Abstract
The TAL1 (or SCL) gene, originally discovered through its involvement by a chromosomal translocation in T-cell acute lymphoblastic leukemia, encodes a basic helix-loop-helix (bHLH) transcription factor essential for hematopoietic and vascular development. To identify its interaction partners, we expressed a tandem epitope-tagged protein in murine erythroleukemia (MEL) cells and characterized affinity-purified Tal1-containing complexes by liquid chromatography-tandem mass spectrometry analysis. In addition to known interacting proteins, two proteins related to the Eight-Twenty-One (ETO) corepressor, Eto2/Mtg16 and Mtgr1, were identified from the peptide fragments analyzed. Tal1 interaction with Eto2 and Mtgr1 was verified by coimmunoprecipitation analysis in Tal1, Eto2-, and Mtgr1-transfected COS-7 cells, MEL cells expressing V5 epitope-tagged Tal1 protein, and non-transfected MEL cells. Mapping analysis with Gal4 fusion proteins demonstrated a requirement for the bHLH domain of Tal1 and TAF110 domain of Eto2 for their interaction, and transient transfection and glutathione S-transferase pull-down analysis showed that Mtgr1 and Eto2 enhanced the other’s association with Tal1. Enforced expression of Eto2 in differentiating MEL cells inhibited the promoter of the Protein 4.2 (P4.2) gene, a direct target of TAL1 in erythroid progenitors, and transduction of Eto2 and Mtgr1 augmented Tal1-mediated gene repression. Finally, chromatin immunoprecipitation analysis revealed that Eto2 occupancy of the P4.2 promoter in MEL cells decreased with differentiation, in parallel with a decline in Eto2 protein abundance. These results identify Eto2 and Mtgr1 as authentic interaction partners of Tal1 and suggest they act as heteromeric corepressors of this bHLH transcription factor during erythroid differentiation.
Keywords: ETO2, MTGR1, TAL1, transcription, erythroid progenitors
Introduction
The basic helix-loop-helix (bHLH) transcription factor TAL1 (or SCL) is required for hematopoietic and vascular development and is mixexpressed in up to 60% of children with T-cell acute lymphoblastic leukemia (T-ALL) [1–3]. Gene targeting and overexpression studies have shown that Tal1 also has important functions in erythroid and megakaryocytic differentiation [4–8].
As for other tissue-restricted bHLH proteins, TAL1 interacts with a specific DNA sequence element, the E box, as a heterodimer with one of a more widely expressed family of E proteins, including E2-2, HEB/HTF4, and the E12 and E47 products of the E2A gene [7,9]. We established previously that a ternary complex containing Tal1, E47, Gata-1, LIM-only protein LMO2, and LIM-binding protein Ldb1 occupies and transactivates the Protein 4.2 (P4.2) promoter in erythroid progenitors [10], and this or a similar complex also regulates the GATA-1, α-globlin, and glycophorin A (GPA) genes in cells of this lineage [11–13].
TAL1 functions as a transcriptional activator or repressor, depending on its interaction with specific coactivators and corepressors. TAL1’s transcriptional potency is increased, for example, by p300/CBP [14] and p300/CBP-associated factor [15] and decreased by the corepressor mSin3A [16], SWI/SNF protein Brg1 [17], and histone deacetylases HDAC1 [16] and HDAC2 [17]. Given the number of proteins already known to interact with TAL1 and the importance of this transcription factor in normal and leukemic hematopoiesis, we set out to comprehensively identify Tal1-interacting proteins in the murine erythroleukemia (MEL) cell line used in our previous studies. A tandem epitope-tagged protein was expressed in these cells and the composition of affinity-purified Tal1-containing complexes was determined by mass spectrometry analysis. Two proteins related to the Eight-Twenty-One (ETO) corepressor, Eto2/Mtg16 and Mtgr1, were found and evidence was obtained for their acting as heteromeric corepressors of TAL1.
Materials and methods
Cell lines and reagents
All cells were cultured in Dulbecco’s modified Eagle medium with 10% fetal bovine serum and 0.1 mM non-essential amino acids.
Plasmid constructs and antibodies
pZome1-C-Tal1TAP was prepared by subcloning a full-length Tal1 cDNA minus its stop codon into vector pZome1-C (from European Saccharomyces Cerevisiae Archive for Functional Analysis). Tal1-TAP coding sequences were transferred to MSCV-IRES-GFP [18] to create MSCV-IRES-Tal1TAP-GFP and pEF-IRES-puro [10] to generate pEF-IRES-Tal1TAP-puro, respectively. A full-length mouse Tal1 cDNA absent its stop codon was subcloned into pTracer-EF/Bsd to create pTracer/Tal1V5 and then into pEF-IRES-puro to construct pEF-IRES-Tal1V5. Gal4-Tal1 fusion constructs were described previously [10,14] and the Gal4-Eto2, pCMV5-Eto2, and pCMV5-Mtgr1 plasmids obtained from Joseph Amann (Vanderbilt University). Eto2 coding sequence was transferred from pCMV5-Eto2 to pEF-IRES-puro to create pEF-IRES-Eto2.
Antibodies to V5 and Gal4 were from Invitrogen and Upstate Biotechnology, respectively. Tal1 antibody was prepared as described [19] and purified Mtgr1 and Eto2 antibodies were obtained from Joseph Amann.
Transient transfection analysis and luciferase reporter assays
COS-7 cells were tranfected using Lipofectamine 2000 with pcDNA3.1-Tal1 (2 μg) and pCMV5-Eto2 (1 μg), pCMV5-Mtgr1 (1 μg), and/or pCMV5-Mtgr1 (1 μg) in the indicated combinations. DNA mass was adjusted to 4 μg with pCMV4 as needed. Cell extracts were prepared 48 hr after transfection. NIH 3T3 cells were transfected with pGal4-tk-Luc (100 ng), Renilla luciferase transfection control (3 ng), and pGal4-Tal1 (50 ng), together with pCMV5-Eto2 (10 ng), pCMV5-Mtgr1 (10 ng), or both using Polyfect. Finally, MEL cells were transduced with pGL2-P4.2p1700-luc (80 ng), Renilla luciferase vector, and increasing amounts of pEF-IRES-Eto2 (0, 0.125, 0.25, 0.5, and 1 μg) using DMRIE-C. Cells were incubated with 1.5% DMSO for 24 hr before and 48 hr after transfection. Luciferase activity was measured 48 hr following transfection with the Dual-Luciferase Reporter Assay System and reporter activities normalized to Renilla luciferase activity. Each transfection was carried out in triplicate and repeated three or more times.
Preparation of stably transduced cells
pEF-IRES-Tal1TAP-puro was introduced into MEL cells with DMRIE-C [15] and cells selected in puromycin (2 μg/ml) starting 48 hr after transfection. The concentration of puromycin was increased to 10 μg/ml five days later and maintained.
Affinity purification of TAP-tagged Tal1 protein
MEL cells (400 ml) stably expressing Tal1-TAP protein were washed in PBS and transferred to hypotonic buffer (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 M DTT, and 0.2 mM PMSF). A 21-gauge syringe was used to disrupt cells, the resulting lysates centrifuged at 3300 × g for 15 min at 4°C, and the cell pellet dissolved in 6 mM Na2HPO4, 4 mM NaH2PO4, 1% NP-40, 150 mM NaCl, 2 mM EDTA, 50 mM NaF, 4 μg/μl leupeptin, 0.1 mM Na3VO4, 1 mM PMSF, 1 mM DTT, 50 μg/ml aprotinin, and 25% glycerol. Sepharose 6 Fast Flow, tobacco etch virus protease, and calmodulin-Sepharose were used in affinity purification according to a published method [20].
Immunoprecipitation and Western blot analysis
Cell lysates were prepared and incubated for 30 min at 4°C with Protein G beads pretreated with preimmune immunoglobulin and then the indicated antibodies for 2 hr at 4°C. Immune complexes were recovered with Protein G beads and washed with 10 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 0.1% NP-40. Western blot analysis was performed as previously described [15] using the antibodies listed above.
Mass spectrometry analysis
Tal1-associated proteins eluted from the calmodulin-Sepharose resin were digested with trypsin and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) in a Thermo Finnigan LTQ mass spectrometer in the Vanderbilt University Mass Spectrometry Shared Resource. Protein identities were determined using SEQUEST [21,22].
In vitro binding assays
35S-labeled E47, Mtgr1, and Eto2 proteins were synthesized using coupled in vitro transcription-translation in reticulocyte lysates and unlabeled E47 and Eto2 proteins synthesized similarly. Glutathione-S-transferase (GST) and GST-Tal1 fusion proteins were expressed in E. coli and purified on glutathione-Sepharose 4B. Equivalent amounts of GST fusion proteins were incubated with radiolabeled proteins in 200 μl of lysis buffer, to which 0.1% NP-40 and 10% glycerol were added. Bound proteins were washed three times with buffer and analyzed by SDS-PAGE.
Results and Discussion
To comprehensively characterize its interaction partners, we stably expressed a tandem epitope-tagged mouse Tal1 fusion protein in MEL cells and determined the identities of all associated proteins by multidimensional mass spectrometry. A number of proteins that associate with TAL1 in erythroid cells were found, including multiple E proteins, the zinc finger transcription factor GATA-1, LIM-only protein LMO2, and LIM domain-binding protein Ldb1. In addition to already described proteins [17,23], polypeptides from Mtgr1 and Eto2 were also recovered. 13 peptides were obtained from Mtgr1, representing 27% of its protein coding sequence (Supplemental Fig.), while 9 peptides were identified from Eto2, encompassing 17% of its coding region (Supplemental Fig.). The isolation of multiple peptides from these proteins was highly suggestive of their incorporation into TAL1-containing complexes.
The ETO/MTG family has three members–ETO (or MTG8 or CBFA2T1), ETO2 (or MTG16 or CBFA2T3) and MTGR1 (or CBFA2T2)–that display ~85% sequence similarity [24] and up to 95% identity in four conserved subdomains [25,26]. To confirm the specificity of our antibodies, we introduced expression vectors for Eto, Eto2, and Mtgr1 into COS-7 cells and carried out Western blot analysis on transductants. Importantly, they identified only their cognate proteins (Fig. 1A). Next, coimmunoprecipitation analysis of Tal1 interaction with Eto2 and Mtgr1 was carried out. V5 epitope-tagged Tal1 and wild-type Eto2 or Mtgr1 were expressed in COS-7 cells and V5 immune precipitates subjected to Western blot analysis (Fig. 1B). Eto2 and Mtgr1 could both be precipitated by the Tal1 antibody but not by rabbit IgG. Next, extracts of MEL cells expressing V5-tagged Tal1 protein, confirmed by Western blot analysis with antibodies to V5 or Tal1, were incubated with antibody to the V5 tag and these immune precipitates subjected to Western blot analysis with antibodies to Eto, Eto2, or Mtgr1. Similar to COS cell transductants, Eto2 and Mtgr1, but not Eto, precipitated with V5-Tal1 protein (not shown). Finally, to substantiate Tal1 interaction with Eto2 and Mtgr1 at endogenous levels, coimmunoprecipitation analysis was carried out using non-transfected MEL cells. Again, Eto2 and Mtgr1 were specifically precipitated with Tal1 (Fig. 1C), establishing Tal1 interacts with both ETO-related corepressors in erythroid progenitors.
Fig. 1.
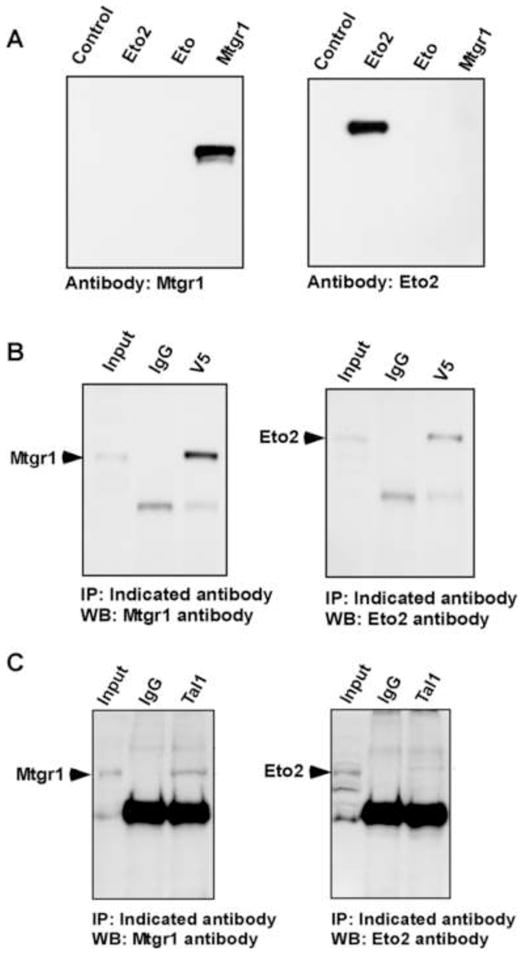
Tal1 interacts with Mtgr1 and Eto2 in cells. (A) Specificity of polyclonal antibodies (Mtgr1, left; Eto2, right) against extracts of COS-7 cells transfected with control plasmid or expression vectors for Mtgr1, Eto, or Eto2. (B) Coimmunoprecipitation analysis with extracts of COS-7 cells transiently transfected with pEF-IRES-Tal1V5 and pCMV5-Mtgr1 (left) or pCMV5-Eto2 (right). Lysates were immunoprecipitated (IP) with normal mouse IgG or murine anti-V5 antibody and these immune precipitates subjected to immunblotting (WB) with antibodies specific for Mtgr1 (left) or Eto2 (right). (C) Coimmunoprecipitation analysis with extracts of MEL cells. Lysates were subjected to IP with normal rabbit IgG or affinity-purified antibody to Tal1 and the resulting immune precipitates subjected to WB with antibodies specific for Mtgr1 (left) or Eto2 (right).
To explore the physiological ramifications of these findings, we investigated whether the abundance of Eto2 and Mtgr1 changed with differentiation in two experimental models, an established line of erythroleukemia cells and in explanted splenic proerythroblasts from mice infected with the anemia-inducing strain of Friend erythroleukemia virus (FVA cells). In MEL cells treated with dimethylsulfoxide and FVA cells cultured with erythropoietin, Eto2 expression decreased within 48 and 24 hr, respectively, in advance of a change in Tal1 abundance, with Mtgr1 declining with slightly slower kinetics in both systems (Fig. 2A and 2B). Eto protein was not detectable in MEL cells and was therefore not studied further (not shown). Likewise, Tal1 interaction with Eto2 and Mtgr1 decreased with differentiation, roughly paralleling the decline in Eto2 protein abundance (Fig. 2A and 2B). Taken together, these data indicate that Eto2 and Mtgr1 can associate with Tal1 in erythroid cells, that these interactions are influenced by the extent of differentiation, and that Eto2 levels may be limiting for interaction of both ETO-related proteins with Tal1.
Fig. 2.
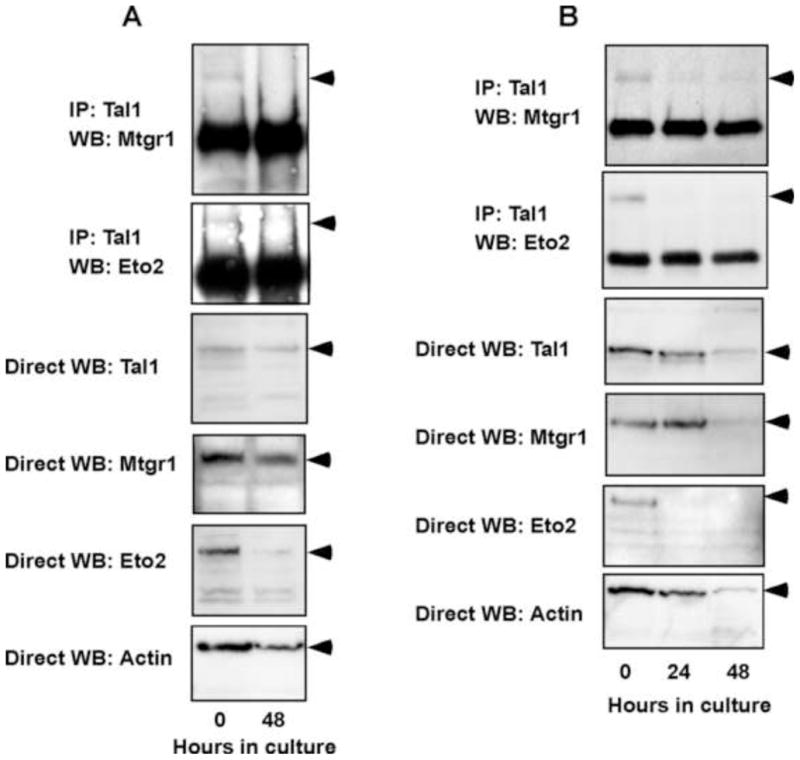
Tal1 interaction with Mtgr1 and Eto2 in erythroid cells declines with cellular differentiation. Total cellular lysates were prepared from DMSO-induced MEL cell (A) and Epo-cultured FVA cells (B) at the indicated times, and immune precipitates obtained with rabbit anti-Tal1 antibody were subjected to WB with antibodies specific to Mtgr1 or Eto2 as indicated. Direct WB analysis was also carried out on the same samples using antibodies to Tal1, Mtgr1, Eto2, or β-actin.
Published studies have shown Eto2 and Mtgr1 interact when overexpressed [27,28]. To investigate whether Tal1 interacts with a heteromeric complex of the two, COS-7 cells were transfected with expression vectors for Tal1, Eto2, and/or Mtgr1 and cellular extracts subjected to immunoprecipitation-immunoblot analysis. Although Eto2 and Mtgr1 were precipitated by antibody to Tal1 in cells transfected with these cDNAs individually, significantly greater amounts of Eto2 and Mtgr1 were precipitated in cells transduced with both proteins (Fig. 3A, top). Precipitation of Mtgr1 with Tal1, in particular, depended on Eto2 coexpression. Since expression studies ruled out an effect of one ETO protein on the concentration of the other, these results suggest that Tal1 interacts with hetero-oligomers of Eto2 and Mtgr1. To further validate this conclusion, GST pull-down assays were carried out with in vitro translated proteins. First, we confirmed the strong interaction reported between E47 and Tal1 [29,30] using radiolabeled E47 and bacterially expressed GST-Tal1 (Fig. 3B, lane 3). Second, measurable, although weak, interaction was observed between radiolabeled Eto2 and GST-Tal1 (Fig. 3B, Lane 4), with Eto2 and GST-Tal1 interaction enhanced by the presence of E47 (Fig. 3B, lane 5). Similarly, while interaction between radiolabeled Mtgr1 and GST-Tal1 was detectable (Fig. 3C), it was significantly increased by addition of Eto2 (Fig. 3C, lane 3). Together, these results are suggestive of a heterodimer of Tal1 and an E protein interacting with an Eto2-Mtgr1 heterodimer.
Fig. 3.
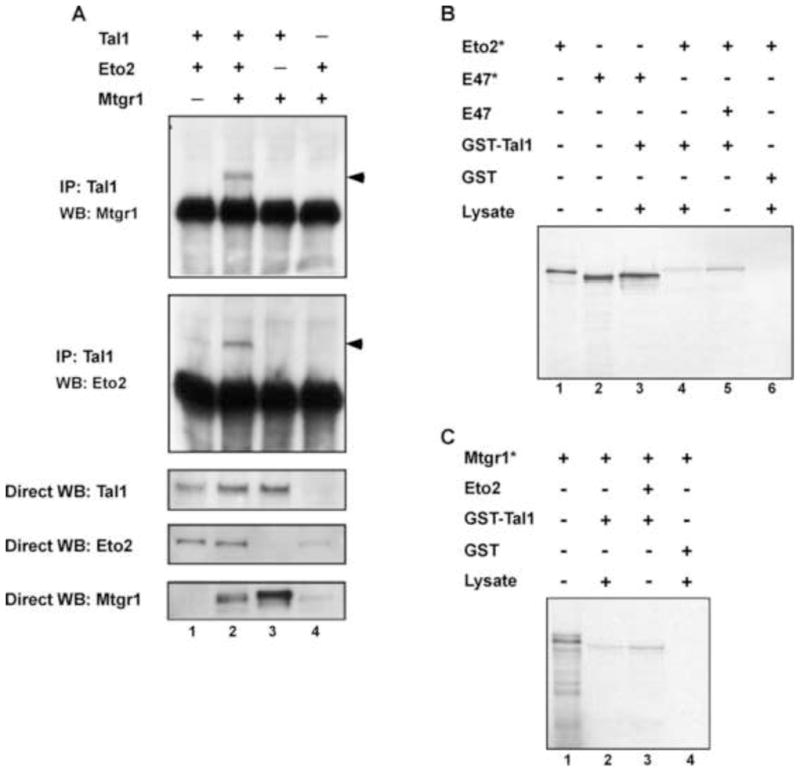
Eto2 and Mtgr1 enhance each other’s interaction with Tal1. (A) For coimmunoprecipitation analysis, COS-7 cells were transfected with the indicated expression constructs, lysates prepared, and cellular proteins immunoprecipitated with Tal1 antibody. Immune precipitates were then subjected to Western blot analysis with anti-Mtgr1 antibody (top panel) or anti-Eto2 antibody (second panel). Total cellular lysates were also directly immunoblotted with antibodies to Tal1 (third panel), Eto2 (fourth panel), and Mtgr1 (fifth panel). Arrows denote the proteins of interest. (B and C) For GST pull-down assays, radiolabeled Eto2 (Eto2*), radiolabeled E47 (E47*), radiolabeled Mtgr1 (Mtgr1*), and unlabeled E47 and Eto2, all prepared by in vitro transcription-translation. Reticulocyte lysates (lysate) were mixed in the indicated combinations and incubated with glutathione-Sepharose charged with GST-Tal1 or GST. Eluates from washed beads were fractionated by SDS-polyacrylamide gel electrophoresis and analyzed by autoradiography.
To characterize the region(s) of Tal1 involved in binding the ETO-related proteins, COS-7 cells were transiently transfected with vectors for Gal4-Tal1 fusion proteins and Eto2 or Mtgr1, and cellular extracts were subjected to coimmunoprecipitation analysis. When expressed individually, neither Eto2 nor Mtgr1 precipitated to any extent with Gal4-Tal1 (Fig. 4A, lanes 3, 4). In contrast, Gal4-Tal1, but not Gal4, precipitated significant amounts of Eto2 and Mtgr1 when both were transfected. Mapping analysis showed that Tal1 fusions containing, or even limited to, the bHLH domain (Gal4-Tal1 construct 6, lane 6, Figure 4A) interacted with Eto2 and Mtgr1, whereas fusions with the N-terminal 144 or C-terminal 88 amino acids did not. Thus, the bHLH domain of Tal1 is necessary and sufficient for Tal1 interaction with Eto2 and Mtgr1.
Fig. 4.
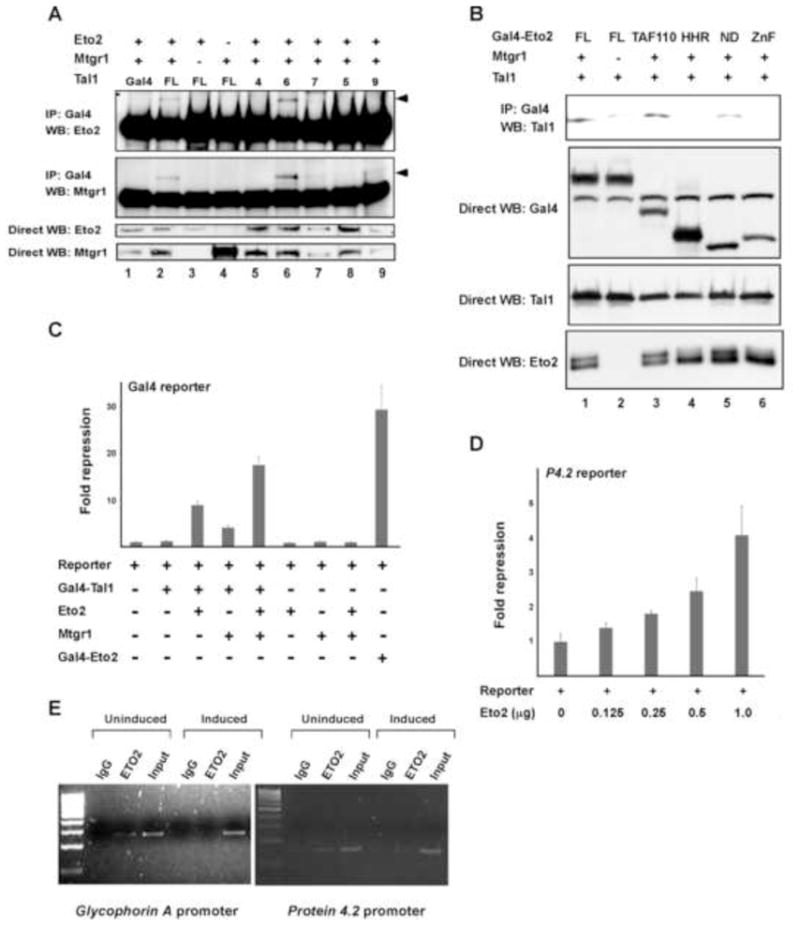
Structural and functional characteristics of Eto2 and Mtgr1 interaction with Tal1. (A) Results of coimmunoprecipitation analysis of interaction between Gal4-Tal1 fusion proteins and Eto2 and Mtgr1 in transfected COS-7 cells. Construct FL contains the full-length Tal1 coding sequence (1–329), while Gal4-Tal1 constructs 4, 6, 7, 5, and 9 contain amino acids 1–144, 142–329, 185–329, 242–329 and 185–240, respectively, of Tal1 coding sequence. Gal4-Tal1 construct 6 (lane 6) includes the entire bHLH domain. (B) Results of reciprocal coimmunoprecipitation analysis of Gal4-Eto2 fusion protein interaction with Tal1 in transfected COS-7 cells. Construct FL contains full-length Eto2 coding sequence (1–620), while Gal4-Eto2 constructs TAF110, HHR, ND, and ZnF contain amino acids 46–242, 371–402, 461–510, and 533–572 respectively, of Eto2 protein coding sequence. (C) Effect of different combinations of Eto2 and Mtgr1 expression vectors Gal4-Tal1-directed repression of a luciferase reporter gene linked to multiple copies of the Gal4 binding site. After 48 h, cellular lysates were prepared and luciferase activities were determined. Plotted is the mean repression ± SD of luciferase activity from three independent experiments. (D) Effect of the indicated amounts of an Eto2 expression vector on expression of a luciferase reporter gene linked to the proximal promoter of the mouse P4.2 gene. The two plasmids were transiently transfected in MEL cells. Plotted is the mean repression ± SD of luciferase enzymatic activity. (E) ChIP analysis of Eto2 occupancy of the indicated TAL1 target genes in differentiating MEL cells. Cells were incubated with (induced) or without (uninduced) 1.5% DMSO for 3 days and nuclear proteins cross-linked to DNA by addition of formaldehyde to culture. Chromatin fragments generated by sonication were immunoprecipitated with anti-Eto2 antibody or normal rabbit IgG and used in PCR analysis with primers flanking the characterized TAL1 binding sites.
To resolve the region(s) of the two ETO-related proteins responsible for their interaction with Tal1, we expressed a series of Eto2-Gal4 fusion proteins with full-length Tal1 in COS-7 cells, in the presence and absence of Mtgr1 (Fig. 4B). Coimmunoprecipitation analysis with an antibody to Gal4 showed that the TAF110 domain of Eto2 was required for Tal1 interaction, although the nervy-like domain (ND) also appeared to contribute (Fig. 4B). In contrast, the HHR domain mediating oligomerization of ETO family members [28] did not. This indicates that the TAF110 domain is involved in Eto2-Tal1 interaction, while a separate region likely mediates dimerization of Eto2 and Mtgr1.
To investigate the effects of Eto2 and Mtgr1 on TAL1-directed transcription, transient transfection analysis was carried out with a Gal4 reporter construct. While Gal4-Tal1 is capable of stimulating luciferase activity from this construct, albeit weakly [14], it repressed luciferase activity 8.9- and 4.1-fold, respectively, when cotransfected with Eto2 or Mtgr1, with somewhat greater than additive repression (17.6-fold) seen with expression of both ETO-related proteins (Fig. 4C). Thus, while Eto2 and Mtgr1 can separately interact with Tal1, greater repression is achieved when they are recruited together. To characterize Eto2’s effect on transcription in a more physiologic context, we transfected MEL cells with the Eto2 expression vector and a promoter-luciferase reporter for a well-characterized TAL1 target gene, P4.2 [10]. Luciferase assays in these Mtgr1- and Eto2-expressing cells showed that excess Eto2 inhibited P4.2 promoter activity in a dose-related manner (Fig. 4D).
Finally, chromatin immunoprecipitation (ChIP) analysis was used to investigate whether Eto2 was recruited to the promoters of TAL1 target genes in erythroid cells and whether its occupancy of these sites changed with differentiation. Chromatin from formaldehyde-treated MEL cells was fragmented by sonication and precipitated with the Eto2 antibody. PCR analysis was then carried out with primers flanking the Tal1 binding sites in the P4.2 [10] and GPA [13] promoters. DNA taken prior to immunoprecipitation was used as a positive control (input), and a region of the glyceraldehyde-3-phosphate dehydrogenase gene in input and immunoprecipitated DNA served as positive and negative control, respectively. Chromatin fragments from the P4.2 and GPA promoters were precipitated in undifferentiated but not DMSO-differentiated cells (Fig. 4E), consistent with the coimmunoprecipitation results. These findings demonstrate that Eto2 is recruited to TAL1 target genes in erythroid progenitors and that its occupancy of those genes correlates inversely with their transcription during terminal differentiation.
In summary, multi-dimensional mass spectrometry identified every member of Tal1-containing DNA-binding complexes described in erythroid cells [10], including multiple E proteins, GATA-1, LMO2, and Ldb1, and proteins known to interact with GATA-1 (FOG1), LMO2 (ELF2A2), and Ldb1 (SSBP2 and SSBP3). A number of previously unrecognized components were also discovered, including Brg1 [17] and ETO2 and MTGR1 (this work). Using similar approaches, three groups have recently reported that ETO2 and/or MTGR1 interact with TAL1 in erythroid cells to augment TAL1-directed gene repression [29,31,32]. Our studies extend their work by demonstrating that a hetero-oligomer of ETO2 and MTGR1 constitutes the active corepressor species.
Erythroid progenitors were reduced and erythropoiesis impaired in Eto2 knockout mice [33], whereas no hematopoietic abnormalities were detectable in Mtgr1−/− animals [34]. Even if Eto2 abundance is limiting for Mtgr1 interaction with Tal1, the finding that they act as heteromeric corepressors of this transcription factor in erythroid cells predicts that deletion of Eto2 and Mtgr1 would have greater phenotypic consequences than of Eto2 alone. Studies to investigate this issue are underway.
Supplementary Material
Supplemental Fig. 1. Sequences of Eto2- and Mtgr1-derived peptides deduced from LC-MS/MS (shaded) in digests of affinity-purified Tal1-containing complexes from MEL cells. Longer polypeptide sequences derive from contiguous tryptic fragments. Mtgr1 sequence was identified in 13 peptides, with total coverage of 27%. Eto2 sequence was identified in 9 peptides, with total coverage of 17%.
Acknowledgments
We thank Joseph Amann (Vanderbilt University Medical Center) for purifying Eto2 and Mtgr1 antibodies and Salisha Hill in the Vanderbilt Proteomics Shared Resource for important assistance in mass spectrometry analysis. This work was supported in part by NIH grants R01 HL49118 (to S.J.B.) and 5P30CA068485 (to Vanderbilt-Ingram Cancer Center) and a Merit Review Award from the Department of Veterans Affairs (to S.J.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Begley CG, Aplan PD, Denning SM, Haynes BF, Waldmann TA, Kirsch IR. The gene SCL is expressed during early hematopoiesis and encodes a differentiation-related DNA-binding motif. Proc Natl Acad Sci USA. 1989;86:10128–10132. doi: 10.1073/pnas.86.24.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Q, Cheng JT, Tasi LH, Schneider N, Buchanan G, Carroll A, Crist W, Ozanne B, Siciliano MJ, Baer R. The tal gene undergoes chromosome translocation in T cell leukemia and potentially encodes a helix-loop-helix protein. EMBO J. 1990;9:415–424. doi: 10.1002/j.1460-2075.1990.tb08126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finger LR, Kagan J, Christopher G, Kurtzberg J, Hershfield MS, Nowell PC, Croce CM. Involvement of the TCL5 gene on human chromosome 1 in T-cell leukemia and melanoma. Proc Natl Acad Sci U S A. 1989;86:5039–5043. doi: 10.1073/pnas.86.13.5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Condorelli G, Vitelli L, Valtieri M, Marta I, Montesoro E, Lulli V, Baer R, Peschle C. Coordinate expression and developmental role of Id2 protein and TAL1/E2A heterodimer in erythroid progenitor differentiation. Blood. 1995;86:164–175. [PubMed] [Google Scholar]
- 5.Elwood NJ, Zogos H, Pereira DS, Dick JE, Begley CG. Enhanced megakaryocyte and erythroid development from normal human CD34(+) cells: consequence of enforced expression of SCL. Blood. 1998;91:3756–3765. [PubMed] [Google Scholar]
- 6.Aplan PD, Nakahara K, Orkin SH, Kirsch IR. The SCL gene product: a positive regulator of erythroid differentiation. EMBO J. 1992;11:4073–4081. doi: 10.1002/j.1460-2075.1992.tb05500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shivdasani RA, Mayer EL, Orkin SH. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature. 1995;373:432–434. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- 8.Robb L, Lyons I, Li R, Hartley L, Ktgen F, Harvey RP, Metcalf D, Begley CG. Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proc Natl Acad Sci USA. 1995;92:7075–7079. doi: 10.1073/pnas.92.15.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quong MW, Romanow WJ, Murre C. E protein function in lymphocyte development. Annu Rev Immunol. 2002;20:301–322. doi: 10.1146/annurev.immunol.20.092501.162048. [DOI] [PubMed] [Google Scholar]
- 10.Xu Z, Huang S, Chang LS, Agulnick AD, Brandt SJ. Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol Cell Biol. 2003;23:7585–7599. doi: 10.1128/MCB.23.21.7585-7599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valverde-Garduno V, Guyot B, Anguita E, Hamlett I, Porcher C, Vyas P. Differences in the chromatin structure and cis-element organization of the human and mouse GATA1 loci: implications for cis-element identification. Blood. 2004;104:3106–3116. doi: 10.1182/blood-2004-04-1333. [DOI] [PubMed] [Google Scholar]
- 12.Anguita E, Hughes J, Heyworth C, Blobel GA, Wood WG, Higgs DR. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J. 2004;23:2841–2852. doi: 10.1038/sj.emboj.7600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lahlil R, Luyer E, Herblot S, Hoang T. SCL assembles a multifactorial complex that determines glycophorin A expression. Mol Cell Biol. 2004;24:1439–1452. doi: 10.1128/MCB.24.4.1439-1452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang S, Qiu Y, Stein RW, Brandt SJ. p300 functions as a transcriptional coactivator for the TAL1/SCL oncoprotein. Oncogene. 1999;18:4958–4967. doi: 10.1038/sj.onc.1202889. [DOI] [PubMed] [Google Scholar]
- 15.Huang S, Qiu Y, Shi Y, Xu Z, Brandt SJ. P/CAF-mediated acetylation regulates the function of the basic helix-loop-helix transcription factor TAL1/SCL. EMBO J. 2000;19:6792–6803. doi: 10.1093/emboj/19.24.6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang S, Brandt SJ. mSin3A regulates murine erythroleukemia cell differentiation through association with the TAL1 (or SCL) transcription factor. Mol Cell Biol. 2000;20:2248–2259. doi: 10.1128/mcb.20.6.2248-2259.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Z, Meng X, Cai Y, Koury MJ, Brandt SJ. Recruitment of the SWI/SNF protein Brg1 by a multiprotein complex effects transcriptional repression in murine erythroid progenitors. Biochem J. 2006;399:297–304. doi: 10.1042/BJ20060873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Persons DA, Allay JA, Allay ER, Smeyne RJ, Ashmun RA, Sorrentino BP, Nienhuis AW. Retroviral-mediated transfer of the green fluorescent protein gene into murine hematopoietic cells facilitates scoring and selection of transduced progenitors in vitro and identification of genetically modified cells in vivo. Blood. 1997;90:1777–1786. [PubMed] [Google Scholar]
- 19.Kallianpur AR, Jordan JE, Brandt SJ. The SCL/TAL-1 gene is expressed in progenitors of both the hematopoietic and vascular systems during embryogenesis. Blood. 1994;83:1200–1208. [PubMed] [Google Scholar]
- 20.Tasto JJ, Carnahan RH, McDonald WH, Gould KL. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast. 2001;18:657–662. doi: 10.1002/yea.713. [DOI] [PubMed] [Google Scholar]
- 21.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR. Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 22.Ducret A, Van Oostveen I, Eng JK, Yates JR, Aebersold R. High throughput protein characterization by automated reverse-phase chromatography/electrospray tandem mass spectrometry. Protein Sci. 1998;7:706–719. doi: 10.1002/pro.5560070320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Z, Meng X, Cai Y, Liang H, Nagarajan L, Brandt SJ. Single-stranded DNA-binding proteins regulate the abundance of LIM domain and LIM domain-binding proteins. Genes Dev. 2007;21:942–955. doi: 10.1101/gad.1528507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calabi F, Cilli V. CBFA2T1, a gene rearranged in human leukemia, is a member of a multigene family. Genomics. 1998;52:332–341. doi: 10.1006/geno.1998.5429. [DOI] [PubMed] [Google Scholar]
- 25.Davis JN, McGhee L, Meyers S. The ETO (MTG8) gene family. Gene. 2003;303:1–10. doi: 10.1016/s0378-1119(02)01172-1. [DOI] [PubMed] [Google Scholar]
- 26.Feinstein PG, Kornfeld K, Hogness DS, Mann RS. Identification of homeotic target genes in Drosophila melanogaster including nervy, a proto-oncogene homologue. Genetics. 1995;140:573–586. doi: 10.1093/genetics/140.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoogeveen AT, Rossetti S, Stoyanova V, Schonkeren J, Fenaroli A, Schiaffonati L, van Unen L, Sacchi N. The transcriptional corepressor MTG16a contains a novel nucleolar targeting sequence deranged in t (16; 21)-positive myeloid malignancies. Oncogene. 2002;21:6703–6712. doi: 10.1038/sj.onc.1205882. [DOI] [PubMed] [Google Scholar]
- 28.Lindberg SR, Olsson A, Persson AM, Olsson I. Interactions between the leukaemia-associated ETO homologues of nuclear repressor proteins. Eur J Haematol. 2003;71:439–447. doi: 10.1046/j.0902-4441.2003.00166.x. [DOI] [PubMed] [Google Scholar]
- 29.Schuh AH, Tipping AJ, Clark AJ, Hamlett I, Guyot B, Iborra FJ, Rodriguez P, Strouboulis J, Enver T, Vyas P, Porcher C. ETO-2 associates with SCL in erythroid cells and megakaryocytes and provides repressor functions in erythropoiesis. Mol Cell Biol. 2005;25:10235–10250. doi: 10.1128/MCB.25.23.10235-10250.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Kalkum M, Yamamura S, Chait BT, Roeder RG. E protein silencing by the leukemogenic AML1-ETO fusion protein. Science. 2004;305:1286–1289. doi: 10.1126/science.1097937. [DOI] [PubMed] [Google Scholar]
- 31.Goardon N, Lambert JA, Rodriguez P, Nissaire P, Herblot S, Thibault P, Dumenil D, Strouboulis J, Romeo PH, Hoang T. ETO2 coordinates cellular proliferation and differentiation during erythropoiesis. EMBO J. 2006;25:357–366. doi: 10.1038/sj.emboj.7600934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meier N, Krpic S, Rodriguez P, Strouboulis J, Monti M, Krijgsveld J, Gering M, Patient R, Hostert A, Grosveld F. Novel binding partners of Ldb1 are required for haematopoietic development. Development. 2006;133:4913–4923. doi: 10.1242/dev.02656. [DOI] [PubMed] [Google Scholar]
- 33.Chyla BJ, Moreno-Miralles I, Steapleton MA, Thompson MA, Bhaskara S, Engel M, Hiebert SW. Deletion of Mtg16, a target of t(16;21), alters hematopoietic progenitor cell proliferation and lineage allocation. Mol Cell Biol. 2008;28:6234–6247. doi: 10.1128/MCB.00404-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amann JM, Chyla BJ, Ellis TC, Martinez A, Moore AC, Franklin JL, McGhee L, Meyers S, Ohm JE, Luce KS, Ouelette AJ, Washington MK, Thompson MA, King D, Gautam S, Coffey RJ, Whitehead RH, Hiebert SW. Mtgr1 is a transcriptional corepressor that is required for maintenance of the secretory cell lineage in the small intestine. Mol Cell Biol. 2005;25:9576–9585. doi: 10.1128/MCB.25.21.9576-9585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1. Sequences of Eto2- and Mtgr1-derived peptides deduced from LC-MS/MS (shaded) in digests of affinity-purified Tal1-containing complexes from MEL cells. Longer polypeptide sequences derive from contiguous tryptic fragments. Mtgr1 sequence was identified in 13 peptides, with total coverage of 27%. Eto2 sequence was identified in 9 peptides, with total coverage of 17%.


