SUMMARY
Macrolide antibiotics are a class of valuable anti-infective agents that include a macrolactone ring, at least one appended sugar unit, and in most cases, additional functionalization in the form of hydroxyl and/or epoxide groups. There is a significant body of work to understand assembly of the polyketide derived aglycone for this class of compounds, particularly for erythromycin and pikromycin that are generated through the action of a modular polyketide synthases. In addition, the mode of assembly of deoxysugars and aminosugars as well as their transfer to the aglycone has been reported over the past decade. However, much less information exists for the final tailoring reactions, typically mediated by P450 enzymes that are capable of regio- and stereospecific oxidations to add hydroxyl or epoxide functionality. Herein, we have characterized in vitro two P450 enzymes from the mycinamicin biosynthetic gene cluster of Micromonospora griseorubida. Cloning, overexpression and purification of MycCI revealed its selectivity for C21 methyl group hydroxylation. The natural substrate for this P450 enzyme is mycinamicin VIII, the earliest macrolide form in the post-PKS tailoring pathway appended with desosamine at C5 OH. Moreover, we found the optimal activity of MycCI is dependent on the native ferredoxin MycCII. The second and third oxidation reactions including hydroxylation and epoxidation respectively are mediated by MycG with mycinamicin IV as initial substrate. This reaction requires prior dimethylation of the second deoxysugar residue (e.g., 6-deoxyallose to mycinose) for effective conversion by the dual function MycG P450, the first natural product biosynthetic monooxygenase characterized with an ability to catalyze both hydroxylation and epoxidation steps.
INTRODUCTION
The cytochrome P450 enzymes (P450s) form a very large family of oxidative hemeproteins that are responsible for a diversity of oxidative transformations across most life forms (Coon, 2005; Guengerich, 2001). These reactions typically involve modification of physiologic and xenobiotic compounds, and include the biosynthesis of various bioactive compounds (e.g. steroids, antibiotics, signaling molecules). Recent bacterial genome sequencing efforts have uncovered an unexpected large number of genes encoding P450 enzymes. For example, the model actinomycete Streptomyces coelicolor A3(2) that produces actinorhodin and undecylprodigiosin revealed the presence of 18 different P450 genes (Bentley et al., 2002), whereas Streptomyces avermitilis MA-4680, an avermectin producer, contains 33 (Ikeda et al., 2003), and Saccharopolyspora erythraea NRRL 23338, the erythromycin producing bacterium encodes 36 P450s (Oliynyk et al., 2007). In secondary metabolic pathways, it is typical that P450 genes are integrated within the biosynthetic cluster where their products catalyze regio- and stereospecific oxidation of precursors leading to structural diversity as well as improved bioactivities of these molecules (Lamb et al., 2003; Rix et al., 2002). Thus, cytochrome P450 enzymes EryF (Andersen and Hutchinson, 1992) and EryK (Stassi et al., 1993) that are encoded within the erythromycin biosynthetic gene cluster are involved in the biosynthesis of erythromycin A. Specifically, EryF hydroxylates the macrolactone precursor 6-deoxyerthronolide B, whereas EryK is a macrolide hydroxylase resulting in formation of erythromycin D. As prototypic P450 hydroxylases involved in secondary metabolism, EryF and EryK exhibit strict substrate specificity. In contrast, PikC cytochrome P450 involved in the methymycin/neomethymycin and pikromycin biosynthetic pathway of Streptomyces venezuelae has broader substrate tolerance (Xue et al., 1998). PikC catalyzes the final hydroxylation step toward the 12-membered ring macrolide YC-17 and the 14-membered ring macrolide narbomycin to produce methymycin/neomethymycin and pikromycin as major products.
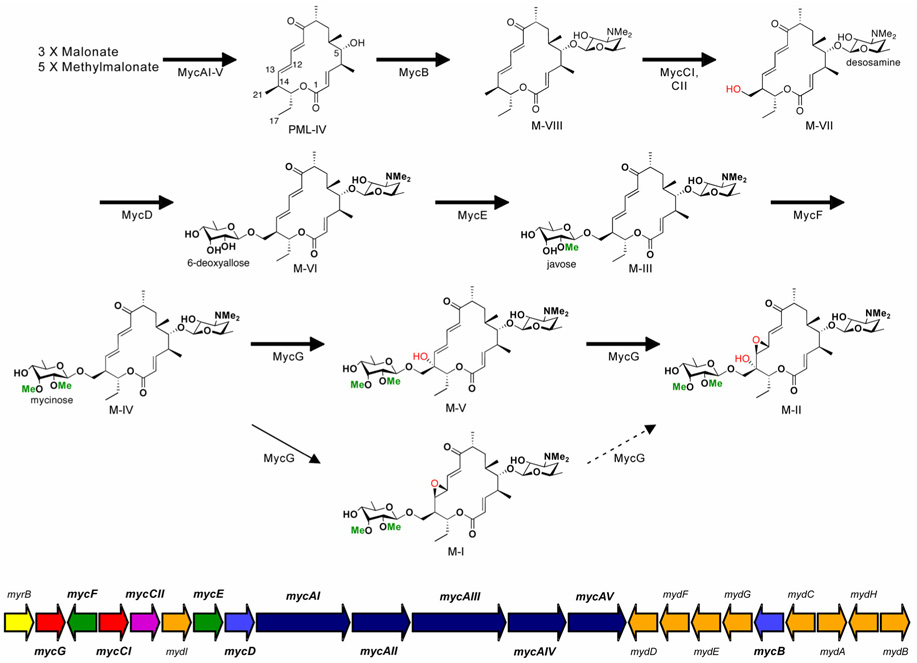
Mycinamicins, a series of macrolide antibiotics produced by the rare actinomycete Micromonospora griseorubida, have shown impressive activities against a spectrum of Gram-positive strains, especially some antibiotic-resistant human pathogens (Kinoshita et al., 1988; Satoi et al., 1980; Suzuki et al., 1990). Stucturally, the major mycinamicin products of wild type strain M. griseorubida A11725 including mycinamicin I (M-I), II (M-II), IV (M-IV), and V (M-V) (Figure 1) consist of a 16-membered ring polyketide macrolactone substituted with 6-deoxyhexose sugars desosamine and mycinose. Partial characterization of the biosynthetic pathway for mycinamicins has been obtained through analysis of blocked mutants and corresponding bioconversion studies (Kinoshita, 1991a; Suzuki et al., 1990). More recently, the nucleotide sequence of the complete mycinamicin biosynthetic gene cluster has been reported (Anzai et al., 2003), wherein two putative P450 genes mycCI and mycG were identified (Figure 1).
Figure 1. Mycinamicin post-PKS biosynthetic pathway and organization of the mycinamicin biosynthetic gene cluster.
Methyl groups introduced by O-methyltransferases, and oxidation steps catalyzed by cytochrome P450 enzymes are highlighted in green and red, respectively. Bold arrows represent the main pathway, whereas thin arrows indicate a shunt pathway. The dashed arrow indicates low-level conversion as previously reported (Inouye et al., 1994). Color codes in the myc gene cluster are: Red, cytochrome P450 genes; purple, ferredoxin gene; indigo blue, polyketide synthetase genes; blue, glycosyltransferase genes; orange, O-methyltransferase genes; green, deoxysugar biosynthetic genes; yellow, self-resistance gene (rRNA methyltransferase gene).
Analysis of the 5’ region of the myc gene cluster upstream from the PKS locus revealed that mycCI is located adjacent to mycCII, which encodes a putative ferredoxin (Anzai et al., 2003). Since the deduced amino acid sequences of mycCI and mycCII show high sequence similarities to TylHI and TylHII (Figure 2), respectively, that are likely responsible for hydroxylation at the C23 methyl group of tylactone (Baltz and Seno, 1981), the function of MycCI and MycCII was accordingly proposed to mediate hydroxylation at the analogous C21 methyl group of protomycinolide IV (PML-IV) (Anzai et al., 2003) (Figure 1). Based on genetic complementation analysis of a targeted mutant strain of M. griseorubida, mycG was presumed to encode a P450 enzyme that catalyzes both hydroxylation and epoxidation at C14 and C12/13 on the macrolactone ring of mycinamicin (Inouye et al., 1994; Suzuki et al., 1990). In the current study, mycCI and mycG genes were overexpressed in Escherichia coli, and the functions of purified MycCI and MycG proteins were determined in vitro using natural substrates derived from semi-synthesis or isolated from wild type or engineered strains of M. griseorubida that accumulate key mycinamycin intermediates. Moreover, specific roles have been proposed for both the desosamine, and mycinose sugar residues in the oxidative cascade leading to M-II, the final product in the pathway.
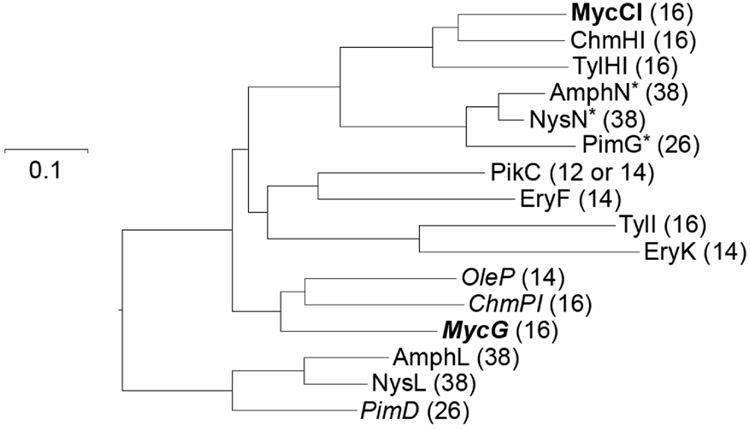
Figure 2. Phylogenetic tree of macrolide biosynthetic P450 monooxygenases.
The selected cytochrome P450s include OleP (accession # AAA92553) [oleandomycin pathway], ChmHI and ChmPI (accession # AAS79447 and AAS79453) [chalcomycin pathway], MycCI and MycG [mycinamicin pathway] (accession # BAC57023 and BAA03672), AmphL and AmphN (accession # AAK73504 and AAK73509) [amphotericin pathway], NysL and NysN (accession # AAF71769 and AAF71771) [nystatin pathway], PimD and PimG (accession # CAC20932 and CAC20928) [pimaricin pathway], TylHI and TylI (accession # AAD41818 and AAA21341) [tylosin pathway], EryF and EryK (accession # AAA26496 and YP-001102980) [erythromycin pathway], and PikC (accession # AAC68886) [pikromycin pathway]. The numbers in parentheses indicate the macrolactone ring size of the corresponding P450 substrate. Unless otherwise specified, all selected P450 enzymes are hydroxylases. The epoxidases are noted in italics. The enzymes marked with an asterisk are presumed to mediate carboxylic acid formation. MycCI and MycG are highlighted in bold.
RESULTS
Protein Sequence Analysis of MycCI and MycG
Comparison of the deduced amino acid sequences of MycCI and MycG showed relatively low sequence identity (33%). In the phylogenetic tree (Figure 2) of selected bacterial macrolide biosynthetic P450 enzymes, they were clustered in distinct branches, suggesting these two unrelated P450 genes may have been integrated into the mycinamicin biosynthetic gene cluster sequentially from different ancestors, as opposed to being derived from divergent evolution following duplication of a parental gene. Specifically, MycG, as the first biosynthetic monooxygenase characterized with an ability to catalyze both hydroxylation and epoxidation steps, is clustered with two other epoxidases OleP (Rodriguez et al., 1995) and ChmPI (Ward et al., 2004), whereas MycCI is closely related to ChmHI (Ward et al., 2004) and TylHI (Baltz and Seno, 1981), both of which are responsible for methyl group hydroxylations of 16-membered ring macrolides. Interestingly, we revealed that the clustering of macrolide biosynthetic P450s correlated with not only the functionalities (i.e. hydroxylases, epoxidases, or ones responsible for carboxylic acid formation), but also the substrate macrolactone ring size, suggesting that selection based on product structure derived from the upstream PKS biosynthetic system might be among the significant factors directing evolution of secondary metabolic P450s.
Heterologous Expression of MycCI, MycCII and MycG
The mycCI, mycCII and mycG genes were overexpressed in E. coli BL21(DE3) and the resulting proteins were purified (see Supplemental Data). MycCI was overexpressed as either a C- or N-terminal 6 × His tagged protein. After purification using Ni-NTA agarose chromatography, the individual polypeptides showed molecular weights of approximately 44 and 45 kDa, corresponding to the estimated masses of C- and N-terminal 6 × His tagged MycCI. The wild type MycCII (MycCII-wt, m.w. ≈ 8 kDa) and N-terminal 6 × His-tagged ferredoxin MycCII (MycCII-NH, m.w. ≈ 10 kDa) were purified to homogeneity (see Experimental Procedures). MycG expression was placed under the control of a T7 promoter including a 6 × His-tag introduced at its amino terminus. Similarly, one-step purification was performed using a Ni-NTA agarose column to obtain homogeneous protein with expected molecular weight of 46 kDa. Subsequently, the CO-bound reduced difference spectra confirmed the identity of both MycCI and MycG as cytochrome P450 enzymes (see Supplemental Data).
Synthesis of Mycinamicin VIII (M-VIII) from Protomycinolide IV (PML-IV)
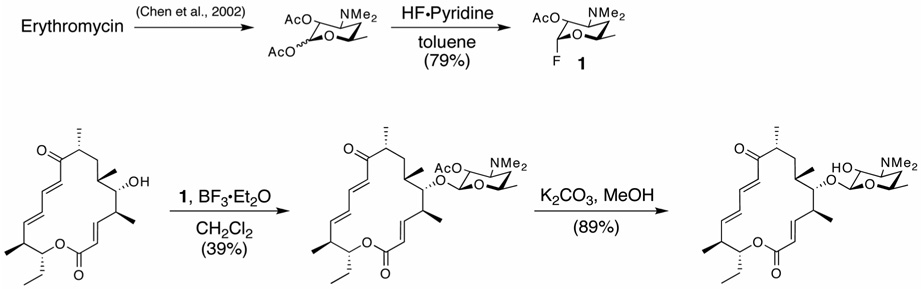
Due to the limited quantity (< 0.1 mg) of totally available M-VIII isolated from M. griseorubida fermentation culture, the presumed substrate of MycCI, we developed an effective chemical glycosylation strategy to obtain this important intermediate by coupling desosamine as sugar donor to the readily available PML-IV aglycone precursor (Hayashi et al., 1981). Glycosylation with desosamine has previously employed either thioglycosides or glycosyl fluorides, with a methyl carbonate as the C2 protecting group (Martin et al., 1997; Matsumoto et al., 1988; Suzuki et al., 1988; Toshima et al., 1995; Woodward et al., 1981a; Woodward et al., 1981b; Woodward et al., 1981c). After examining several combinations of protecting groups and anomeric leaving groups, we found that the C2 acetoxy-protected glycosyl fluoride of desosamine was conveniently obtained, easily purified, stable to storage, and effective in the transfer of desosamine to a range of aglycones. In this procedure (Figure 3), erythromycin hydrate was hydrolyzed under acidic conditions to obtain a crude sample of desosamine hydrochloride that was converted to the bis(acetate) of desosamine in 88% isolated yield as previously reported (Chen et al., 2002). Further treatment of this material with HF·pyridine afforded the C2 acetoxy glycosyl fluoride of desosamine 1, which serves as a stable and convenient precursor for chemical glycosylations. Direct treatment of PML-IV (Figure 1) with the glycosyl fluoride and BF3·Et2O in CH2Cl2 afforded C2 acetoxy mycinamycin VIII as a 2:1 mixture of β and α anomers in 39% (not optimized) isolated yield. Higher yielding desosamine glycosylations with other mycinamicins have been reported (Matsumoto et al., 1988), however, these alternative procedures with different glycosyl donors were not explored for comparison. Purification by preparative HPLC (SiO2) afforded the pure β-anomer (see Supplemental Data), which was deprotected with K2CO3 in methanol to afford semi-synthetic M-VIII in 89% isolated yield. The 1H NMR spectrum of this synthetic material was coincident with previously reported data (Kinoshita et al., 1989).
Figure 3. Synthetic scheme for M-VIII.
Functional Analysis of MycCI in vitro
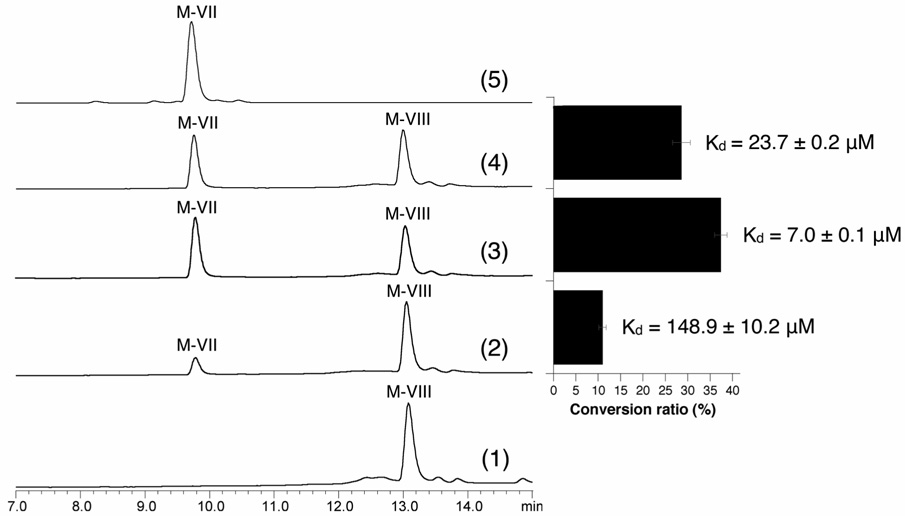
In order to establish the role of each mycinamicin P450, we first tested whether MycCI is capable of hydroxylating the predicted substrate M-VIII using a typical heterologous spinach ferredoxin and ferredoxin reductase system (Xue et al., 1998). As expected, M-VIII was converted to the corresponding C21 hydroxymethyl product M-VII (Figure 4), albeit inefficiently. Notably, the C- and N-terminal His-tagged MycCI showed similar activities (data not shown), indicating the location of His-tag has no significant impact on MycCI activity. We selected the C-terminal His-tagged MycCI to perform biochemical studies and refer to this form as MycCI for convenience. The low conversion ratio of M-VIII (11.0 ± 0.8 %) might be due to the higher activation energy required to functionalize the primary C-H bond. Alternatively, the fact that a specific ferredoxin gene mycCII is clustered with mycCI suggests that MycCI might possess higher ferredoxin specificity than MycG. Thus, the capability of MycCII (together with spinach ferredoxin reductase since the native ferredoxin reductase for MycCI remains unknown) to reconstitute the in vitro M-VIII hydroxylation activity of MycCI was assessed and compared to spinach ferredoxin. Thus, when partnered by MycCII-wt and MycCII-NH, MycCI converted 28.6 ± 2.0 % and 37.4 ± 1.4 % of M-VIII to M-VII respectively, suggesting MycCII mediates the electron transfer from spinach ferredoxin reductase to MycCI more efficiently than spinach ferredoxin (Figure 4). However, it is currently unclear why the activity of the His-tagged MycCII is higher than MycCII-wt. Considering spinach ferredoxin and ferredoxin reductase are natural partners, we reason that the higher activity of MycCII could derive from a more favorable interaction with the MycCI P450 compared to the spinach ferredoxin reductase. Therefore, the binding affinities of different ferredoxins toward MycCI were compared with one another. As expected, the most active form (MycCII-NH) exhibited the lowest dissociation constant Kd of 7.0 ± 0.1 µM. In contrast, the binding affinity of spinach ferredoxin to MycCI (Kd = 148.9 ± 10.2 µM) is much lower than MycCII-wt or MycCII-NH. Interestingly, we found MycCII is unable to support in vitro activity of MycG (see Supplemental Data), the other P450 enzyme involved in mycinamicin biosynthesis. These results strongly suggest MycCII has evolved to selectively serve MycCI catalysis.
Figure 4. In vitro M-VIII conversions catalyzed by MycCI (all LC traces were analyzed at 280 nm).
(1) negative control, M-VIII + boiled MycCI; (2) M-VIII + MycCI + spinach ferredoxin; (3) M-VIII + MycCI + MycCII-NH; (4) M-VIII + MycCI + MycCII-wt; (5) M-VII standard. The right diagram shows M-VIII conversion ratios calculated by using equation 1-AUCunreacted M-VIII/AUCtotal M-VIII (AUC: area under curve) based on the corresponding LC-traces in parallel. In addition, dissociation constants (Kd) of the spinach and MycCII ferredoxins toward MycCI are shown.
In addition, similar MycCI reactions were assessed with the PML-IV, M-VI, M-III, M-IV, and M-V (Figure 1), but no products were observed and starting materials were quantitatively recovered (data not shown). These results establish that MycCI catalyzes the first oxidation step, converting the C21 methyl group to the corresponding hydroxymethyl that is subsequently functionalized with 6-deoxyallose, followed by two methylation steps to mycinose. Moreover, the substrate requires desosamine modification at the C5 hydroxyl group in order to be accepted as a substrate by MycCI. This is analogous to PikC, shown through co-crystal structure analysis with natural 12- and 14-membered ring macrolides to involve desosamine mediated substrate anchoring (Sherman et al., 2006).
Functional Analysis of MycG in vitro
Previous genetic studies on the mycinamicin biosynthetic pathway identified a gene fragment that complemented a M-II non-producing mutant of M. griseorubida (Inouye et al., 1994). DNA sequence analysis revealed an open reading frame whose translated product showed high level amino acid sequence similarity to cytochrome P450 enzymes. The mycinamicin biosynthetic intermediate isolated from the M-II non-producing M. griseorubida strain lacked both C14 hydroxyl and C12-C13 epoxide functionalities and the subcloned fragment bearing the putative P450 gene was able to restore production of M-II (Inouye et al., 1994). These data provided strong but indirect evidence that the mycG gene product is capable of catalyzing both oxidation steps at adjacent positions on the macrolactone ring (Figure 1).
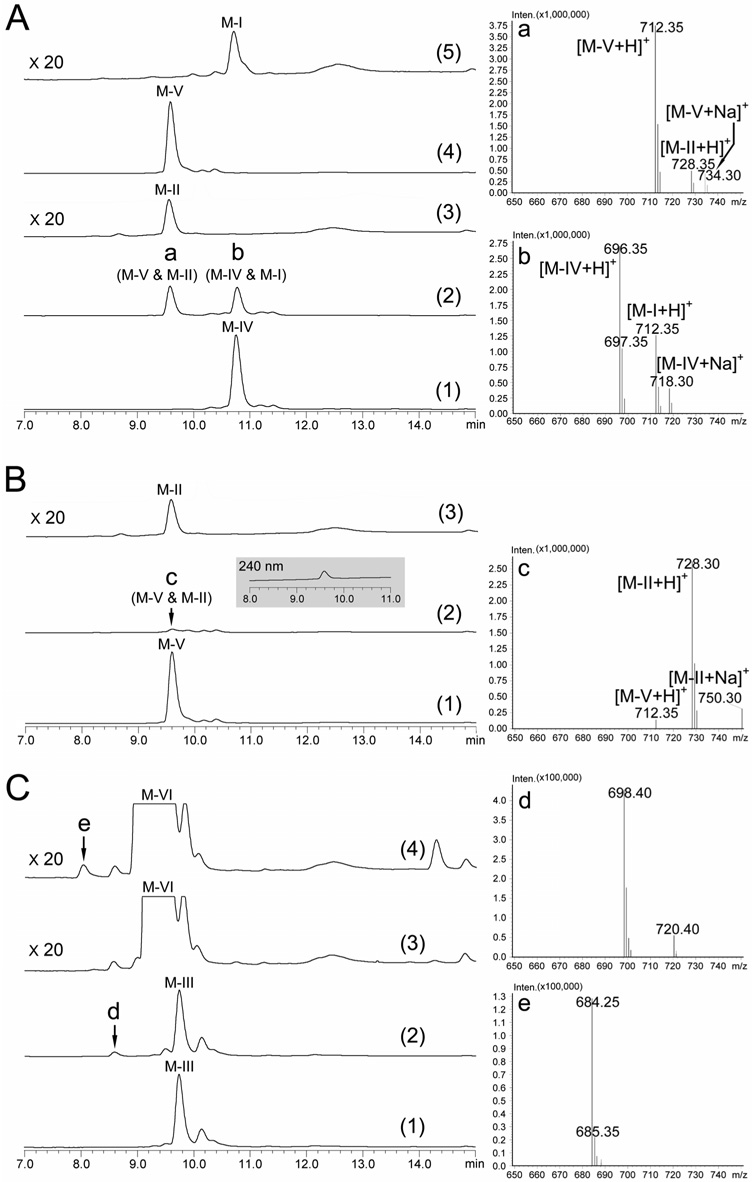
To confirm the dual function of MycG, our in vitro analysis was initiated by testing directly the ability of MycG to convert the putative substrate M-IV. As shown in Figure 5A, the LC trace of the reaction extract (2) at 280 nm showed two peaks a and b. The corresponding mass spectra indicate M-V ([M+H]+ = 712.35) and M-II ([M+H]+ = 728.35) co-eluted as peak a, and peak b includes M-IV ([M+H]+ = 696.35) and M-I ([M+H]+ = 712.35). This LC-MC analysis clearly demonstrates the dual function of MycG as hydroxylase and epoxidase. However, despite efforts to optimize reverse-phase HPLC conditions, we were unable to separate M-IV and M-V from their epoxidized products.
Figure 5. LC-MS analysis of in vitro conversions catalyzed by MycG (all LC traces were analyzed at 280 nm unless otherwise specified).
(A) MycG reaction using M-IV as substrate: (1) negative control, M-IV + boiled MycG; (2) M-IV + MycG. The right panel shows the mass spectra of peak a and b; (3) M-II standard amplified 20 × due to its poor absorbance at 280 nm; (4) M-V standard; (5) M-I standard amplified 20 ×. (B) MycG reaction using M-V as substrate: (1) negative control, M-V + boiled MycG; (2) M-V + MycG. The inset shows the 240 nm LC trace, under which M-II has better absorbance. The right panel shows the mass spectrum of peak c; (3) M-II standard amplified 20 ×. (C) MycG reactions using M-III and M-VI as alternative substrates: (1) negative control, M-III + boiled MycG; (2) M-III + MycG; (3) negative control, M-VI + boiled MycG; (4) M-VI + MycG. LC traces (3) and (4) amplified 20 × compared to (1) and (2) to visualize the peak e in trace amount. The right panel shows the mass spectra of peak d and e.
To determine the precursor of M-II, M-V and M-I were incubated with MycG in separate reactions. When M-V was used as substrate, it was almost completely converted into M-II. Since M-II (and M-I) lacks strong absorbance at 280 nm (Satoi et al., 1980), only a minor peak c was observed in the LC trace although substantial M-II was formed based on mass spectral analysis (Figure 5B; notably, the M-II product was visible at 240 nm). However, M-I was not converted into M-II in vitro (see Supplemental Data), indicating that M-II can only be generated from M-V as substrate. Epoxidation of M-IV prior to hydroxylation evidently results in termination of this post-PKS tailoring pathway (Figure 1).
To assess further enzyme selectivity and ability to catalyze alternative oxidative reactions against the mycinamicin aglycone, and several early glycosylated intermediates (PML-4, M-VIII, M-VII, M-VI, and M-III), MycG-mediated reactions were performed. No products were observed when using PML-4, M-VIII, and M-VII (see Supplemental Data), but two new compounds appeared at low levels in LC-MS analysis of the M-VI and M-III reactions (Figure 5C). When M-III was used as a substrate, a minor new peak d was detected with m/z = 698.40, which is 16 Da higher than the molecular weight of M-III. Moreover, in the MycG reaction with M-VI, a new peak e (m/z = 684.25), presumed to be oxidized M-VI was detected at an even lower conversion level. Both peak d and e were visible under 280 nm and their retention times were faster than corresponding starting materials strongly suggesting they are hydroxylated instead of epoxidized products. Moreover, the MS-MS analysis (data not shown) of d and e indicated that both hydroxylations occurred on the macrolactone ring although we could not further determine the site of modification due to limited amounts of each compound. Interestingly, the C14 hydroxylated forms of M-III and M-VI have already been reported as minor components of the mycinamicin pathway designated as mycinamicin IX (M-IX) and mycinamicin XV (M-XV), respectively (Kinoshita et al., 1992). Accordingly, it is likely that d and e correspond to M-IX and M-XV, providing direct evidence for the in vivo origin of these two minor compounds. More importantly, we established that efficient catalysis by MycG requires mycinose, derived from dimethylation of the second sugar residue as C21 OH linked 6-deoxyallose.
Measurement of Substrate Dissociation Constants
To understand why mycinamicin P450 enzymes (especially MycG) behave differently toward compounds with subtle structural differences, we carried out spectrophotometric substrate binding assays to determine dissociation constants (Kd). As shown in Table 1, M-VIII binds to MycCI with a Kd value of 28.1 ± 3.2 µM, which is significantly higher than 0.7 ± 0.1 µM, the Kd value of M-IV toward MycG. Evidently, the substrate binding to MycG is much tighter than MycCI, which is likely attributed to additional interactions of mycinose within the P450 substrate binding pocket. Interestingly, the C14 hydroxylated M-V showed approximately 14 times lower binding affinity toward MycG than M-IV, suggesting M-V is a less suitable substrate for MycG. However, this is inconsistent with the results from in vitro assays (described in Figure 4C above) in which M-V was converted to oxidized product more effectively than M-IV (kinetic analysis (see below) provided insights into this apparent paradox). In contrast to M-V, the epoxidized product M-I was not able to function as a substrate since we did not observe a Type-I difference spectra even at high substrate concentrations. This result is consistent with a lack of MycG activity toward M-I in vitro. It was particularly surprising that the final product M-II bound to MycG with a dissociation constant of 71.5 ± 8.4 µM, indicating the compromising effect of the epoxide moiety toward substrate binding could be overridden by an adjacent C14 hydroxyl group. The reason for this unusual finding will await substrate co-crystallographic analysis of MycG, which is currently in progress.
Table 1.
Binding and steady-state kinetic analysis of MycCI and MycG
| Enzyme | Substrate | Kd (µM) | Km (µM) | kcat (min−1) | kcat/Km (µM−1·min−1) |
|---|---|---|---|---|---|
| MycCI | M-VIII | 28.1 ± 3.2 | 34.4 ± 5.5a | 71.7 ± 3.2a | 2.1a |
| 5.8 ± 0.7b | 104.1 ± 1.8b | 17.9b | |||
| MycG | M-IV | 0.7 ± 0.1 | NDc | NDc | NDc |
| M-V | 10.4 ± 0.2 | 16.2 ± 3.1a | 415.7 ± 22.9a | 25.7a | |
| M-I | NBd | - | - | - | |
| M-II | 71.5 ± 8.4 | - | - | - |
Kinetic parameters were determined when spinach ferredoxin was employed
Kinetic constants were determined when MycCII-NH ferredoxin was employed
ND: The kinetic parameters were not determined because multiple reactions occur at the same time
NB: No binding
-: No reaction
Steady-State Kinetic Analysis of MycCI and MycG
Assuming a 1:1 stoichiometric relationship between NADPH consumption and substrate oxidation, the steady-state kinetic parameters of MycCI were determined using purified synthetic M-VIII as substrate. When partnered with spinach ferredoxin, MycCI demonstrated a Km of 34.5 ± 5.5 µM, kcat of 71.7 ± 3.2 min−1 for the C21 methyl group hydroxylation step. In contrast, with MycCII-NH coupled with MycCI catalysis became more efficient with a decreased Km of 5.8 ± 0.7 µM and an improved kcat of 104.1 ± 1.8 min−1. This suggests that MycCII not only induces better substrate binding of MycCI, but also improves electron transfer efficiency as reflected by increased turnover number (kcat).
Although MycG is capable of catalyzing three different reactions (M-IV to M-V; M-IV to M-I; M-V to M-II), we limited our study to kinetic constants for M-V to M-II conversion (Km = 16.2 ± 3.1 µM, kcat = 415.7 ± 22.9 min−1). This was due to the complexity of the analysis when M-IV was used as substrate, since multiple reactions (M-IV to M-V or M-I and M-V to M-II) did not allow fitting of data to the Michaelis-Menten equation. However, it is noteworthy that we observed the overall NADPH consumption rate at high M-IV concentration (e.g. 100 µM) to be considerably reduced compared to low M-IV concentration (e.g. 10 µM), suggesting strong substrate inhibition. This could explain the above mentioned contradiction between the conversion ratio and binding affinity. According to the in vitro analysis of MycG activity (Figure 5A and B), M-V appears to be a better substrate than M-IV based on the conversion ratios. But this result might be misleading since M-IV has better binding affinity toward MycG (Table 1). Thus, based on in vitro reactions using substrate concentrations at 500 µM, we believe the lower conversion of M-IV is a reflection of substrate inhibition.
DISCUSSION
Mycinamicins represent a large family of macrolide antibiotics with more than twenty members (Kinoshita et al., 1992; Kinoshita, 1991b). The structural diversity is derived primarily from post-PKS tailoring modifications including glycosylation, oxidation, and methylation steps. Therefore, this biosynthetic pathway represents an important system to explore the mechanism and significance of secondary metabolite diversification.
Here we provide a detailed analysis of two cytochrome P450 enzymes MycCI and MycG and their role in late-stage chemical modifications. By gene cloning, protein expression, purification and reconstitution assays with heterologous redox partners, we unambiguously confirmed their physiological role in vitro. To the best of our knowledge, the functional and kinetic analysis of MycCI represents the first enzymatic characterization of a biosynthetic monooxygenase responsible for methyl group hydroxylation. In addition, we established that MycCII is a specific ferredoxin, whose corresponding gene (mycCII) is immediately adjacent to mycCI, and capable of effectively supporting MycCI activity. Compared to spinach ferredoxin, MycCII binds to MycCI more tightly, presumably leading to more efficient electron transfer. On the other hand, we established that C21 hydroxylation by MycCI depends on C5 linked desosamine, which is analogous to PikC (Xue et al., 1998), another cytochrome P450 involved in pikromycin biosynthesis. Recent co-crystallographic analysis of PikC (Sherman et al., 2006) and its natural substrates (YC-17 and narbomycin) revealed that desosamine acts as an indispensable anchor responsible for productive binding, and proper positioning of substrate in the active site. Accordingly, we surmise that desosamine plays a similar role in hydroxylation of precursor macrolide M-VIII.
The current study has revealed that MycG is a more versatile enzyme than MycCI providing the first example of a biosynthetic P450 involved physiologically in catalyzing both hydroxylation and epoxidation reactions. In addition, MycG represents the primary basis for structural diversification in the mycinamicin pathway since it is solely responsible for generating multiple products including M-V, M-I, M-II as well as some minor components including M-IX and M-XV. This work has also demonstrated that MycG function is dependent on the presence of both desosamine and the second sugar mycinose in the substrate. It is also noteworthy that both deoxysugars are essential for mycinamicin bioactivity (Kinoshita et al., 1989). Although the precise role of the second sugar for MycG substrate recognition remains obscure due to lack of x-ray structural information, based on the high binding affinity of M-IV toward MycG (Kd = 0.7 ± 0.1 µM), we predict there might be a specific active site binding pocket in the polypeptide to accommodate mycinose, thereby significantly improving substrate affinity. Moreover, of particular interest, the activity of MycG against diglycosylated substrates (M-VI, M-III, and M-IV) is related to the extent of methylation mediated by sequential O-methyltransfer by MycE and MycF. In the absence of methylether groups installed on the second sugar molecule, M-VI bearing 6-deoxyallose appears to be a very poor substrate for MycG. This situation can be improved by the first methylation to form javose in M-III, although its conversion is low (Figure 5C). Subsequently, the second methylation catalyzed by MycF imparts a pronounced effect upon MycG-mediated binding of M-IV. It appears likely that these stepwise methylations decrease the polarity of 6-deoxyallose in M-VI, thus enabling it to be accepted by a putative hydrophobic MycG binding pocket.
Substrate inhibition occurs in about 20% of all known enzymes (Copeland, 2000). In biosynthetic P450 enzymes, this phenomenon has been observed in previous kinetic analysis of PikC (Li et al., 2007), EryK (Lambalot and Cane, 1995), and PimD (Mendes et al., 2005). In this study, M-IV inhibition of MycG was also detected indirectly even though the kinetic parameters were not obtained due to complexity of the multiple-reaction system. Thus, the mechanism of this substrate inhibition cannot be further elucidated without generating detailed kinetic curves. However, we realize this behavior of MycG might be physiologically significant for maintaining the chemical diversity within the mycinamicin pathway. In the event that M-IV inhibition does not occur, MycG would convert all M-IV into final products M-I and M-II, instead of a mixture of M-I, II, IV, and V as shown in Figure 5A, and observed in the fermentation culture of M. griseorubida strain A11725 (Satoi et al., 1980). This would ultimately limit the spectrum of its metabolic output, and perhaps compromise the ability of the microorganism to adapt to a variable and competitive environment.
SIGNIFICANCE
This report provides detailed information about the oxidative cascade that introduces structural diversity into the mycinamicin class of macrolide antibiotics. We have established in vitro that MycCI is a cytochrome P450 enzyme responsible for the C21 methyl hydroxylation of M-VIII, the starting substrate of the mycinamicin post-PKS tailoring pathway. In addition, we have demonstrated that the MycG monooxygenase catalyzes sequential hydroxylation and epoxidation steps with M-IV or M-V as substrates. MycCI represents the first biosynthetic methyl group hydroxylase characterized in vitro. Its optimal activity depends on MycCII, a ferredoxin whose gene is encoded within the mycinamycin biosynthetic gene cluster. MycG is the first biosynthetic monooxygenase to be characterized whose physiological function includes hydroxylation and epoxidation reactions. Moreover, the unprecedented requirement of the second deoxysugar with essential methylether modifications for activity of MycG reveals the important interplay between three typical post-tailoring modifications in biosynthesis including glycosylation, methylation and oxidation. Through detailed functional analysis of these two P450 enzymes, we have unambiguously established the mycinamicin post-PKS oxidation pathway.
EXPERIMENTAL PROCEDURES
Materials
Unless otherwise specified, all chemical reagents were ordered from Sigma-Aldrich. DNA oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA). Molecular cloning used New England Biolabs (Ipswich, MA) restriction enzymes, Stratagene (La Jolla, CA) Pfu Turbo DNA polymerase, Novagen (Madison, WI) pET vectors, Invitrogen (Carlsbad, CA) T4 DNA ligase and Z-Competent™ Escherichia Coli Transformation Buffer set from Zymo Research (Orange, CA). Protein purification used Qiagen (Valencia, CA) Ni-NTA resin, Millipore (Billerica, MA) Amicon Ultra centrifugal filter, PD-10 desalting columns from GE Healthcare (Piscataway, NJ), and Thrombin restriction grade Kit from Novagen (Madison, WI). LB Broth was from EM Sciences (Gibbstown, NJ). All of mycinamicins except for M-VIII were obtained from the fermentation broth of M. griseorubida A11725. M-VIII was synthesized from PML-IV using the method described above.
DNA Manipulation, Cloning, and PCR
Using cosmid pMR01 (Anzai et al., 2003) as template, mycCI, mycCII and mycG genes were amplified by PCR under standard conditions with primers as follows: forward, 5’-CAGCATATGGTGGTCTGGCCCATGGACCGCACCTG-3’ for mycCI, 5’-GTGCCATATGCGGATAGTCCTGGACGCCGAAC-3’ for mycCII and 5’-CGGTCATATGACTTCAGCTGAACCTAGGGCGTATCC-3’ for mycG (the underlined bases represent the introduced NdeI site for further cloning); reverse, 5’-TCGTAAGCTTCCACTCGACCAGCAGCTCGTCGATG-3’ for cloning mycCI gene without a stop codon, 5’-TCCAAAGCTTTCCGCATACCGCACCCCCATTCGTC-3’ for amplifying the mycCI gene retaining a stop codon, 5’-TGACAAGCTTACTCCTGTTGGCCCACCTGTCCCGTG-3’ for mycCII and 5’-GGCAAAGCTTCTCCGACGAGATCGTCGAGATCGAC-3’ for mycG (the italic letters indicate a HindIII restriction site for later cloning). The gel purified cDNAs were rescued by double digestion of NdeI and HindIII. Then, the fragment containing the mycCI gene with stop codon removed was ligated into NdeI/HindIII treated pET21b to generate recombinant plasmid pET21b-mycCI for expression of C-terminal His-tagged MycCI. Genes including mycCI, mycCII, and mycG retaining stop codons were ligated into previously NdeI/HindIII-digested pET28b to generate recombinant plasmids for expression of N-terminal His-tagged proteins. After confirming the identity of inserted genes by DNA sequencing, the constructs were used to transform E. coli BL21(DE3) for protein overexpression.
Protein Overexpression and Purification
MycCI and MycG (P450s) overexpression and purification followed previously developed procedures with minor modifications (Li et al., 2007; Xue et al., 1998). The E. coli BL21 (DE3) transformants carrying certain plasmids were grown at 37°C in 1 liter of LB broth containing thiamine (1 mM), 5% glycerol, 50 µg/ml of selective antibiotics (ampicillin for pET21b and kanamycin for pET28b), and a rare salt solution (6750 µg/l FeCl3, 500 µg/l ZnCl2, CoCl2, Na2MoO4, 250 µg/l CaCl2, 465 µg/l CuSO4, and 125 µg/l H3BO3) until OD600 reached 0.6~0.8. Then isopropyl β-D-thiogalactoside (IPTG, 0.1mM) and δ-aminolevulinic acid (1mM) were added, and the cells were cultured at 18°C overnight. After harvesting cells by centrifugation, 40 ml of lysis buffer (50mM NaH2PO4, pH8.0, 300 mM NaCl, 10% glycerol, 10 mM imidazole) was used to resuspend the cell pellet. Lysis was accomplished on a Model 500 Sonic Dismembrator (ThermoFisher Scientific). The insoluble material was separated by centrifugation (35,000 × g, 30 min at 4°C). The soluble fraction was collected and incubated for 1 h at 4°C after addition of 1ml Ni-NTA resin. The slurry was loaded onto an empty column, and the column was then washed with 40 to 80 ml of wash buffer (50mM NaH2PO4, pH8.0, 300 mM NaCl, 10% glycerol, 20 ~ 30 mM imidazole). The elution buffer (50mM NaH2PO4, pH8.0, 300 mM NaCl, 10% glycerol, 250 mM imidazole) was added onto the column, and eluted protein fraction was concentrated with Amicon Ultra 4, Ultracel – 30K. Subsequent desalting was attained by buffer exchange into desalting buffer (50mM NaH2PO4, pH7.3, 1 mM EDTA, 0.2 mM dithioerythritol, 10% glycerol) with a PD-10 column.
Overexpression and purification of N-terminal 6 × His-tagged ferredoxin MycCII-NH are similar to above procedures with minor modifications as follows: 1) δ-aminolevulinic acid, the precursor for heme biosynthesis, was omitted in the culture broth; 2) The protein obtained from elution buffer was concentrated using Amicon Ultra 4, Ultracel – 5K due to the low molecular weight of MycCII (~ 10 kDa). To obtain MycCII-wt lacking a His-tag, 2.4 mg of MycCII-NH in desalting buffer was digested by 2 units of thrombin at 4°C overnight. The cleaved His-tag and residual thrombin were removed by Ni-NTA resin and a 30K size exclusion filter sequentially. The concentration of purified ferredoxins was determined by Coomassie protein assay using spinach ferredoxin as standard.
CO-bound reduced difference spectra
The identification of MycCI and MycG as active P450 enzymes was performed through getting the CO-bound reduced difference spectra using a UV-visible spectrophotometer 300 Bio (Cary). First, the P450 enzyme in desalting buffer was reduced by adding several milligrams of sodium dithionite (Na2S2O4) and a spectrum was recorded from 350 to 600 nm. Then, after CO bubbling of the solution for 30~60 s, the spectrum of CO-bound reduced P450 species was recorded using previous reduced spectrum as reference. This assay was also employed to determine the functional P450 concentration using the extinction coefficient of 91,000 M−1·cm−1 (Omura and Sato, 1964).
Functional Analysis of In Vitro Activities of MycCI and MycG
The standard conversion was accomplished by combining 1 µM of desalted MycCI or MycG, whose functional concentrations were determined using UV-visible absorption spectrum method (Omura and Sato, 1964), 0.5 mM mycinamicin biosynthetic intermediate, 3.5 µM spinach ferredoxin or MycCII ferredoxin, 0.1 Unit/ml spinach ferredoxin-NADP+ reductase and 0.5 mM NADPH in 100 µl of desalting buffer. The reaction with boiled P450 enzyme was used as negative controls. The reaction was carried out for 2 h at 30°C and was terminated by extraction, using 3 × 200 µl of CHCl3. The resulting organic extraction was dried and redissolved in 150 µl of methanol. The LC-MS analysis of reaction extract was performed on LCMS-2010 EV (Shimadzu) by using an XBridge™ C18 3.5 µm 150 mm reverse-phase HPLC column under following conditions: mobile phase, 20~100% solvent B over 18 min (A = deionized water + 0.1% formic acid, B = acetonitrile + 0.1% formic acid); flow rate: 0.2 ml/min; UV wavelength: 240 and 280 nm.
Spectral Substrate Binding Assay
Spectral substrate binding assay was carried out on UV-visible spectrophotometer 300 Bio (Cary) at room temperature by titrating substrate DMSO solution (blank DMSO for reference group) into 1 ml of 0.5~1 µM P450 solution in 1 µl aliquots. The series of Type I difference spectra were used to deduce ΔA (Apeak – Atrough). Then, the data from duplicated experiments were fit to Michaelis-Menten equation to obtain the dissociation constant Kd.
Spectral Ferredoxin Binding Assay
Spectral ferredoxin titrations were performed as described (Coghlan and Vickery, 1991) using 0.7 µM MycCI and 10 µM M-VIII in desalting buffer at room temperature. Ferredoxin solutions in appropriate concentrations were used to titrate the P450 solution. Binding of ferredoxins to MycCI induced increased M-VIII binding, thus leading to a larger absorbance difference ΔA (Apeak – Atrough). The data from duplicated experiments were linearized by using a Hanes-Wolf plot to deduce dissociation constant Kd.
Steady-State Kinetics of MycCI and MycG
The standard reactions contain 0.6 µM of MycCI (when partnered with spinach ferredoxin), 0.1 µM of MycCI (when partnered with MycCII-NH), or 0.1 µM of MycG, 1.9 µM spinach or MycCII ferredoxins, 0.02 Unit/ml spinach ferredoxin-NADP+ reductase, 5~200 µM M-VIII for MycCI and 2~160 µM M-V for MycG in 90 µl of desalting buffer. After pre-incubation in 96-well plate at room temperature for 5 min, the reactions with different substrate concentrations were initiated spontaneously by adding 10 µl of 2 mM NADPH with a multichannel pipette. The rate of NADPH consumptions were monitored continuously over 2 min under 340 nm by SpectraMax M5 spectrophotometer (Molecular Devices). The initial velocities of NADPH consumptions were deduced from the absorbance curves within the linear range (0~20 s). Then, assuming a 1:1 stoichiometric relationship between NADPH consumption and substrate oxidation, the initial velocities of hydroxylation reactions were calculated by using the millimolar absorption coefficient 6.22 mM−1·cm−1 of NADPH at 340 nm (Greenbaum et al., 1972). Finally, the results from duplicated experiments were fit to Michaelis-Menten equation to obtain steady-state kinetic parameters.
Supplementary Material
Supplemental data include protein purification SDS-PAGE gel results, LC-MS analysis of selected P450 reactions, P450 CO-bound reduced spectra, substrate binding affinity measurements, steady-state kinetic curves, and experimental details for the synthesis of M-VIII.
Acknowledgements
We thank Mr. Yousong Ding for assistance with LC-MS analysis. Anonymous reviewers are gratefully acknowledged for helpful comments. This work was supported by NIH grant GM078553, the Hans W. Vahlteich Professorship (to D.H.S.), and NIH grant GM57014 (to J. M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andersen JF, Hutchinson CR. Characterization of Saccharopolyspora erythraea cytochrome P-450 genes and enzymes, including 6-deoxyerythronolide B hydroxylase. J. Bacteriol. 1992;174:725–735. doi: 10.1128/jb.174.3.725-735.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzai Y, Saito N, Tanaka M, Kinoshita K, Koyama Y, Kato F. Organization of the biosynthetic gene cluster for the polyketide macrolide mycinamicin in Micromonospora griseorubida. FEMS Microbiol. Lett. 2003;218:135–141. doi: 10.1111/j.1574-6968.2003.tb11509.x. [DOI] [PubMed] [Google Scholar]
- Baltz RH, Seno ET. Properties of Streptomyces fradiae mutants blocked in biosynthesis of the macrolide antibiotic tylosin. Antimicrob. Agents Chemother. 1981;20:214–225. doi: 10.1128/aac.20.2.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- Chen H, Yamase H, Murakami K, Chang C-w, Zhao L, Zhao Z, Liu H-w. Expression, purification, and characterization of two N,N-dimethyltransferases, TylM1 and DesVI, involved in the biosynthesis of mycaminose and desosamine. Biochemistry. 2002;41:9165–9183. doi: 10.1021/bi020245j. [DOI] [PubMed] [Google Scholar]
- Coghlan VM, Vickery LE. Site-specific mutations in human ferredoxin that affect binding to ferredoxin reductase and cytochrome P450scc. J. Biol. Chem. 1991;266:18606–18612. [PubMed] [Google Scholar]
- Coon MJ. Cytochrome P450: Nature's most versatile biological catalyst. Ann. Rev. Pharmacol. Toxicol. 2005;45:1–25. doi: 10.1146/annurev.pharmtox.45.120403.100030. [DOI] [PubMed] [Google Scholar]
- Copeland RA. Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis. 2nd Ed. New York: Wiley; 2000. [Google Scholar]
- Greenbaum E, Austin RH, Frauenfelder H, Gunsalus IC. Photoreduction of NADP+ sensitized by synthetic pigment systems. Proc. Natl. Acad. Sci. USA. 1972;69:1273–1276. doi: 10.1073/pnas.69.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich FP. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem. Res. Toxicol. 2001;14:611–650. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Ohara H, Ohno M, Sakakibara H, Satoi S. Mycinamicins, new macrolide antibiotics. V. Isolation and structures of new 16-membered aglycones, mycinolide IV and protomycinolide IV. J. Antibiot. 1981;34:1075–1077. doi: 10.7164/antibiotics.34.1075. [DOI] [PubMed] [Google Scholar]
- Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- Inouye M, Takada Y, Muto N, Beppu T, Horinouchi S. Characterization and expression of a P-450-like mycinamicin biosynthesis gene using a novel Micromonospora-Escherichia coli shuttle cosmid vector. Mol. Gen. Genet. 1994;245:456–464. doi: 10.1007/BF00302258. [DOI] [PubMed] [Google Scholar]
- Kinoshita K, Imura Y, Takenaka S, Hayashi M. Mycinamicins, new macrolide antibiotics XI. Isolation and structure elucidation of a key intermediate in the biosynthesis of the mycinamicins, mycinamicin VIII. J. Antibiot. 1989;44:1869–1872. doi: 10.7164/antibiotics.42.1869. [DOI] [PubMed] [Google Scholar]
- Kinoshita KST, Hayashi M. Isolation of proposed intermediates in the biosynthesis of mycinamicins. J. Chem. Soc. Chem. Commun. 1988;5:943–945. [Google Scholar]
- Kinoshita K, Takenaka S, Suzuki H, Morohoshi T, Hayashi M. Mycinamicins, new macrolide antibiotics. XIII. Isolation and structures of novel fermentation products from Micromonospora griseorubida (FERM BP-705) J. Antibiot. 1992;45:1–9. doi: 10.7164/antibiotics.45.1. [DOI] [PubMed] [Google Scholar]
- Kinoshita K, Takenaka S, Hayashi M. Mycinamicin biosynthesis: Isolation and structural elucidation of mycinonic acids, proposed intermediates for formation of mycinamicins. X-ray molecular structure of p-bromophenacyl 5-hydroxy-4-methylhept-2-enoate. J. Chem. Soc. Perkin Trans. 1. 1991a;1:2547–2553. [Google Scholar]
- Kinoshita K, Takenaka S, Hayashi M. Mycinamicins, new macrolide antibiotics. XII. Isolation and structural elucidation of mycinamicin X and XI. J. Antibiot. 1991b;44:1270–1273. doi: 10.7164/antibiotics.44.1270. [DOI] [PubMed] [Google Scholar]
- Lamb DC, Ikeda H, Nelson DR, Ishikawa J, Skaug T, Jackson C, Omura S, Waterman MR, Kelly SL. Cytochrome p450 complement (CYPome) of the avermectin-producer Streptomyces avermitilis and comparison to that of Streptomyces coelicolor A3(2) Biochem. Biophys. Res. Commun. 2003;307:610–619. doi: 10.1016/s0006-291x(03)01231-2. [DOI] [PubMed] [Google Scholar]
- Lambalot RH, Cane DE. Overproduction and characterization of the erythromycin C-12 hydroxylase, EryK. Biochemistry. 1995;34:1858–1866. doi: 10.1021/bi00006a006. [DOI] [PubMed] [Google Scholar]
- Li S, Podust LM, Sherman DH. Engineering and analysis of a self-sufficient biosynthetic cytochrome P450 PikC fused to the RhFRED reductase domain. J. Am. Chem. Soc. 2007;129:12940–12941. doi: 10.1021/ja075842d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SF, Hida T, Kym PR, Loft M, Hodgson A. The asymmetric synthesis of erythromycin B. J. Am. Chem. Soc. 1997;119:3193–3194. [Google Scholar]
- Matsumoto T, Maeta H, Suzuki K, Tsuchihashi G. First total synthesis of mycinamicin IV and VII. Successful application of new glycosidation reaction. Tetrahedron Lett. 1988;29:3575–3578. [Google Scholar]
- Mendes MV, Anton N, Martin JF, Aparicio JF. Characterization of the polyene macrolide P450 epoxidase from Streptomyces natalensis that converts de-epoxypimaricin into pimaricin. Biochem. J. 2005;386:57–62. doi: 10.1042/BJ20040490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliynyk M, Samborskyy M, Lester JB, Mironenko T, Scott N, Dickens S, Haydock SF, Leadlay PF. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat. Biotechnol. 2007;25:447–453. doi: 10.1038/nbt1297. [DOI] [PubMed] [Google Scholar]
- Omura T, Sato R. The Carbon Monoxide-Binding Pigment of Liver Microsomes. Ii. Solubilization, Purification, and Properties. J. Biol. Chem. 1964;239:2379–2385. [PubMed] [Google Scholar]
- Rix U, Fischer C, Remsing LL, Rohr J. Modification of post-PKS tailoring steps through combinatorial biosynthesis. Nat. Prod. Rep. 2002;19:542–580. doi: 10.1039/b103920m. [DOI] [PubMed] [Google Scholar]
- Rodriguez AM, Olano C, Mendez C, Hutchinson CR, Salas JA. A cytochrome P450-like gene possibly involved in oleandomycin biosynthesis by Streptomyces antibioticus. FEMS Microbiol. Lett. 1995;127:117–120. doi: 10.1111/j.1574-6968.1995.tb07459.x. [DOI] [PubMed] [Google Scholar]
- Satoi I, Muto N, Hayashi M, Fujii T, Otani M. Mycinamicins, new macrolideantibiotics. I. Taxonomy, production, isolation, characterization and properties. J. Antibiot. 1980;33:364–376. doi: 10.7164/antibiotics.33.364. [DOI] [PubMed] [Google Scholar]
- Sherman DH, Li S, Yermalitskaya LV, Kim Y, Smith JA, Waterman MR, Podust LM. The structural basis for substrate anchoring, active site selectivity, and product formation by P450 PikC from Streptomyces venezuelae. J. Biol. Chem. 2006;281:26289–26297. doi: 10.1074/jbc.M605478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stassi D, Donadio S, Staver MJ, Katz L. Identification of a Saccharopolyspora erythraea gene required for the final hydroxylation step in erythromycin biosynthesis. J. Bacteriol. 1993;175:182–189. doi: 10.1128/jb.175.1.182-189.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Takenaka S, Kinoshita K, Morohoshi T. Biosynthesis of mycinamicins by a blocked mutant of Micromonospora griseorubida. 1990;43:1508–1511. doi: 10.7164/antibiotics.43.1508. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Maeta H, Matsumoto T, Tsuchihashi G. New glycosidation reaction. 2. Preparation of 1-fluoro-D-desosamine derivative and its efficient glycosidation by the use of Cp2HfCl2-AgClO4 as the activator. Tetrahedron Lett. 1988;29:3571–3574. [Google Scholar]
- Toshima K, Nozaki Y, Mukaiyama S, Tamai T, Nakata M, Tatsuta K, Kinoshita M. Application of highly stereocontrolled glycosidations employing 2,6-anhydro-2-thio sugars to the syntheses of erythromycin A and olivomycin A trisaccharide. J. Am. Chem. Soc. 1995;117:3717–3727. [Google Scholar]
- Ward SL, Hu Z, Schirmer A, Reid R, Revill WP, Reeves CD, Petrakovsky OV, Dong SD, Katz L. Chalcomycin biosynthesis gene cluster from Streptomyces bikiniensis: Novel features of an unusual ketolide produced through expression of the chm polyketide synthase in Streptomyces fradiae. Antimicrob. Agents Chemother. 2004;48:4703–4712. doi: 10.1128/AAC.48.12.4703-4712.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward RB, Logusch E, Nambiar KP, Sakan K, Ward DE, Au-Yeung BW, Balaram P, Browne LJ, Card PJ, Chen CH. Asymmetric total synthesis of erythromcin. 1. Synthesis of an erythronolide A secoacid derivative via asymmetric induction. J. Am. Chem. Soc. 1981a;103:3210–3213. [Google Scholar]
- Woodward RB, Logusch E, Nambiar KP, Sakan K, Ward DE, Au-Yeung BW, Balaram P, Browne LJ, Card PJ, Chen CH. Asymmetric total synthesis of erythromycin. 2. Synthesis of an erythronolide A lactone system. J. Am. Chem. Soc. 1981b;103:3213–3215. [Google Scholar]
- Woodward RB, Logusch E, Nambiar KP, Sakan K, Ward DE, Au-Yeung BW, Balaram P, Browne LJ, Card PJ, Chen CH. Asymmetric total synthesis of erythromycin. 3. Total synthesis of erythromycin. J. Am. Chem. Soc. 1981c;103:3215–3217. [Google Scholar]
- Xue Y, Wilson D, Zhao L, Liu H-w, Sherman DH. Hydroxylation of macrolactones YC-17 and narbomycin is mediated by the pikC-encoded cytochrome P450 in Streptomyces venezuelae. Chem. Biol. 1998;5:661–667. doi: 10.1016/s1074-5521(98)90293-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental data include protein purification SDS-PAGE gel results, LC-MS analysis of selected P450 reactions, P450 CO-bound reduced spectra, substrate binding affinity measurements, steady-state kinetic curves, and experimental details for the synthesis of M-VIII.