (pAc), GFP-Ci CORD (UAS) [11, 13], GFP-Smo (UAS) [56], Klp10A-EGFP (pMT) [26], pAc-mCherry-tubulin [57]. GFP-Ci CORD was a gift from Dr. J. Jiang (UTSW). GFP-Smo was a gift from Dr. M. Scott (Stanford). Klp10A-EGFP and mCherry-tubulin were gifts from the Vale Lab (UCSF). Cos2-GFP, Cos2-S182N-GFP, GFP-Fu-tail and HA-Fu were engineered into pAc 5.1 (or pUAS) vector using standard molecular biology techniques. pUAS-3xHA Cos2 was a gift from Dr. K. Nybakken (Harvard University). dsRNA against fu, smo and cos2 were produced as previously described [58]. Plasmids used to express hh contained either the full-length cDNA (hh) or only the biologically active amino-terminal portion of the cDNA (hhN), both of which can induce a response in S2 cells. The cos2-GFP, cos2-RFP, ci-RFP, smo-RFP and Myc-fu contructs used in Fig. S6 were constructed by the Gateway recombination method (Invitrogen). The full coding sequences (without the termination codon) of smo, fu, cos2 or ci cDNAs were amplified by PCR, and cloned in the entry vector pENTR/D-TOPO by directional TOPO Cloning. The resulting plasmids were checked by sequencing. The destination vectors pAWG (pAct5C-GW-EGFP), pAWR (pAct5C-GW -mRFP), pAMW (pAct5C-Myc-GW) (where GW is the recombination cassette) were a gift from T. Murphy (Carnegie Institute). All cloning steps were performed according to the manufacturer's instructions. Luciferase assays were performed as previously described [3, 59], using a firefly luciferase construct containing a portion of the ptc promoter responsive to Ci dependent activation, in conjunction with a standard Renilla luciferase (coupled to the actin promoter). Plasmids containing ci, hh and the luciferase cDNA were transiently transfected into S2 cells where indicated, and processed for luciferase activity using a Dual-luciferase assay kit (Promega) 40–48 hours later. Drosophila S2 cells were cultured and transfected under standard conditions in serum-containing (Drosophila Schneider, Invitrogen) or non-serum optimized media (Drosophila SFM, Invitrogen). In Fig. S2, S2 cells were transiently transfected using Effectene (Qiagen) as previously described [60]. A stable S2 cell line expressing GFP-tagged α-tubulin was a gift from the Sharp laboratory (Albert Einstein College of Medicine).
Cellular lysates and immunoblot analyses
Simple subcellular fractionation of S2 cells, by separation of cytoplasmic extracts from total membranes, were performed as previously reported [13]. Briefly, the various treated cells were dounce homogenized (type B pestle) in hypotonic HK buffer (20 mM HEPES, 10 mM KCl, pH 7.9) and cleared of nuclei (centrifuged 2,000 × g for 15 minutes at 4°C). Post-nuclear supernatants were centrifuged at 100,000 × g for 30 min. at 4°C. The resulting supernatant was separated from the membrane enriched pellet, then supplemented to 1% Nonidet P-40 (NP-40). The membrane enriched pellet was resuspended in HK containing 1% NP-40 buffer. The samples were normalized to volume, resolved by SDS-PAGE and analyzed by immunoblotting using the following antibodies: mouse anti-HA.11 (Covance), mouse anti-Cos2 5D6 [40], rabbit anti-Fu [7], rabbit anti-Kinesin (Kin01-KHC subunit, Cytoskeleton, Inc.). Rat anti-Smo antibodies were generated against residues 1000–1030 of the Drosophila Smo carboxyl-terminal tail and affinity purified over a peptide column.
Immunoprecipitation from Drosophila lysate
S2 cells were lysed in 1% Nonidet P-40 lysis buffer (1% NP-40, 150 mM NaCl, 50 mM Tris, 50 mM NaF, pH 8.0) 48 hours after transfection. Postnuclear lysates were precleared for 30 minutes with Protein A/G Plus agarose beads (Santa Cruz) and then incubated at 4°C for 90 minutes with rabbit anti-HA (Genetex) or rabbit IgG (Jackson Labs). Immune complexes were then collected on Protein A/G beads for 45 minutes at 4°C, then washed three times with 1% NP-40 lysis buffer. Complexes were resuspended in 2x Laemmli buffer, boiled for 5 minutes and then analyzed by SDS-PAGE and immunoblotting as described previously [16].
Microscopy
Indirect immunofluorescence experiments were performed essentially as previously described [60, 61], using a Leica Confocal laser scanning microscope. Primary antibodies used were as follows: mouse anti-Cos2 mAb 5D6 [40], rabbit anti-Fu [7], rat anti-Rab11 (a gift from R. Cohen); rabbit anti-Rab5 (a gift from M. Gonzales-Gaitan), mouse anti-LBPA (lysobisphosphatidic acid, a late endosome specific lipid) (a gift from J. Gruenberg), guinea pig anti-Hrs (a gift from H. Bellen) [62]. Secondary antibodies were from the Jackson Immunology Research Laboratory and were all used at 1:200. Transfected cells were plated on concanavalin A (ConA) coated cover glasses for 15 minutes and then fixed for 30 minutes with 4% paraformaldehyde at room temperature (RT). Cells were then washed 3 times for 5 minutes and permeabilized in PBS + 0.1% Tween (PBST). Cells were incubated for 1 hour in PBST + 2% BSA with the primary antibody at RT, washed 3 times for 5 minutes with PBST, then incubated with the secondary antibody in PBST + 2% BSA for 1 hour at RT. They were then rinsed 3 times for 5 minutes in PBST before being mounted in Cytifluor. In Fig. S6d, the anti-Fu primary antibodies were incubated with cells in PBS-Tween 0.5%, followed by incubation with secondary antibodies overnight. For labeling of cells with Lysotracker (Invitrogen), cells were incubated with the Lysotracker for 1 hour at 29°C, before being plated on ConA, and fixed. GFP-α-tubulin, Cos2-GFP, Cos2-RFP, Cos2-S182N-GFP, COS2ΔMotor-GFP, Klp10A-EGFP, mCherry-tubulin, Ci-RFP, Smo-RFP, Smo-GFP, GFP-Ci CORD and GFP Fu-tail were detected with the appropriate settings to visualize GFP or RFP/Cherry fluorescence. Live imaging studies were performed using a Zeiss LSM-510 inverted confocal microscope, fitted with a heating device to maintain the temperature of the slide and objective at 25°C. 24 to 48 hours after transfection with the appropriate plasmids, S2 cells were transferred to 4-well chamber slides with a coverglass bottom (Nunc) pre-treated with ConA to promote cells spreading. The following treatments were then performed: Nocodazole (10 µg/ml) for 0–6 hours, DMSO as vehicle control [63], sodium-azide (5 mM) and 2-deoxyglucose (1 mM) for 30 minutes [31], 5 µM cytochalasin D for 18–24 hours [35],or PBS as vehicle control. For time-lapse studies, serial images were taken every 1–3 seconds for 10 minutes to 6 hours, depending on the experiment. Time-lapse images shown in figures, or movies, correspond to a relevant fraction of the total acquisition time. Images and movies, from the Leica or Zeiss confocal microscopes, were further processed using Zeiss Image Browser software and Adobe Photoshop 6. Supplementary movies were generated by conversion of exported and uncompressed AVI files from Zeiss Image Browser to MOV files using Apple Quicktime Pro 7.1.5. The playback speed for all movies was set at 8 frames per second, with each frame representing approximately 3–5 seconds of real time.
Quantification of particle velocities
All velocities were calculated and analyzed using the Zeiss LSM Image Browser software. Particle movement was defined as visible movement of a single particle that 1) spanned at least two frames of time lapse imaging and 2) appeared linear. Measurements of velocity were quantified by measuring from the center of a puncta’s starting position to where it stopped moving, using the LSM software overlay line tool. If a single particle stopped and then restarted, only the moving sections were quantified. If a particle fused with another particle, then only the non-fusing frames were quantified. If these movement criteria were not fulfilled, then the diameter of the particle was recorded and a speed of 0 was entered. An immotile particle was defined as a particle that has a composite speed of less than 5 nm/sec, which accounted for puncta that may "shake" or "drift", but lack directed movement over the course of imaging. If a particle exhibited any movement, as defined above, at any time during the recording window, then it was not considered an immotile particle and its speed was quantified. All statistics and determinations of significance were done using a two-tailed Student’s T-test. More details regarding our quantification procedure are available upon request.
Supplementary Material
(a) Hh exposure results in a more diffuse localization of endogenous Cos2. Wide-field images of S2 cells with (+) and without hh were fixed and processed for indirect immunofluorescent staining of endogenous Cos2. The bulk of Cos2 is found in puncta in the absence of Hh, as quantified in Table 1. (b) S2 cells were transfected with plasmids expressing hh, or vector alone, then treated with smo, fu or control (Ctrl) dsRNA. The cells were then fractionated into a cytosolic enriched fraction (S) and a membrane enriched fraction (P). Membrane enrichment of Cos2 is not lost upon knockdown of Smo or Fu. However, knockdown of Smo or Fu prevents Hh-induced Cos2 relocalization. (c) S2 cells were transfected with plasmids expressing hh, or vector alone, then treated with fu or Ctrl dsRNA. The cells were then fixed and processed for indirect immunofluorescent staining of endogenous Cos2. Knockdown of Fu prevents the Hh induced relocalization of endogenous Cos2 from puncta (bottom panels). (d) S2 cells were treated with cos2 or Ctrl dsRNA. The cells were then fixed and processed for indirect immunofluorescent staining of endogenous Cos2. Knockdown of Cos2 demonstrates the specificity of the Cos2 monoclonal antibody used here, as Cos2 knockdown leads to a loss of the Cos2 puncta. Note that the apparent large nuclear enrichment of Cos2 is not affected by Cos2 knockdown, and is presumably a non-specific signal. Image for Ctrl dsRNA represents a higher magnification version of the image shown in the Ctrl dsRNA –Hh condition in panel (c). Images in panels (c) and (d) were all acquired at the same time from a single experiment. (e) Live images of S2 cells expressing cos2-GFP in the presence (+) and absence (−) of exogenous fu. In the presence of exogenous Fu, an increased fraction of Cos2-GFP appears more diffuse, similar to what is observed upon Hh exposure. The effect of Fu on Cos2-GFP localization was dependent on the level of Fu expression (data not shown). (f) A Hh dependent reporter assay was used to determine Ci activity in the presence of Fu and Cos2. Expression of ci in S2 cells leads to a net activation of the luciferase reporter. Co-expression of ci with cos2 can attenuate this activation. Addition of exogenous fu, but not the kinase-dead fu mutant, fu-K33R, can neutralize the effects of Cos2 on Ci activity. Thus, exogenous Cos2 can actively attenuate Ci activity and this negative effect of Cos2 is reversed by exogenous Fu. (g) S2 cells expressing vector alone (Con), wt cos2-GFP (Cos2) or cos2-S182N-GFP (Cos2-S182N), in the presence (+) and absence (−) of hh, were lysed, resolved by SDS-PAGE and subjected to immunoblotting using the indicated antisera. Antibodies to kinesin heavy-chain were used to verify normalization.
Cos2-RFP co-localizes with Smo-GFP in motile puncta. Movie 13 corresponds to the image seen in Fig. 4e and shows an S2 cell expressing both cos2-RFP and smo-GFP. A subset of puncta appear to be co-localized (yellow) and appear to be highly motile. A single, yellow puncta shown in the boxed region moves out of the box in a quick, downward trajectory.
Movie 14 complements Fig. S4a and shows two Cos2-GFP puncta fusing together to become a single punctate structure. This puncta remains stable, and relatively immobile, before a puncta separates away.
Movie 15 corresponds to Fig. S4b and shows a S2 cell expressing cos2-GFP that was photobleached to reduce the bulk of the fluorescence signal within a square region (see lower left side of the screen). The movie represents approximately 2.5 minutes of real-time and demonstrates that the intensity of fluorescence in the larger photobleached puncta was recovered in the same puncta by the end of the movie. The arrow highlights a single, photobleached puncta that recovers its fluorescence.
Movie 1A complements the image seen in Fig. 2a, showing that a S2 cell expressing cos2-GFP exhibits mobile puncta. These puncta appear to move throughout the cytoplasm. Note the single puncta in the boxed region that rapidly moves downward toward the center of the cell in the beginning of the movie.
Movie 1B shows another S2 cell expressing cos2-GFP and represents an enlarged area of approximately one-quarter of a cell. Movement of a single Cos2-GFP puncta can be tracked over time. Note the single puncta in the boxed region that moves upward, pauses, then restarts, ultimately fusing with another large puncta, in addition to the movements of small, faint puncta at the bottom of the screen.
Movie 2 corresponds to the image seen in Fig. 2b, showing S2 cells expressing klp10A-GFP. Klp10A also enriches in puncta that appear to move throughout the cytoplasm. The puncta outlined by the box moves quickly out, and other puncta pass through this same region over the course of the movie.
Movie 3 corresponds to the image seen in Fig. 2c, showing that Cos2ΔMotor localizes to relatively immobile puncta. Puncta appear to accumulate near the plasma membrane, as shown in the boxed region. Residual movement seen here is likely due to the presence of another motor on a small subset of puncta (as seen in Fig. S3d), as the cargo of KLPs typically associate with both an anterograde motor and a retrograde motor.
Movie 4 corresponds to the image seen in Fig. 3a, showing a single S2 cell depleted of ATP. Cos2-GFP localizes to puncta, but these puncta are immotile in the absence of ATP.
Movie 5 corresponds to the image seen in Fig. 3b, showing the same S2 cell that was depleted of ATP in Movie 4, then allowed to recover for 30 minutes in fresh media. While some Cos2-GFP puncta remain immotile, many small puncta have regained ability to move in this recovery time frame. Note one smaller, dim puncta moving quickly into the boxed area.
cos2-RFP was expressed in a S2 cell line stably expressing GFP-α-tubulin and imaged in live cells. Movie 6 represents an enlarged area of approximately one-quarter of a cell and corresponds to the images seen in Fig. S5b, which is also representative of similarly transfected cells seen in Fig. 3c. Note the single puncta that quickly moves in a linear trajectory from approximately the eight o’clock position toward the center of the cell.
Drosophila S2 cells expressing cos2-GFP were probed by indirect immunofluorescence for endogenous vesicular markers: Rab5, Rab11, Hrs, lysobisphosphatidic acid (LBPA), or stained with Lysotracker fluorescent dye. Cos2 co-localized with each vesicular marker to various degrees, and exhibited the highest percentage of co-localization (∼30%) with Rab5. The number of fluorescently labeled pixels was quantified for Cos2 and each marker, respectively, and then the percent of those Cos2 fluorescently pixels that were co-labeled with each vesicular marker were quantified. (See Table S1 for quantification of co-labeling.)
Movie 7A shows live images of S2 cells expressing cos2-GFP and mCherry-tubulin, treated with 5 µM cytochalasin for 18 hours. Cytochalasin disrupts the actin cytoskeleton, allowing formation of long MT projections, along which Cos2-GFP puncta appear to localize and move. The brightness of the GFP signal was artificially increased using the LSM Image Browser brightness/contrast control to highlight the many small, dim puncta moving along the MT projections.
Movie 7B shows an enlarged area of a single cellular projection from live S2 cells expressing cos2-GFP and mCherry-tubulin, treated with 5 µM cytochalasin for 18 hours. This movie corresponds to the images seen in Fig. 3e. The puncta shown moves away from the upper right corner along the visible MT projection, traveling approximately 8 µm over the course of the movie.
Movie 8 corresponds to the image seen in Fig. 3f and shows an S2 cell expressing cos2-S182N-GFP, which exhibits a punctate staining pattern. Cos2-S182-GFP puncta do not exhibit significant motility.
Movie 9 corresponds to the image seen in Fig. 4b, showing that Fu-tail is recruited to motile puncta in the presence of untagged, exogenous wt Cos2. Note the puncta that moves around the boxed region and then quickly downward.
Drosophila S2 cells expressing cos2-GFP were probed by indirect immunofluorescence for endogenous vesicular markers; Rab5, Rab11, Hrs, and lysobisphosphatidic acid (LBPA). Rab5 is a marker of early endosomes, Rab11 marks recycling endosomes, Hrs and LBPA are markers of late endosomes, and Lysotracker is a fluorescent dye that labels lysosomes. The number of fluorescently labeled pixels was quantified for Cos2 and each marker and then the percent of those fluorescently labeled pixels that exhibited co-localization were quantified. To calculate the percentage of random co-localization, images of cells expressing cos2-GFP were rotated by ninety degrees respective to their matching vesicular marker images, and then reassessed for co-localization with markers. Note that under these conditions, less than five percent of Cos2 randomly co-localizes with any given marker.
Velocities were measured as described in Methods section. Puncta were divided into three size categories; small (0.10–0.40 µm), medium (0.41–0.60 µm) and large (> 0.60 µm), as described in Fig. S3. Significance was calculated by comparing each velocity to Cos2 within the same size category (i.e. small Cos2ΔMotor puncta are significantly slower than small Cos2 puncta). Standard error of the mean (SE) is shown in the rightmost column and ** indicates values shown are significantly different from wt Cos2 puncta (p ≤ 0.02). Velocities that were not determined (indicated by “ND”) were due to lack of data points within a particular size category.
(a) Puncta velocities plotted according to their diameter. Velocities of puncta were separated into three arbitrary size categories, as indicated by the red doubleheaded arrows; small (0.10–0.40 µm), medium (0.41–0.60 µm), and large (> 0.60 µm). Symbols shown are as follows; ◆: wtCos2,  : Ci-CORD co-localized with Cos2,
: Ci-CORD co-localized with Cos2,  : Fu-tail co-localized with Cos2,
: Fu-tail co-localized with Cos2,  : Smo co-localized with Cos2,
: Smo co-localized with Cos2,  : Klp10A. (b) Average velocities for individual or co-localized Cos2 puncta, or the control kinesin Klp10A, categorized by various conditions or treatment and pooled for all sizes of puncta. Values for Ci-CORD, Fu-tail, and Smo were calculated from puncta co-localized with Cos2-RFP. Error bars represent standard error of the mean and * indicates that values shown are significantly different than the velocity of Cos2 (p ≤ 0.02). (c) Average puncta velocities separated into size categories. Division of mean velocities into size categories illustrates the difference in small puncta velocity versus large puncta velocity. Cos2ΔMotor, Cos2-S182N, and ATP depletion all show significantly slower velocity than Cos2, while Cos2 co-localized with Smo exhibits a markedly faster velocity. (c’) Average puncta velocities for Klp10A separated into size categories, shown on a different y-axis scale. Klp10A exhibits a faster average velocity than Cos2. Error bars represent standard error of the mean and * indicates that values shown are significantly different than the velocity of Cos2 in the same size category (p ≤ 0.02). (d) Cos2-ΔMotor associates with full-length Cos2. Drosophila S2 cells expressing vector alone, Myc-cos2, HA-cos2-ΔMotor, or a combination of both were lysed, immunoprecipitated using anti-HA antibodies or control IgG, and then resolved by SDS-PAGE and subjected to immunoblotting using the indicated antisera. Immunoprecipitation of HA-Cos2-ΔMotor specifically co-precipitates Myc-Cos2, consistent with them forming dimers. (d’) Corresponding S2 cell lysates expressing vector alone, Myc-cos2, HA-cos2-ΔMotor, or a combination of each, were lysed, resolved by SDS-PAGE, and subjected to immunoblotting using the indicated antisera. An antibody to kinesin heavy-chain was used to verify normalization.
: Klp10A. (b) Average velocities for individual or co-localized Cos2 puncta, or the control kinesin Klp10A, categorized by various conditions or treatment and pooled for all sizes of puncta. Values for Ci-CORD, Fu-tail, and Smo were calculated from puncta co-localized with Cos2-RFP. Error bars represent standard error of the mean and * indicates that values shown are significantly different than the velocity of Cos2 (p ≤ 0.02). (c) Average puncta velocities separated into size categories. Division of mean velocities into size categories illustrates the difference in small puncta velocity versus large puncta velocity. Cos2ΔMotor, Cos2-S182N, and ATP depletion all show significantly slower velocity than Cos2, while Cos2 co-localized with Smo exhibits a markedly faster velocity. (c’) Average puncta velocities for Klp10A separated into size categories, shown on a different y-axis scale. Klp10A exhibits a faster average velocity than Cos2. Error bars represent standard error of the mean and * indicates that values shown are significantly different than the velocity of Cos2 in the same size category (p ≤ 0.02). (d) Cos2-ΔMotor associates with full-length Cos2. Drosophila S2 cells expressing vector alone, Myc-cos2, HA-cos2-ΔMotor, or a combination of both were lysed, immunoprecipitated using anti-HA antibodies or control IgG, and then resolved by SDS-PAGE and subjected to immunoblotting using the indicated antisera. Immunoprecipitation of HA-Cos2-ΔMotor specifically co-precipitates Myc-Cos2, consistent with them forming dimers. (d’) Corresponding S2 cell lysates expressing vector alone, Myc-cos2, HA-cos2-ΔMotor, or a combination of each, were lysed, resolved by SDS-PAGE, and subjected to immunoblotting using the indicated antisera. An antibody to kinesin heavy-chain was used to verify normalization.
(a) Time-lapse images of two Cos2-GFP puncta (arrows) that fuse together within 12 seconds. The puncta appear to become a single punctate structure that remains stable, and relatively immobile, for approximately 40 seconds. Eventually, a puncta separates away and moves towards the bottom of the field. These events, of puncta fusion and separation, all occur within 1 minute of total elapsed time. (b) Fluorescence recovery after photobleaching of Cos2-GFP. A section of a S2 cell expressing cos2-GFP (denoted by square) was photobleached (at t = 3.35 sec) to reduce the bulk of the fluorescence signal within the region. Fluorescence recovery was subsequently monitored within the square. In less than 2.5 minutes, the fluorescence intensity of the large photobleached puncta was similar to neighboring non-bleached puncta. In all cases, puncta that were subjected to the high-powered laser eventually recovered all or a significant fraction of their starting fluorescence intensity.
(a) Live images of a S2 cell co-expressing cos2-GFP (green) and mCherry-tubulin (red), then treated with nocodazole for the indicated time period. Images show a single cell imaged every 60 minutes over the course of 6 hours of treatment. Peripheral MT breakdown is visible 60 minutes after nocodazole treatment and correlates with the apparent dissociation of Cos2-GFP puncta. After 6 hours, MT breakdown is nearly complete and Cos2-GFP puncta are barely visible. (b–b’) cos2-RFP was expressed in a S2 cell line stably expressing GFP-α-tubulin in order to visualize Cos2 movement along MTs. For better visualization, Cos2-RFP was pseudo-colored green and GFP-α-tubulin was pseudo-colored red. Cos2-RFP puncta appear to co-localize and move along MTs. The puncta indicated by the arrow traveled along a perinuclear localized MT.
(a–b) Live images of Drosophila S2 cells co-expressing cos2-RFP with GFP-tagged signaling complex members. Single fluorescence images of Fu-tail-GFP (a) and Ci-CORD-GFP (b), alongside the corresponding Cos2-RFP images (a’ and b’), both show a high degree of co-localization with Cos2-RFP when merged (a” and b”). These highly co-localized Fu-tail/Cos2 (a”) or Ci-CORD/Cos2 (b”) puncta also exhibit significant motility (a’” and b’”). The large box in panels (a”) and (b”) indicates the enlarged area depicted in the corresponding time series (a’” and b’”). The smaller boxes in (a’”) and (b’”) indicate the origin of a single particle (arrow) that is tracked over time. (c) Single fluorescence images of Smo-GFP (c) and Cos2-RFP (c’), which correspond to co-localization’ images shown in Fig. 4e and in panel (c”). (d) Co-localization of multiple signaling components in S2 cells. Myc-Fu was imaged by indirect immunofluorescence, alongside fluorescent fusion proteins Cos2-GFP and Ci-RFP, in order to observe the localization of three signaling components simultaneously.
Movie 10 corresponds to the image seen in Fig. S6a and shows an S2 cell expressing both cos2-RFP and fu-tail-GFP. Puncta appear to be highly co-localized (yellow) and appear to move together. Note that one puncta moves quickly out of the boxed region, followed by another small puncta that enters the box shortly after.
Movie 11 corresponds to the image seen in Fig. 4d, showing that Ci-CORD is recruited to motile puncta when untagged, exogenous wt cos2 is co-expressed with ci-CORD-GFP in live S2 cells. Note the puncta that move in and out of the boxed region.
Movie 12 corresponds to the image seen in Fig. S6b and shows an S2 cell expressing both cos2-RFP and ci-CORD-GFP. Cos2-RFP and Ci-CORD-GFP puncta appear to be highly co-localized (yellow) and appear to move together. Note the two puncta that appear in the lower box and then as one moves up to the higher box.
ACKNOWLEDGEMENTS
We thank Dr. David Casso (UCSF) and Dr. Antoine Guichet (IJM) for helpful discussions. We also thank Kenneth Orndorff (Dartmouth NCCC), Eric Griffis (UCSF), Tristan Piolot and Christophe Chamot (IJM), for expert advice and technical assistance for our imaging studies. This work was supported by a NIH grant CA8628 (D.J.R.), a NIH training grant CA09658 (M.A.), a Ryan fellowship (S.F.F.), a MRT fellowship (A.B), the CNRS, the Universities Paris Diderot et UPMC and by the ARC (4010). The IJM imaging facilities were partially funded by the ARC, the region Ile de France (SESAME) and the University Paris 7.
REFERENCES
- 1.Wang G, Jiang J. Multiple Cos2/Ci interactions regulate Ci subcellular localization through microtubule dependent and independent mechanisms. Dev Biol. 2004;268:493–505. doi: 10.1016/j.ydbio.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 2.Stegman MA, Vallance JE, Elangovan G, Sosinski J, Cheng Y, Robbins DJ. Identification of a tetrameric hedgehog signaling complex. J Biol Chem. 2000;275:21809–21812. doi: 10.1074/jbc.C000043200. [DOI] [PubMed] [Google Scholar]
- 3.Ogden SK, Ascano M, Jr, Stegman MA, Suber LM, Hooper JE, Robbins DJ. Identification of a functional interaction between the transmembrane protein Smoothened and the kinesin-related protein Costal2. Curr Biol. 2003;13:1998–2003. doi: 10.1016/j.cub.2003.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruel L, Rodriguez R, Gallet A, Lavenant-Staccini L, Therond PP. Stability and association of Smoothened, Costal2 and Fused with Cubitus interruptus are regulated by Hedgehog. Nat Cell Biol. 2003;5:907–913. doi: 10.1038/ncb1052. [DOI] [PubMed] [Google Scholar]
- 5.Lum L, Zhang C, Oh S, Mann RK, von Kessler DP, Taipale J, Weis-Garcia F, Gong R, Wang B, Beachy PA. Hedgehog signal transduction via Smoothened association with a cytoplasmic complex scaffolded by the atypical kinesin, Costal-2. Mol Cell. 2003;12:1261–1274. doi: 10.1016/s1097-2765(03)00426-x. [DOI] [PubMed] [Google Scholar]
- 6.Jia J, Tong C, Jiang J. Smoothened transduces Hedgehog signal by physically interacting with Costal2/Fused complex through its C-terminal tail. Genes Dev. 2003;17:2709–2720. doi: 10.1101/gad.1136603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robbins DJ, Nybakken KE, Kobayashi R, Sisson JC, Bishop JM, Therond PP. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell. 1997;90:225–234. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- 8.Apionishev S, Katanayeva NM, Marks SA, Kalderon D, Tomlinson A. Drosophila Smoothened phosphorylation sites essential for Hedgehog signal transduction. Nat Cell Biol. 2005;7:86–92. doi: 10.1038/ncb1210. [DOI] [PubMed] [Google Scholar]
- 9.Zhang W, Zhao Y, Tong C, Wang G, Wang B, Jia J, Jiang J. Hedgehog-regulated Costal2-kinase complexes control phosphorylation and proteolytic processing of Cubitus interruptus. Dev Cell. 2005;8:267–278. doi: 10.1016/j.devcel.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Monnier V, Ho KS, Sanial M, Scott MP, Plessis A. Hedgehog signal transduction proteins: contacts of the Fused kinase and Ci transcription factor with the Kinesin-related protein Costal2. BMC Dev Biol. 2002;2:4. doi: 10.1186/1471-213X-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G, Amanai K, Wang B, Jiang J. Interactions with Costal2 and suppressor of fused regulate nuclear translocation and activity of cubitus interruptus. Genes Dev. 2000;14:2893–2905. doi: 10.1101/gad.843900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lefers MA, Wang QT, Holmgren RA. Genetic dissection of the Drosophila Cubitus interruptus signaling complex. Dev Biol. 2001;236:411–420. doi: 10.1006/dbio.2001.0345. [DOI] [PubMed] [Google Scholar]
- 13.Stegman MA, Goetz JA, Ascano M, Jr, Ogden SK, Nybakken KE, Robbins DJ. The Kinesin-related protein Costal2 associates with membranes in a Hedgehog-sensitive, Smoothened-independent manner. J Biol Chem. 2004;279:7064–7071. doi: 10.1074/jbc.M311794200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho KS, Suyama K, Fish M, Scott MP. Differential regulation of Hedgehog target gene transcription by Costal2 and Suppressor of Fused. Development. 2005;132:1401–1412. doi: 10.1242/dev.01689. [DOI] [PubMed] [Google Scholar]
- 15.Aza-Blanc P, Ramirez-Weber FA, Laget MP, Schwartz C, Kornberg TB. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- 16.Therond PP, Knight JD, Kornberg TB, Bishop JM. Phosphorylation of the fused protein kinase in response to signaling from hedgehog. Proc Natl Acad Sci U S A. 1996;93:4224–4228. doi: 10.1073/pnas.93.9.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hooper JE. Smoothened translates Hedgehog levels into distinct responses. Development. 2003;130:3951–3963. doi: 10.1242/dev.00594. [DOI] [PubMed] [Google Scholar]
- 18.Alcedo J, Ayzenzon M, Von Ohlen T, Noll M, Hooper JE. The Drosophila smoothened gene encodes a seven-pass membrane protein, a putative receptor for the hedgehog signal. Cell. 1996;86:221–232. doi: 10.1016/s0092-8674(00)80094-x. [DOI] [PubMed] [Google Scholar]
- 19.van den Heuvel M, Ingham PW. smoothened encodes a receptor-like serpentine protein required for hedgehog signalling. Nature. 1996;382:547–551. doi: 10.1038/382547a0. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Struhl G. In vivo evidence that Patched and Smoothened constitute distinct binding and transducing components of a Hedgehog receptor complex. Development. 1998;125:4943–4948. doi: 10.1242/dev.125.24.4943. [DOI] [PubMed] [Google Scholar]
- 21.Nybakken KE, Turck CW, Robbins DJ, Bishop JM. Hedgehog-stimulated phosphorylation of the kinesin-related protein Costal2 is mediated by the serine/threonine kinase fused. J Biol Chem. 2002;277:24638–24647. doi: 10.1074/jbc.M110730200. [DOI] [PubMed] [Google Scholar]
- 22.Kwok BH, Kapitein LC, Kim JH, Peterman EJ, Schmidt CF, Kapoor TM. Allosteric inhibition of kinesin-5 modulates its processive directional motility. Nat Chem Biol. 2006;2:480–485. doi: 10.1038/nchembio812. [DOI] [PubMed] [Google Scholar]
- 23.Cai G, Romagnoli S, Moscatelli A, Ovidi E, Gambellini G, Tiezzi A, Cresti M. Identification and characterization of a novel microtubule-based motor associated with membranous organelles in tobacco pollen tubes. Plant Cell. 2000;12:1719–1736. doi: 10.1105/tpc.12.9.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cole DG, Saxton WM, Sheehan KB, Scholey JM. A "slow" homotetrameric kinesin-related motor protein purified from Drosophila embryos. J Biol Chem. 1994;269:22913–22916. [PMC free article] [PubMed] [Google Scholar]
- 25.Song H, Golovkin M, Reddy AS, Endow SA. In vitro motility of AtKCBP, a calmodulin-binding kinesin protein of Arabidopsis. Proc Natl Acad Sci U S A. 1997;94:322–327. doi: 10.1073/pnas.94.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mennella V, Rogers GC, Rogers SL, Buster DW, Vale RD, Sharp DJ. Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat Cell Biol. 2005;7:235–245. doi: 10.1038/ncb1222. [DOI] [PubMed] [Google Scholar]
- 27.Goshima G, Vale RD. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol Biol Cell. 2005;16:3896–3907. doi: 10.1091/mbc.E05-02-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogers SL, Tint IS, Fanapour PC, Gelfand VI. Regulated bidirectional motility of melanophore pigment granules along microtubules in vitro. Proc Natl Acad Sci U S A. 1997;94:3720–3725. doi: 10.1073/pnas.94.8.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuznetsov SA, Gelfand VI. Bovine brain kinesin is a microtubule-activated ATPase. Proc Natl Acad Sci U S A. 1986;83:8530–8534. doi: 10.1073/pnas.83.22.8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Platani M, Goldberg I, Lamond AI, Swedlow JR. Cajal body dynamics and association with chromatin are ATP-dependent. Nat Cell Biol. 2002;4:502–508. doi: 10.1038/ncb809. [DOI] [PubMed] [Google Scholar]
- 32.Sisson JC, Ho KS, Suyama K, Scott MP. Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell. 1997;90:235–245. doi: 10.1016/s0092-8674(00)80332-3. [DOI] [PubMed] [Google Scholar]
- 33.Woehlke G, Ruby AK, Hart CL, Ly B, Hom-Booher N, Vale RD. Microtubule interaction site of the kinesin motor. Cell. 1997;90:207–216. doi: 10.1016/s0092-8674(00)80329-3. [DOI] [PubMed] [Google Scholar]
- 34.Rogers SL, Rogers GC, Sharp DJ, Vale RD. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J Cell Biol. 2002;158:873–884. doi: 10.1083/jcb.200202032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim H, Ling SC, Rogers GC, Kural C, Selvin PR, Rogers SL, Gelfand VI. Microtubule binding by dynactin is required for microtubule organization but not cargo transport. J Cell Biol. 2007;176:641–651. doi: 10.1083/jcb.200608128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marx A, Muller J, Mandelkow E. The structure of microtubule motor proteins. Adv Protein Chem. 2005;71:299–344. doi: 10.1016/S0065-3233(04)71008-6. [DOI] [PubMed] [Google Scholar]
- 37.Blangy A, Chaussepied P, Nigg EA. Rigor-type mutation in the kinesin-related protein HsEg5 changes its subcellular localization and induces microtubule bundling. Cell Motil Cytoskeleton. 1998;40:174–182. doi: 10.1002/(SICI)1097-0169(1998)40:2<174::AID-CM6>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 38.Meluh PB, Rose MD. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- 39.Matthies HJ, Baskin RJ, Hawley RS. Orphan kinesin NOD lacks motile properties but does possess a microtubule-stimulated ATPase activity. Mol Biol Cell. 2001;12:4000–4012. doi: 10.1091/mbc.12.12.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ascano M, Jr, Nybakken KE, Sosinski J, Stegman MA, Robbins DJ. The carboxyl-terminal domain of the protein kinase fused can function as a dominant inhibitor of hedgehog signaling. Mol Cell Biol. 2002;22:1555–1566. doi: 10.1128/mcb.22.5.1555-1566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang QT, Holmgren RA. Nuclear import of cubitus interruptus is regulated by hedgehog via a mechanism distinct from Ci stabilization and Ci activation. Development. 2000;127:3131–3139. doi: 10.1242/dev.127.14.3131. [DOI] [PubMed] [Google Scholar]
- 42.Alves G, Limbourg-Bouchon B, Tricoire H, Brissard-Zahraoui J, Lamour-Isnard C, Busson D. Modulation of Hedgehog target gene expression by the Fused serine-threonine kinase in wing imaginal discs. Mech Dev. 1998;78:17–31. doi: 10.1016/s0925-4773(98)00130-0. [DOI] [PubMed] [Google Scholar]
- 43.Therond P, Alves G, Limbourg-Bouchon B, Tricoire H, Guillemet E, Brissard-Zahraoui J, Lamour-Isnard C, Busson D. Functional domains of fused, a serine-threonine kinase required for signaling in Drosophila. Genetics. 1996;142:1181–1198. doi: 10.1093/genetics/142.4.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang QT, Holmgren RA. The subcellular localization and activity of Drosophila cubitus interruptus are regulated at multiple levels. Development. 1999;126:5097–5106. doi: 10.1242/dev.126.22.5097. [DOI] [PubMed] [Google Scholar]
- 45.Ogden SK, Casso DJ, Ascano M, Jr, Yore MM, Kornberg TB, Robbins DJ. Smoothened regulates activator and repressor functions of Hedgehog signaling via two distinct mechanisms. J Biol Chem. 2006;281:7237–7243. doi: 10.1074/jbc.M510169200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Methot N, Basler K. An absolute requirement for Cubitus interruptus in Hedgehog signaling. Development. 2001;128:733–742. doi: 10.1242/dev.128.5.733. [DOI] [PubMed] [Google Scholar]
- 47.Lum L, Yao S, Mozer B, Rovescalli A, Von Kessler D, Nirenberg M, Beachy PA. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science. 2003;299:2039–2045. doi: 10.1126/science.1081403. [DOI] [PubMed] [Google Scholar]
- 48.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL, Jr, White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM. A Protein Interaction Map of Drosophila melanogaster 10.1126/science.1090289. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 49.Batut J, Howell M, Hill CS. Kinesin-mediated transport of Smad2 is required for signaling in response to TGF-beta ligands. Dev Cell. 2007;12:261–274. doi: 10.1016/j.devcel.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 50.Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci U S A. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 52.Liu A, Wang B, Niswander LA. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132:3103–3111. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- 53.Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han YG, Kwok BH, Kernan MJ. Intraflagellar transport is required in Drosophila to differentiate sensory cilia but not sperm. Curr Biol. 2003;13:1679–1686. doi: 10.1016/j.cub.2003.08.034. [DOI] [PubMed] [Google Scholar]
- 55.Huangfu D, Anderson KV. Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development. 2006;133:3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- 56.Zhu AJ, Zheng L, Suyama K, Scott MP. Altered localization of Drosophila Smoothened protein activates Hedgehog signal transduction. Genes Dev. 2003 doi: 10.1101/gad.1080803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci U S A. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen CH, von Kessler DP, Park W, Wang B, Ma Y, Beachy PA. Nuclear trafficking of Cubitus interruptus in the transcriptional regulation of Hedgehog target gene expression. Cell. 1999;98:305–316. doi: 10.1016/s0092-8674(00)81960-1. [DOI] [PubMed] [Google Scholar]
- 60.Claret S, Sanial M, Plessis A. Evidence for a novel feedback loop in the Hedgehog pathway involving Smoothened and Fused. Curr Biol. 2007;17:1326–1333. doi: 10.1016/j.cub.2007.06.059. [DOI] [PubMed] [Google Scholar]
- 61.Ascano M, Jr, Robbins DJ. An intramolecular association between two domains of the protein kinase Fused is necessary for Hedgehog signaling. Mol Cell Biol. 2004;24:10397–10405. doi: 10.1128/MCB.24.23.10397-10405.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lloyd TE, Atkinson R, Wu MN, Zhou Y, Pennetta G, Bellen HJ. Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell. 2002;108:261–269. doi: 10.1016/s0092-8674(02)00611-6. [DOI] [PubMed] [Google Scholar]
- 63.Rothenberg ME, Rogers SL, Vale RD, Jan LY, Jan YN. Drosophila pod-1 crosslinks both actin and microtubules and controls the targeting of axons. Neuron. 2003;39:779–791. doi: 10.1016/s0896-6273(03)00508-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) Hh exposure results in a more diffuse localization of endogenous Cos2. Wide-field images of S2 cells with (+) and without hh were fixed and processed for indirect immunofluorescent staining of endogenous Cos2. The bulk of Cos2 is found in puncta in the absence of Hh, as quantified in Table 1. (b) S2 cells were transfected with plasmids expressing hh, or vector alone, then treated with smo, fu or control (Ctrl) dsRNA. The cells were then fractionated into a cytosolic enriched fraction (S) and a membrane enriched fraction (P). Membrane enrichment of Cos2 is not lost upon knockdown of Smo or Fu. However, knockdown of Smo or Fu prevents Hh-induced Cos2 relocalization. (c) S2 cells were transfected with plasmids expressing hh, or vector alone, then treated with fu or Ctrl dsRNA. The cells were then fixed and processed for indirect immunofluorescent staining of endogenous Cos2. Knockdown of Fu prevents the Hh induced relocalization of endogenous Cos2 from puncta (bottom panels). (d) S2 cells were treated with cos2 or Ctrl dsRNA. The cells were then fixed and processed for indirect immunofluorescent staining of endogenous Cos2. Knockdown of Cos2 demonstrates the specificity of the Cos2 monoclonal antibody used here, as Cos2 knockdown leads to a loss of the Cos2 puncta. Note that the apparent large nuclear enrichment of Cos2 is not affected by Cos2 knockdown, and is presumably a non-specific signal. Image for Ctrl dsRNA represents a higher magnification version of the image shown in the Ctrl dsRNA –Hh condition in panel (c). Images in panels (c) and (d) were all acquired at the same time from a single experiment. (e) Live images of S2 cells expressing cos2-GFP in the presence (+) and absence (−) of exogenous fu. In the presence of exogenous Fu, an increased fraction of Cos2-GFP appears more diffuse, similar to what is observed upon Hh exposure. The effect of Fu on Cos2-GFP localization was dependent on the level of Fu expression (data not shown). (f) A Hh dependent reporter assay was used to determine Ci activity in the presence of Fu and Cos2. Expression of ci in S2 cells leads to a net activation of the luciferase reporter. Co-expression of ci with cos2 can attenuate this activation. Addition of exogenous fu, but not the kinase-dead fu mutant, fu-K33R, can neutralize the effects of Cos2 on Ci activity. Thus, exogenous Cos2 can actively attenuate Ci activity and this negative effect of Cos2 is reversed by exogenous Fu. (g) S2 cells expressing vector alone (Con), wt cos2-GFP (Cos2) or cos2-S182N-GFP (Cos2-S182N), in the presence (+) and absence (−) of hh, were lysed, resolved by SDS-PAGE and subjected to immunoblotting using the indicated antisera. Antibodies to kinesin heavy-chain were used to verify normalization.
Cos2-RFP co-localizes with Smo-GFP in motile puncta. Movie 13 corresponds to the image seen in Fig. 4e and shows an S2 cell expressing both cos2-RFP and smo-GFP. A subset of puncta appear to be co-localized (yellow) and appear to be highly motile. A single, yellow puncta shown in the boxed region moves out of the box in a quick, downward trajectory.
Movie 14 complements Fig. S4a and shows two Cos2-GFP puncta fusing together to become a single punctate structure. This puncta remains stable, and relatively immobile, before a puncta separates away.
Movie 15 corresponds to Fig. S4b and shows a S2 cell expressing cos2-GFP that was photobleached to reduce the bulk of the fluorescence signal within a square region (see lower left side of the screen). The movie represents approximately 2.5 minutes of real-time and demonstrates that the intensity of fluorescence in the larger photobleached puncta was recovered in the same puncta by the end of the movie. The arrow highlights a single, photobleached puncta that recovers its fluorescence.
Movie 1A complements the image seen in Fig. 2a, showing that a S2 cell expressing cos2-GFP exhibits mobile puncta. These puncta appear to move throughout the cytoplasm. Note the single puncta in the boxed region that rapidly moves downward toward the center of the cell in the beginning of the movie.
Movie 1B shows another S2 cell expressing cos2-GFP and represents an enlarged area of approximately one-quarter of a cell. Movement of a single Cos2-GFP puncta can be tracked over time. Note the single puncta in the boxed region that moves upward, pauses, then restarts, ultimately fusing with another large puncta, in addition to the movements of small, faint puncta at the bottom of the screen.
Movie 2 corresponds to the image seen in Fig. 2b, showing S2 cells expressing klp10A-GFP. Klp10A also enriches in puncta that appear to move throughout the cytoplasm. The puncta outlined by the box moves quickly out, and other puncta pass through this same region over the course of the movie.
Movie 3 corresponds to the image seen in Fig. 2c, showing that Cos2ΔMotor localizes to relatively immobile puncta. Puncta appear to accumulate near the plasma membrane, as shown in the boxed region. Residual movement seen here is likely due to the presence of another motor on a small subset of puncta (as seen in Fig. S3d), as the cargo of KLPs typically associate with both an anterograde motor and a retrograde motor.
Movie 4 corresponds to the image seen in Fig. 3a, showing a single S2 cell depleted of ATP. Cos2-GFP localizes to puncta, but these puncta are immotile in the absence of ATP.
Movie 5 corresponds to the image seen in Fig. 3b, showing the same S2 cell that was depleted of ATP in Movie 4, then allowed to recover for 30 minutes in fresh media. While some Cos2-GFP puncta remain immotile, many small puncta have regained ability to move in this recovery time frame. Note one smaller, dim puncta moving quickly into the boxed area.
cos2-RFP was expressed in a S2 cell line stably expressing GFP-α-tubulin and imaged in live cells. Movie 6 represents an enlarged area of approximately one-quarter of a cell and corresponds to the images seen in Fig. S5b, which is also representative of similarly transfected cells seen in Fig. 3c. Note the single puncta that quickly moves in a linear trajectory from approximately the eight o’clock position toward the center of the cell.
Drosophila S2 cells expressing cos2-GFP were probed by indirect immunofluorescence for endogenous vesicular markers: Rab5, Rab11, Hrs, lysobisphosphatidic acid (LBPA), or stained with Lysotracker fluorescent dye. Cos2 co-localized with each vesicular marker to various degrees, and exhibited the highest percentage of co-localization (∼30%) with Rab5. The number of fluorescently labeled pixels was quantified for Cos2 and each marker, respectively, and then the percent of those Cos2 fluorescently pixels that were co-labeled with each vesicular marker were quantified. (See Table S1 for quantification of co-labeling.)
Movie 7A shows live images of S2 cells expressing cos2-GFP and mCherry-tubulin, treated with 5 µM cytochalasin for 18 hours. Cytochalasin disrupts the actin cytoskeleton, allowing formation of long MT projections, along which Cos2-GFP puncta appear to localize and move. The brightness of the GFP signal was artificially increased using the LSM Image Browser brightness/contrast control to highlight the many small, dim puncta moving along the MT projections.
Movie 7B shows an enlarged area of a single cellular projection from live S2 cells expressing cos2-GFP and mCherry-tubulin, treated with 5 µM cytochalasin for 18 hours. This movie corresponds to the images seen in Fig. 3e. The puncta shown moves away from the upper right corner along the visible MT projection, traveling approximately 8 µm over the course of the movie.
Movie 8 corresponds to the image seen in Fig. 3f and shows an S2 cell expressing cos2-S182N-GFP, which exhibits a punctate staining pattern. Cos2-S182-GFP puncta do not exhibit significant motility.
Movie 9 corresponds to the image seen in Fig. 4b, showing that Fu-tail is recruited to motile puncta in the presence of untagged, exogenous wt Cos2. Note the puncta that moves around the boxed region and then quickly downward.
Drosophila S2 cells expressing cos2-GFP were probed by indirect immunofluorescence for endogenous vesicular markers; Rab5, Rab11, Hrs, and lysobisphosphatidic acid (LBPA). Rab5 is a marker of early endosomes, Rab11 marks recycling endosomes, Hrs and LBPA are markers of late endosomes, and Lysotracker is a fluorescent dye that labels lysosomes. The number of fluorescently labeled pixels was quantified for Cos2 and each marker and then the percent of those fluorescently labeled pixels that exhibited co-localization were quantified. To calculate the percentage of random co-localization, images of cells expressing cos2-GFP were rotated by ninety degrees respective to their matching vesicular marker images, and then reassessed for co-localization with markers. Note that under these conditions, less than five percent of Cos2 randomly co-localizes with any given marker.
Velocities were measured as described in Methods section. Puncta were divided into three size categories; small (0.10–0.40 µm), medium (0.41–0.60 µm) and large (> 0.60 µm), as described in Fig. S3. Significance was calculated by comparing each velocity to Cos2 within the same size category (i.e. small Cos2ΔMotor puncta are significantly slower than small Cos2 puncta). Standard error of the mean (SE) is shown in the rightmost column and ** indicates values shown are significantly different from wt Cos2 puncta (p ≤ 0.02). Velocities that were not determined (indicated by “ND”) were due to lack of data points within a particular size category.
(a) Puncta velocities plotted according to their diameter. Velocities of puncta were separated into three arbitrary size categories, as indicated by the red doubleheaded arrows; small (0.10–0.40 µm), medium (0.41–0.60 µm), and large (> 0.60 µm). Symbols shown are as follows; ◆: wtCos2,  : Ci-CORD co-localized with Cos2,
: Ci-CORD co-localized with Cos2,  : Fu-tail co-localized with Cos2,
: Fu-tail co-localized with Cos2,  : Smo co-localized with Cos2,
: Smo co-localized with Cos2,  : Klp10A. (b) Average velocities for individual or co-localized Cos2 puncta, or the control kinesin Klp10A, categorized by various conditions or treatment and pooled for all sizes of puncta. Values for Ci-CORD, Fu-tail, and Smo were calculated from puncta co-localized with Cos2-RFP. Error bars represent standard error of the mean and * indicates that values shown are significantly different than the velocity of Cos2 (p ≤ 0.02). (c) Average puncta velocities separated into size categories. Division of mean velocities into size categories illustrates the difference in small puncta velocity versus large puncta velocity. Cos2ΔMotor, Cos2-S182N, and ATP depletion all show significantly slower velocity than Cos2, while Cos2 co-localized with Smo exhibits a markedly faster velocity. (c’) Average puncta velocities for Klp10A separated into size categories, shown on a different y-axis scale. Klp10A exhibits a faster average velocity than Cos2. Error bars represent standard error of the mean and * indicates that values shown are significantly different than the velocity of Cos2 in the same size category (p ≤ 0.02). (d) Cos2-ΔMotor associates with full-length Cos2. Drosophila S2 cells expressing vector alone, Myc-cos2, HA-cos2-ΔMotor, or a combination of both were lysed, immunoprecipitated using anti-HA antibodies or control IgG, and then resolved by SDS-PAGE and subjected to immunoblotting using the indicated antisera. Immunoprecipitation of HA-Cos2-ΔMotor specifically co-precipitates Myc-Cos2, consistent with them forming dimers. (d’) Corresponding S2 cell lysates expressing vector alone, Myc-cos2, HA-cos2-ΔMotor, or a combination of each, were lysed, resolved by SDS-PAGE, and subjected to immunoblotting using the indicated antisera. An antibody to kinesin heavy-chain was used to verify normalization.
: Klp10A. (b) Average velocities for individual or co-localized Cos2 puncta, or the control kinesin Klp10A, categorized by various conditions or treatment and pooled for all sizes of puncta. Values for Ci-CORD, Fu-tail, and Smo were calculated from puncta co-localized with Cos2-RFP. Error bars represent standard error of the mean and * indicates that values shown are significantly different than the velocity of Cos2 (p ≤ 0.02). (c) Average puncta velocities separated into size categories. Division of mean velocities into size categories illustrates the difference in small puncta velocity versus large puncta velocity. Cos2ΔMotor, Cos2-S182N, and ATP depletion all show significantly slower velocity than Cos2, while Cos2 co-localized with Smo exhibits a markedly faster velocity. (c’) Average puncta velocities for Klp10A separated into size categories, shown on a different y-axis scale. Klp10A exhibits a faster average velocity than Cos2. Error bars represent standard error of the mean and * indicates that values shown are significantly different than the velocity of Cos2 in the same size category (p ≤ 0.02). (d) Cos2-ΔMotor associates with full-length Cos2. Drosophila S2 cells expressing vector alone, Myc-cos2, HA-cos2-ΔMotor, or a combination of both were lysed, immunoprecipitated using anti-HA antibodies or control IgG, and then resolved by SDS-PAGE and subjected to immunoblotting using the indicated antisera. Immunoprecipitation of HA-Cos2-ΔMotor specifically co-precipitates Myc-Cos2, consistent with them forming dimers. (d’) Corresponding S2 cell lysates expressing vector alone, Myc-cos2, HA-cos2-ΔMotor, or a combination of each, were lysed, resolved by SDS-PAGE, and subjected to immunoblotting using the indicated antisera. An antibody to kinesin heavy-chain was used to verify normalization.
(a) Time-lapse images of two Cos2-GFP puncta (arrows) that fuse together within 12 seconds. The puncta appear to become a single punctate structure that remains stable, and relatively immobile, for approximately 40 seconds. Eventually, a puncta separates away and moves towards the bottom of the field. These events, of puncta fusion and separation, all occur within 1 minute of total elapsed time. (b) Fluorescence recovery after photobleaching of Cos2-GFP. A section of a S2 cell expressing cos2-GFP (denoted by square) was photobleached (at t = 3.35 sec) to reduce the bulk of the fluorescence signal within the region. Fluorescence recovery was subsequently monitored within the square. In less than 2.5 minutes, the fluorescence intensity of the large photobleached puncta was similar to neighboring non-bleached puncta. In all cases, puncta that were subjected to the high-powered laser eventually recovered all or a significant fraction of their starting fluorescence intensity.
(a) Live images of a S2 cell co-expressing cos2-GFP (green) and mCherry-tubulin (red), then treated with nocodazole for the indicated time period. Images show a single cell imaged every 60 minutes over the course of 6 hours of treatment. Peripheral MT breakdown is visible 60 minutes after nocodazole treatment and correlates with the apparent dissociation of Cos2-GFP puncta. After 6 hours, MT breakdown is nearly complete and Cos2-GFP puncta are barely visible. (b–b’) cos2-RFP was expressed in a S2 cell line stably expressing GFP-α-tubulin in order to visualize Cos2 movement along MTs. For better visualization, Cos2-RFP was pseudo-colored green and GFP-α-tubulin was pseudo-colored red. Cos2-RFP puncta appear to co-localize and move along MTs. The puncta indicated by the arrow traveled along a perinuclear localized MT.
(a–b) Live images of Drosophila S2 cells co-expressing cos2-RFP with GFP-tagged signaling complex members. Single fluorescence images of Fu-tail-GFP (a) and Ci-CORD-GFP (b), alongside the corresponding Cos2-RFP images (a’ and b’), both show a high degree of co-localization with Cos2-RFP when merged (a” and b”). These highly co-localized Fu-tail/Cos2 (a”) or Ci-CORD/Cos2 (b”) puncta also exhibit significant motility (a’” and b’”). The large box in panels (a”) and (b”) indicates the enlarged area depicted in the corresponding time series (a’” and b’”). The smaller boxes in (a’”) and (b’”) indicate the origin of a single particle (arrow) that is tracked over time. (c) Single fluorescence images of Smo-GFP (c) and Cos2-RFP (c’), which correspond to co-localization’ images shown in Fig. 4e and in panel (c”). (d) Co-localization of multiple signaling components in S2 cells. Myc-Fu was imaged by indirect immunofluorescence, alongside fluorescent fusion proteins Cos2-GFP and Ci-RFP, in order to observe the localization of three signaling components simultaneously.
Movie 10 corresponds to the image seen in Fig. S6a and shows an S2 cell expressing both cos2-RFP and fu-tail-GFP. Puncta appear to be highly co-localized (yellow) and appear to move together. Note that one puncta moves quickly out of the boxed region, followed by another small puncta that enters the box shortly after.
Movie 11 corresponds to the image seen in Fig. 4d, showing that Ci-CORD is recruited to motile puncta when untagged, exogenous wt cos2 is co-expressed with ci-CORD-GFP in live S2 cells. Note the puncta that move in and out of the boxed region.
Movie 12 corresponds to the image seen in Fig. S6b and shows an S2 cell expressing both cos2-RFP and ci-CORD-GFP. Cos2-RFP and Ci-CORD-GFP puncta appear to be highly co-localized (yellow) and appear to move together. Note the two puncta that appear in the lower box and then as one moves up to the higher box.