Abstract
Arenaviruses are enveloped single-strand RNA viruses that mostly have natural hosts in rodents. Upon infection of humans, several arenaviruses can cause severe hemorrhagic fever diseases, including Lassa fever that is endemic in West Africa. The virulence mechanism of these deadly arenaviruses can be studied in a safe and economical small animal model - guinea pigs infected by a non-pathogenic arenavirus Pichinde virus (PICV), a virulent strain of which can cause similar disease syndromes in guinea pigs as arenaviral hemorrhagic fevers in humans. We have recently developed molecular clones for both the virulent and avirulent strains of PICV. Using the available reverse genetics tools, we are characterizing the molecular determinants of virulent arenavirus infections in vivo.
Keywords: viral hemorrhagic fever, arenavirus, reverse genetics system, animal model, virulence factors, Lassa fever, Pichinde virus, guinea pig
Pathogenic and Non-pathogenic Arenaviruses
Arenaviruses are geographically and phylogenetically divided into two serogroups: the Old World arenaviruses such as Lassa fever virus (LASV) and Lymphocytic Choriomeningitis Virus (LCMV), and the New World arenaviruses such as Pichinde virus (PICV) and Junin virus (JUNV) 1. The natural host reservoir species for these and most other arenaviruses are rodents, except for the Tacaribe virus that has been isolated from bats 1.
Several arenaviruses, once transmitted to humans, can cause deadly hemorrhagic fever diseases (VHFs). These VHFs have similar clinical manifestations. After a long incubation period of 6–21 days, the disease usually starts with fever, general weakness and malaise, which is followed by headache, sore throat, nausea, vomiting, diarrhea, and abdominal pain. In most severe cases, mucosal bleeding (hemorrhaging) normally accompanies shock, seizures, and coma, culminating in death (Reviewed in 1, 2).
Lassa fever, caused by infection of the Old World arenavirus Lassa fever virus (LASV), is endemic in West Africa with estimated 300,000 to 500,000 infections and 5,000 deaths annually 3. Human Lassa fever infection can exhibit a variety of clinical manifestations ranging from asymptomatic to multi-organ system failure and death 4. The case fatality of LASV infection is 1% overall but can be up to 15% in hospitalized patients. A novel Old World arenavirus, Lujo virus (LUJV), was identified to be the cause of a few VHF cases in Zambia and South Africa in late 2008 (http://www.cdc.gov/ncidod/dvrd/spb/outbreaks/). In South America, several New World arenaviruses, such as Junin (JUNV), Machupo (MACV), Guanarito (GTOV), Sabia (SABV), and a recently identified Chapare virus (CHPV) 5, are known to cause VHFs, which occur sporadically. It is likely that, with improvements of technology to isolate and identify novel pathogens, more pathogenic arenaviruses will be discovered in the future.
Currently, there are no effective vaccines, with the exception of Junin virus, and limited treatment options for these arenavirus VHFs. Therefore, these highly pathogenic arenaviruses are classified by the Center for Disease Control and Prevention (CDC) in the Category A Pathogen List and can only be handled in the Biosafety Level-4 (BSL-4) facilities.
Due to the highly virulent nature of these viruses, the lack of proper biomedical infrastructures in the endemic areas and the cultural taboos to manipulate corpses, the pathogenesis of arenavirus-induced VHFs is poorly understood. The current knowledge of Lassa fever is based on limited data available from human infections 6 and studies of animal models for VHFs 7–11. A distinct feature of arenavirus VHF is that viremia level is closely associated with disease outcome and can accurately predict mortality 12. Postmortem studies have found high titers of LASV in multiple organs in the body, such as liver, lung, spleen, kidney, heart, placenta, and the mammary gland 6. In contrast, LASV is cleared rapidly in survivors, which is largely due to virus-specific cell-mediated immunity rather than humoral responses to the infection 9, 12, 13. Despite the presence of high virus titers in a wide range of tissues and organs, the pathological lesions in each of the organs are generally not severe enough to explain the cause of death 6.
Arenavirus genome and genes
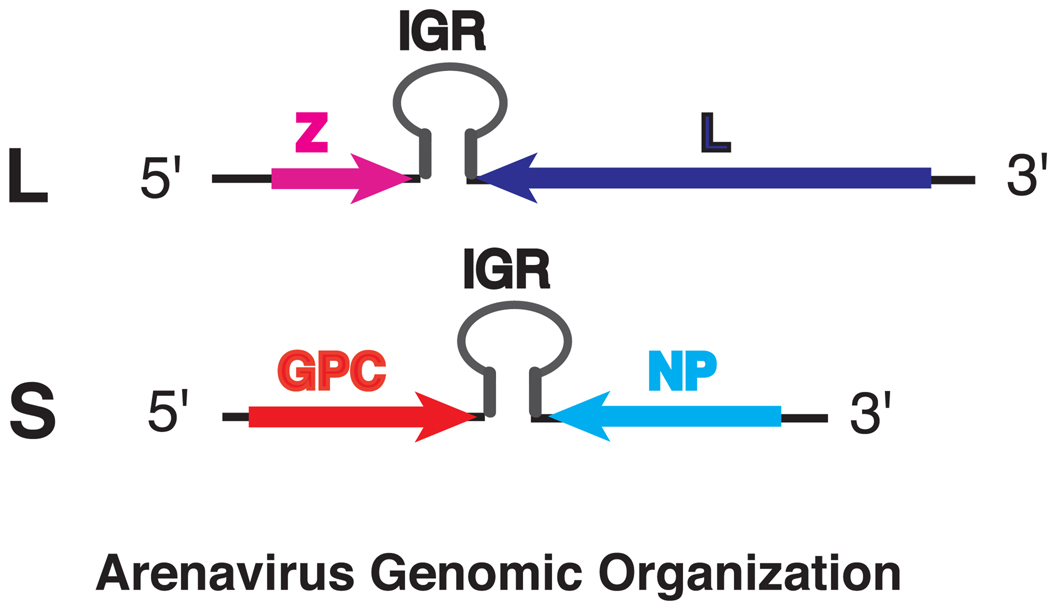
Arenaviruses are enveloped RNA viruses with single-stranded ambi-sense RNA genome that consists of a ~7.2-kb large (L) segment and a ~3.4-kb small (S) segment 1. A total of four open-reading frames (ORFs) are encoded in “ambisense” orientation 1. The L RNA segment encodes the viral polymerase L gene in negative orientation and a small multifunctional Z protein in positive orientation (Fig. 1). The S RNA segment encodes the nucleoprotein NP in negative orientation and the envelope glycoprotein precursor GPC in positive orientation (Fig. 1).
Fig. 1.
Schematic illustration of arenavirus genomic organization. The L segment encodes the Z gene in positive sense and L in negative sense. The S segment encodes the GPC gene in positive sense and NP in negative sense. Between the two ORFs on each segment is the intergenic region (IGR).
The terminal 19 nucleotides from the 5’ and 3’ ends are almost identical between the S and L segments, conserved among all arenaviruses, and are imperfectly complementary to each other, which are predicted to form the panhandle structures that serve as the cis-acting elements required for viral RNA transcription and replication 1. A unique feature among the arenavirus genomic RNAs is the noncoding intergenic regions (IGRs) located between the two ORFs on each segment (Fig. 1) 1. The IGRs, ranging from 59 to 217 nts in length, are predicted to form one to three energetically stable stem-loop structures in both the genomic and antigenomic RNAs 1. The IGRs are proposed to function in transcriptional termination as well as virus assembly and/or budding 14.
The viral L polymerase is a large ~200-kDa protein that functions as an RNA-dependent RNA polymerase. Sequence alignments of all known arenaviral L proteins have revealed multiple conserved domains across this protein 15. In the central region, there are four conserved motifs (A, B, C, and D) identified in all RNA-dependent RNA polymerases (RdRp), which constitute the “polymerase module” implicated in template recognition and polymerizing activity 16.
The nucleoprotein NP of ~65-kDa is the major structural protein and associates with virion RNA. Both the L and NP proteins are required for viral transcription and replication 17. Recently, a novel function of the NP protein in down-regulating interferon (IFN)- β expression has been revealed 18, 19, suggesting that the NP protein, in addition to its function in RNA synthesis, may play an important role in blunting the natural antiviral response of the infected hosts.
Z protein is a small (~11-kDa) multifunctional protein that is involved in virus structure, RNA replication 20–22, viral particle assembly 23, and possibly other unknown functions. The central RING-finger domain of Z proteins is highly conserved across the arenaviruses and is required for inhibiting viral RNA synthesis 22. An N-terminal hydrophobic domain includes a myristylation site required for membrane association, and the C-terminal part contains the Late or L motifs required for virus budding 23, 24.
The viral glycoprotein (GPC) is expressed as a single polypeptide with a long conserved 58-aa stable signal peptide (SSP) and is post-translationally processed by the S1P cellular protease into GP1 and GP2 25, 26. The unique SSP is an essential component of the envelope glycoprotein complex and plays important roles in not only regulating the intracellular trafficking and proteolytic maturation of the GPC complex, but also is involved in the pH-induced membrane fusion 27–29. GP1 is a peripheral membrane protein that participates in receptor binding and GP2 is a transmembrane protein and involved in fusion activity 1.
Pichinde virus infection of guinea pig as a small animal model for arenavirus hemorrhagic fevers
Pichinde virus (PICV) is a nonpathogenic New World arenavirus (a BSL-2 agent), which does not cause disease in humans. The low-passaged PICV causes a limited febrile illness in guinea pigs that eventually clear the infection. Serial passages in inbred guinea pigs dramatically increases the pathogenicity of the virus 10. Upon infection of the adapted viruses, guinea pigs developed severe disease characterized by fever, severe weight loss, terminal vascular collapse, and death 10, 30, 31.
The illness in PICV-infected guinea pigs mimics human Lassa fever infection in many aspects. (a) Both induce a fulminating disease course with a terminal vascular leak syndrome in which hemorrhage is not a major component 6, 10, 32. (b) Viral distribution is identical in the infected hosts 6, 10. (c) The viremia level is closely associated with the disease outcome 11, 12. (d) The histopathological findings generally cannot explain the cause of death 6, 10, 30, 33, 34. (e) General immune suppression is evident in both diseases 35, 36.
Genomic sequence variations between avirulent and virulent Pichinde viruses
The low-passaged PICV strain (i.e. P2, passaged twice in the spleen of guinea pigs) causes a limited febrile illness in guinea pigs that eventually clear the infection, whereas the high-passaged strain (i.e. P18, passaged 18 times in spleen) causes a disease in animals that is similar to VHF in humans 10, 37. The two strains are closely related yet induce opposing disease outcomes in guinea pigs, providing an ideal model to identify the virus factors that are responsible for virulence in infected hosts.
Aronson and colleagues have compared the S segment sequences of P2 and P18 viruses and revealed 3 nonsynonymous amino acid changes (S119T, K140E, and I164V) in the GP1 subunit of the viral glycoprotein and 2 changes (R374K and A35T) in the NP protein 37. We have recently provided the full-length genomic sequences of both the S and L segments for P2 and P18 viruses 15. A total of 48 nucleotide changes were identified, all of which are localized to the protein coding regions. Most of the sequence changes produce synonymous mutations. In comparison to published works 15, 37, we confirmed the 3 missense mutations in the GP1 protein and the R374K mutation (but not the A35T change) in the NP protein. Within the L segment, we have identified 5 missense mutations (N355D, A1808T, V1839L, D1889N, and D1906N) in the L polymerase protein 15.
The 3 missense mutations in the GP1 subunit of the viral glycoprotein are of great interest, as the GP1 protein binds to host receptor. So far two arenavirus receptors have been identified. Old World arenaviruses (LASV and LCMV) and New World Clade C arenaviruses utilize α-dystroglycan (α-DG) as a major entry receptor 38, 39, whereas pathogenic New World arenaviruses (Clade B) use transferrin receptor 1 (TfR1) as an entry receptor 40. The exact host receptor for PICV, a New World Clade A arenavirus, is unknown. Several studies have shown that only pathogenic Clade B arenaviruses use human TfR1 as a receptor while non-pathogenic Clade B arenaviruses use a human TfR1-independent pathway for entry 41, 42. This suggests that cell entry and GPC-receptor interactions may represent a major virulence mechanism for arenaviruses. It remains to be determined whether the differences between the P2 and P18 GPC proteins are major virulence determinants of PICV infection in guinea pigs.
The NP protein is indispensable for viral RNA synthesis and packaging 17, and may also play an important role in evading host innate immune response by suppressing the IFN-β induction 18, 19. We did not observe a significant difference between the P2 and P18 NP proteins in suppressing Sendai virus-induced IFN-β expression 15, suggesting that the conservative amino acid change (R374K) observed between the virus strains may not significantly affect this function of NP. Nevertheless, we cannot rule out the possibility that the P18 NP protein may function via a different mechanism to evade IFN-induced antiviral activity.
Four out of the five mutations between the P2 and P18 L polymerase proteins are localized to a small 99-residue region (1808–1906) at the C terminus, suggesting that this region may play an important role in viral RNA synthesis.
Reverse genetics systems for Pichinde virus
An invaluable tool to identify the viral virulence factors is a molecular clone for the virus, which allows the generation of infectious recombinant viruses from plasmid DNAs. The development of molecular clones for the prototypic arenavirus LCMV 43, 44 has already allowed an opportunity to study the biological functions of the LCMV proteins and RNA elements in viral replication by introducing the desired mutations in the viral genome. We have recently developed the reverse genetics systems for both the P2 and P18 strains of PICV 45.
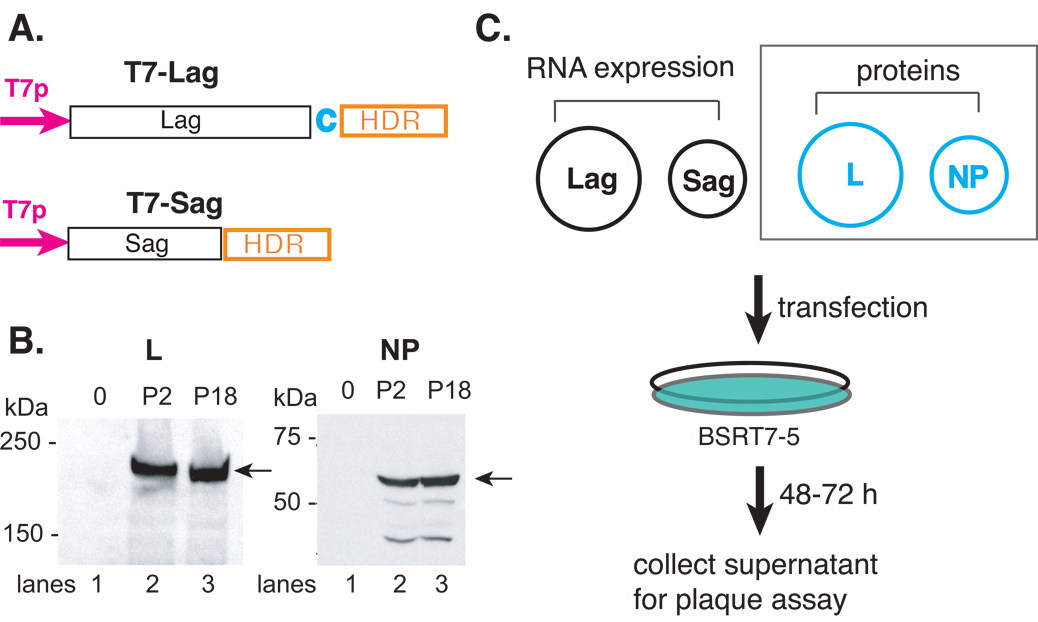
As illustrated in Fig. 2A, we have subcloned the full-length L and S segments, in antigenomic orientation, to immediately follow the T7 promoter sequence. The hepatitis delta ribozyme (HDR) sequence was engineered immediately downstream of the 3’ end of the primary viral RNA transcript to mediate a self-cleavage reaction in order to generate the authentic viral 3’ ends. We have also cloned the L and NP genes into the pCAGGS protein expression vector, to be expressed from the strong viral (CMV) promoter. Expression of the L (~ 200 kDa) and NP (~ 60 kDa) proteins are readily detected using antibodies specific for these proteins (Fig. 2B).
Fig. 2.
Schematic illustration of the reverse genetics systems to generate infectious rP2 and rP18 viruses from plasmid DNA transfection. (A) Schematic illustration of the RNA expression vectors of the full-length antigenomic strands of the S (pT7-Sag) and L (pT7-LagC) segments. The viral RNA segment was cloned in the antigenomic orientation immediately downstream of the T7 promoter (T7p) and upstream of the hepatitis delta ribozyme (HDR) and T7 terminator (T7t) sequences. A nonviral cytosine (in red) was added after the viral L sequence in order to enhance the self-cleavage activity. (B) The expression of the L and NP proteins. The 293T cells were transfected with an empty vector (lane 1) or vector encoding the L (left panel) or NP (right panel) gene of either the P2 (lane 2) or P18 (lane 3) virus. Western blot analyses was conducted using rabbit sera raised against specific short peptides within the L or NP protein. Peptide sequences are available upon request. (C) Schematic illustration of the reverse genetics systems. Two RNA expression plasmids (pT7-Sag and pT7-LagC) and two protein-expression plasmids of the NP and L proteins were used to transfect BSRT7-5 cells, which constitutively express the T7 polymerase protein. Between 48 and 72 hpi, supernatants were collected and plaque assay conducted on Vero cells. The two protein-expression plasmids (NP and L) (shown in dashed square) can be omitted from the reconstitution without compromising the amount of virus production.
To generate recombinant viruses, we transfected BSRT7-5 cells, a stable cell line that constitutively expresses the T7 polymerase, with plasmids encoding the full-length L and S segment RNAs (Lag and Sag) and two other plasmids expressing the L and NP proteins. Infectious viruses were recovered from the supernatants after transfection and quantified by plaque assay on Vero cells (Fig. 2C).
It has been shown previously that the protein expression vectors of L and NP (Fig. 2A) are not necessarily required to generate infectious viruses if the viral RNA segments are expressed as anti-genomic RNA strands 46, 47. This appears to be the case for our PICV reverse genetics systems too. As shown in Table 1, transfection of only the Sag and Lag RNA expression vectors was sufficient to generate infectious rP2 and rP18 viruses at similarly high levels as that of both the RNA and protein expression plasmids, suggesting that the direct translation of the L and NP proteins from the primary RNA transcripts of antigenomic strand is sufficient to support the subsequent viral RNA transcription and replication. Therefore, we used the two-plasmid system that contains only the Lag and Sag plasmids to generate infectious viruses in subsequent experiments. In addition, increasing the amount of the RNA expression vectors from 2 to 4 µg each was shown to significantly increase the production of the recombinant viruses (Table 1). This is particularly true for the rP2 reverse genetics system, in which the recombinant virus titer increased by as many as 500 folds at 72 h post-transfection. The increase was less significant in the rP18 reverse genetics system, which varies from 2 to 6 folds at different time points. The reason for this variation is still unknown.
Table 1.
Titers of recombinant viruses at different hours post plasmid transfection (hpt) into the BSRT7-5 cells.
| virus | Plasmids | Plasmid quantity |
48 hpt | 72 hpt | 96 hpt | 120 hpt |
|---|---|---|---|---|---|---|
| rP2 | Sag + Lag + L + NP |
2 µg each | <20 | <200 | 2.8E4 | 4.0E5 |
| Sag + Lag | 2 µg each | 20 | <200 | 4.0E4 | 5.4E5 | |
| Sag + Lag | 4 µg each | 3.0E3 | 1.0E5 | 3.6E6 | 1.8E7 | |
| rP18 | Sag + Lag + L + NP |
2 µg each | 2.8E2 | 1.0E3 | 1.4E6 | 3.5E7 |
| Sag + Lag | 2 µg each | 4.0E2 | 4.4E4 | 1.4E6 | 5.1E7 | |
| Sag + Lag | 4 µg each | 2.2E3 | 8.0E4 | 5.8E6 | 7.1E7 |
We compared the growth kinetics between the recombinant and parental viruses in cell cultures. The recombinant viruses adopted similar growth kinetics as their parental viruses, with the virulent viruses (P18 and rP18) grow faster by 0.5–1 log than the avirulent ones (P2 and rP2). Similar observations were made in different cell types including Vero, guinea pig cell lines, and primary peritoneal macrophages isolated from guinea pigs 45.
To examine the degrees of virulence caused by PICV infection in vivo, we used outbred Hartley guinea pigs, because the distinct disease phenotypes in these animals infected with the avirulent P2 and virulent P18 viruses 37 were found to be similar to those in inbred strain 13 guinea pigs 10, which are not commercially available. Our data showed that these recombinant viruses could reproduce the virulence phenotypes in vivo as the parental viruses, in which the virulent viruses (P18 and rP18) led to higher mortality rates, shortened survival times, earlier onset of fever, longer duration of fevers, and more weight loss than the avirulent viruses (P2 and rP2) 45.
Identification of molecular determinants of virulent Pichinde virus infection in vivo
Using the reverse genetics systems, we generated recombinant viruses with segment reassortants, i.e., by swapping the L and S segments between the P2 and P18 viruses. The resulted virus reassortants, rS2L18 and rS18L2, were tested for replication in vitro, and compared with the control viruses rP2 (aka rS2L2) and rP18 (aka rS18L18). Growth curve analysis of all recombinant viruses was conducted in Vero cells at moi = 0.01. As shown in Fig. 3A, the rS2L18 virus showed similar growth kinetic as the rP18 virus and rS18L2 was similar to rP2, the former group growing to higher titers than the latter one by 0.5–1 log. These data indicate that the P18 L segment accounts for increased virus replication in vitro.
Fig. 3.
Comparison of growth kinetics in vitro and the virulence in vivo of the different PICV strains. (A) Comparison of the growth kinetics between the parental viruses (P2 and P18), the recombinant viruses (rP2 and rP18), and the recombinant reassortant viruses (rS2L18 and rS18L2) in Vero cells. (B) The daily rectal temperature of animals injected with the different PICV strains or PBS as a control. (C) The average duration of fever (temp > 39.5 °C). (D) The daily body weight as percentage of the original body weight at day 0.
When these reassortant viruses were used to infect guinea pigs, no animals died from the infection as compared to 100% mortality rate in those infected with rP18 virus (Table 2), suggesting that the reassortant viruses exhibited an attenuated phenotype in vivo.
Table 2.
Mortality associated with terminal points in guinea pigs infected by the different PICV strains *
| PBS | P2 | P18 | rP2 | rP18 | rS18L2 | rS2L18 | |
|---|---|---|---|---|---|---|---|
| Mortality | 0/3 | 0/6 | 5/6 | 0/6 | 6/6 | 0/6 | 0/6 |
the number of animals reached terminal points (25% weight loss and rectal termperature < 39.8 °C) within the course of 18-day infection.
Both reassortant viruses, however, caused much higher and longer duration of fever in the infected animals than the rP2 virus at days 6 – 10 (Fig. 3Band 3C). On average, the reassortant viruses caused 4 – 5 days of fever, which was longer than that in the rP2-infected animals but shorter than that in the rP18-infected ones (Fig. 3C). Consistently, the reassortants led to a slight body-weight loss, more than what rP2- but much less than rP18 caused in animals from days 9 to 11, after which the infected animals appeared to recover from the infection (Fig. 3D). Taken together, these data suggest that the P18 L and P18 S segments each contributes to virulence in vivo and that both segments are required to generate the complete virulence phenotype. Similar conclusions have been reached previously in a study, where reassortant viruses were selected by a traditional method of virus co-infection in cell culture 48.
As noted in previous studies, the viremia level is closely associated with the degree of virulence in infected humans or animals 9, 12, 13. In order to determine whether this was also the case in PICV infection, we determined the virus titers in blood drawn from animals infected by different PICV strains every 3 days after infection until day 18. As shown in Table 3, the viremia level in the infected hosts could accurately predict the disease outcomes. For the group of animals that were lethally infected with the virulent virus P18 or rP18, viremia levels were readily detected (up to 106 pfu/ml virus titers at terminal points), whereas it was below the threshold of detection (< 200 pfu/ml) in animals infected with the avirulent (P2 or rP2) or with the reassortant virus (rS2L18 or rS18L2), all of which led to non-lethal infections. Our observations once again reconfirm the close association between viremia level and the disease severity as an important feature of pathogenic arenavirus infection.
Table 3.
Viremia level (PFU/ml) at different days post infection by different virus strains1.
| Time points (dpi) |
P2 | P18 | rP2 | rP18 | rS18L2 | rS2L18 |
|---|---|---|---|---|---|---|
| 3 | < 200 | < 200 | < 200 | < 200 | < 200 | < 200 |
| 6 | < 200 | 590 | < 200 | 200 | < 200 | < 200 |
| 9 | < 200 | 3.3E3 | < 200 | 1.4E4 | < 200 | < 200 |
| 12 | < 200 | 1.8E5 | < 200 | 4.0E5 | < 200 | 200 |
| 15 | < 200 | 1.8E6 (TP)2 | < 200 | 2.2E6 (TP)2 | < 200 | < 200 |
| 18 | < 200 | < 200 | < 200 | < 200 |
Blood was drawn from saphenous vein at different times post-infection. The results were the average from at least three animals.
The P18 and rP18-infected animals reached terminal points (>25% weight loss and rectal temperature lower than 39.8 °C) at various time points ranging from days 11 to 18 and were subsequently euthanized. The average viremia levels from 5 of the P18-infected and 6 of the rP18-infected animals at their terminal points (TP) are shown.
Summary
The development of the reverse genetics systems for both the avirulent and virulent strains of PICV allows for characterization of the molecular determinants of virulent PICV infection in guinea pigs, the gained knowledge has the potential to shed important lights on arenavirus-caused hemorrhagic fevers in humans. A detailed characterization of reassortant viruses generated from these novel reverse genetics systems has suggested that the molecular determinants on both the L and S segments are required to cause mortality in animals. Our data suggest that unique sequences located in the L polymerase coding sequence on the L segment of the virulent strain of PICV are responsible for the increased virus replication capacity in vitro and enhanced virulence in vivo. For the virulence factors on the S segment, it remains to be determined whether GPC, NP or both are required.
Acknowledgements
We thank Dr. Judy Aronson (University of Texas Medical Branch) for providing the stock P2 and P18 viruses, Dr. Karl-Klaus Conzelmann (Ludwig-Maximilians-Universität, Germany) for BSRT7-5 cells, and Drs. Aftab A. Ansari and Tristram G. Parslow (Emory University) for advice and consultation. Lisa McLay Schelde, Naveen Kumar and Jialong Wang are acknowledged for excellent technical assistance. This research was supported in part by the University Research Committee of Emory University, the pilot project component of Dr. Rafi Ahmed’s U 19 grant RFA-AI-02-042, and the new directions award from SERCEB (Southeast Regional Center of Excellence for Emerging Infections and Biodefense) (3U54AI057157-06S10032).
Reference
- 1.Buchmeier MJ, Bowen MD, Peters CJ. Arenaviridae: The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Fields Virology. Vol. 2. Philadelphia, PA: Lippincott-Raven Publisher; 2001. pp. 1635–1668. [Google Scholar]
- 2.McCormick JB, Fisher-Hoch SP. Lassa fever. Curr Top Microbiol Immunol. 2002;262:75–109. doi: 10.1007/978-3-642-56029-3_4. [DOI] [PubMed] [Google Scholar]
- 3.McCormick JB. Epidemiology and control of Lassa fever. Curr Top Microbiol Immunol. 1987;134:69–78. doi: 10.1007/978-3-642-71726-0_3. [DOI] [PubMed] [Google Scholar]
- 4.McCormick JB. Clinical, epidemiologic, and therapeutic aspects of Lassa fever. Med Microbiol Immunol (Berl) 1986;175:153–155. doi: 10.1007/BF02122438. [DOI] [PubMed] [Google Scholar]
- 5.Delgado S, et al. Chapare virus, a newly discovered arenavirus isolated from a fatal hemorrhagic fever case in Bolivia. PLoS Pathog. 2008;4:e1000047. doi: 10.1371/journal.ppat.1000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker DH, et al. Pathologic and virologic study of fatal Lassa fever in man. Am J Pathol. 1982;107:349–356. [PMC free article] [PubMed] [Google Scholar]
- 7.Jahrling PB, et al. Lassa virus infection of rhesus monkeys: pathogenesis and treatment with ribavirin. J Infect Dis. 1980;141:580–589. doi: 10.1093/infdis/141.5.580. [DOI] [PubMed] [Google Scholar]
- 8.Carrion R, Jr, et al. Lassa Virus Infection in Experimentally Infected Marmosets: Liver Pathology and Immunophenotypic Alterations in Target Tissues. J Virol. 2007 doi: 10.1128/JVI.02876-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters CJ, et al. Experimental studies of arenaviral hemorrhagic fevers. Curr Top Microbiol Immunol. 1987;134:5–68. doi: 10.1007/978-3-642-71726-0_2. [DOI] [PubMed] [Google Scholar]
- 10.Jahrling PB, et al. Pathogenesis of a pichinde virus strain adapted to produce lethal infections in guinea pigs. Infect Immun. 1981;32:872–880. doi: 10.1128/iai.32.2.872-880.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aronson JF, Herzog NK, Jerrells TR. Pathological and virological features of arenavirus disease in guinea pigs. Comparison of two Pichinde virus strains. Am J Pathol. 1994;145:228–235. [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson KM, et al. Clinical virology of Lassa fever in hospitalized patients. J Infect Dis. 1987;155:456–464. doi: 10.1093/infdis/155.3.456. [DOI] [PubMed] [Google Scholar]
- 13.Bausch DG, et al. Diagnosis and clinical virology of Lassa fever as evaluated by enzyme-linked immunosorbent assay, indirect fluorescent-antibody test, and virus isolation. J Clin Microbiol. 2000;38:2670–2677. doi: 10.1128/jcm.38.7.2670-2677.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinschewer DD, Perez M, de la Torre JC. Dual role of the lymphocytic choriomeningitis virus intergenic region in transcription termination and virus propagation. J Virol. 2005;79:4519–4526. doi: 10.1128/JVI.79.7.4519-4526.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lan S, et al. Genome comparison of virulent and avirulent strains of the Pichinde arenavirus. Arch Virol. 2008;153:1241–1250. doi: 10.1007/s00705-008-0101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poch O, et al. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. Embo J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinschewer DD, Perez M, de la Torre JC. Role of the virus nucleoprotein in the regulation of lymphocytic choriomeningitis virus transcription and RNA replication. J Virol. 2003;77:3882–3887. doi: 10.1128/JVI.77.6.3882-3887.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Sobrido L, et al. Inhibition of the type I interferon response by the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol. 2006;80:9192–9199. doi: 10.1128/JVI.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Sobrido L, et al. Differential inhibition of type I interferon induction by arenavirus nucleoproteins. J Virol. 2007;81:12696–12703. doi: 10.1128/JVI.00882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornu TI, de la Torre JC. RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J Virol. 2001;75:9415–9426. doi: 10.1128/JVI.75.19.9415-9426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez N, Jacamo R, Franze-Fernandez MT. Transcription and RNA replication of tacaribe virus genome and antigenome analogs require N and L proteins: Z protein is an inhibitor of these processes. J Virol. 2001;75:12241–12251. doi: 10.1128/JVI.75.24.12241-12251.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacamo R, et al. Tacaribe virus Z protein interacts with the L polymerase protein to inhibit viral RNA synthesis. J Virol. 2003;77:10383–10393. doi: 10.1128/JVI.77.19.10383-10393.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez M, Craven RC, de la Torre JC. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc Natl Acad Sci U S A. 2003;100:12978–12983. doi: 10.1073/pnas.2133782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strecker T, et al. Lassa virus Z protein is a matrix protein and sufficient for the release of virus-like particles [corrected] J Virol. 2003;77:10700–10705. doi: 10.1128/JVI.77.19.10700-10705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beyer WR, et al. Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. J Virol. 2003;77:2866–2872. doi: 10.1128/JVI.77.5.2866-2872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenz O, et al. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc Natl Acad Sci U S A. 2001;98:12701–12705. doi: 10.1073/pnas.221447598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agnihothram SS, York J, Nunberg JH. Role of the stable signal peptide and cytoplasmic domain of G2 in regulating intracellular transport of the Junin virus envelope glycoprotein complex. J Virol. 2006;80:5189–5198. doi: 10.1128/JVI.00208-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.York J, Nunberg JH. Role of the stable signal peptide of Junin arenavirus envelope glycoprotein in pH-dependent membrane fusion. J Virol. 2006;80:7775–7780. doi: 10.1128/JVI.00642-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.York J, Nunberg JH. Intersubunit interactions modulate pH-induced activation of membrane fusion by the Junin virus envelope glycoprotein GPC. J Virol. 2009 doi: 10.1128/JVI.02410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lucia HL, et al. The effect of an arenavirus infection on liver morphology and function. Am J Trop Med Hyg. 1990;43:93–98. doi: 10.4269/ajtmh.1990.43.93. [DOI] [PubMed] [Google Scholar]
- 31.Qian C, et al. Cardiovascular and pulmonary responses to Pichinde virus infection in strain 13 guinea pigs. Lab Anim Sci. 1994;44:600–607. [PubMed] [Google Scholar]
- 32.Katz MA, Starr JF. Pichinde virus infection in strain 13 guniea pigs reduces intestinal protein reflection coefficient with compensation. J Infect Dis. 1990;162:1304–1308. doi: 10.1093/infdis/162.6.1304. [DOI] [PubMed] [Google Scholar]
- 33.Connolly BM, et al. Pathogenesis of Pichinde virus infection in strain 13 guinea pigs: an immunocytochemical, virologic, and clinical chemistry study. Am J Trop Med Hyg. 1993;49:10–24. doi: 10.4269/ajtmh.1993.49.10. [DOI] [PubMed] [Google Scholar]
- 34.Peters CJ, et al. Pathogenesis of viral hemorrhagic fevers: Rift Valley fever and Lassa fever contrasted. Rev Infect Dis. 1989;11 Suppl 4:S743–S749. doi: 10.1093/clinids/11.supplement_4.s743. [DOI] [PubMed] [Google Scholar]
- 35.Fennewald SM, et al. Alterations in NF-kappaB and RBP-Jkappa by arenavirus infection of macrophages in vitro and in vivo. J Virol. 2002;76:1154–1162. doi: 10.1128/JVI.76.3.1154-1162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aronson JF, Herzog NK, Jerrells TR. Tumor necrosis factor and the pathogenesis of Pichinde virus infection in guinea pigs. Am J Trop Med Hyg. 1995;52:262–269. doi: 10.4269/ajtmh.52-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, Marriott K, Aronson JF. Sequence analysis of the small RNA segment of guinea pig-passaged Pichinde virus variants. Am J Trop Med Hyg. 1999;61:220–225. doi: 10.4269/ajtmh.1999.61.220. [DOI] [PubMed] [Google Scholar]
- 38.Spiropoulou CF, et al. New World arenavirus clade C, but not clade A and B viruses, utilizes alpha-dystroglycan as its major receptor. J Virol. 2002;76:5140–5146. doi: 10.1128/JVI.76.10.5140-5146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao W, et al. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science. 1998;282:2079–2081. doi: 10.1126/science.282.5396.2079. [DOI] [PubMed] [Google Scholar]
- 40.Radoshitzky SR, et al. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature. 2007;446:92–96. doi: 10.1038/nature05539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flanagan ML, et al. New world clade B arenaviruses can use transferrin receptor 1 (TfR1)-dependent and -independent entry pathways, and glycoproteins from human pathogenic strains are associated with the use of TfR1. J Virol. 2008;82:938–948. doi: 10.1128/JVI.01397-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radoshitzky SR, et al. Receptor determinants of zoonotic transmission of New World hemorrhagic fever arenaviruses. Proc Natl Acad Sci U S A. 2008;105:2664–2669. doi: 10.1073/pnas.0709254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez AB, de la Torre JC. Rescue of the prototypic Arenavirus LCMV entirely from plasmid. Virology. 2006;350:370–380. doi: 10.1016/j.virol.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 44.Flatz L, et al. Recovery of an arenavirus entirely from RNA polymerase I/II-driven cDNA. Proc Natl Acad Sci U S A. 2006;103:4663–4668. doi: 10.1073/pnas.0600652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lan S, et al. Development of infectious clones for virulent and avirulent Pichinde viruses – a model virus to study arenavirus-induced hemorrhagic fevers. J Virolin revision. 2009 doi: 10.1128/JVI.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lowen AC, et al. Efficient bunyavirus rescue from cloned cDNA. Virology. 2004;330:493–500. doi: 10.1016/j.virol.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 47.Blakqori G, Weber F. Efficient cDNA-based rescue of La Crosse bunyaviruses expressing or lacking the nonstructural protein NSs. J Virol. 2005;79:10420–10428. doi: 10.1128/JVI.79.16.10420-10428.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L, et al. Reassortant analysis of guinea pig virulence of pichinde virus variants. Virology. 2001;290:30–38. doi: 10.1006/viro.2001.1127. [DOI] [PubMed] [Google Scholar]