Abstract
There is increasing evidence that sequence-specific formation of 3-nitrotyrosine (3-NT) may cause functional changes in target proteins. Recently, the nitration of Tyr residues in glycogen phosphorylase b (Ph-b) was implicated in the age-associated decline of protein function (Sharov et al., Exp. Gerontol. 41, 407–416; 2006); in another report, the nitration of one specific residue, Tyr613, located in the allosteric inhibition site was hypothesized as a rationale for peroxynitrite inactivation (Dairou et al., J. Mol. Biol. 372, 1009–1021; 2007). In the present study, we have optimized the analysis of in-gel Ph-b digests by high performance liquid chromatography-electro spray ionization-tandem mass spectrometry, in order to achieve a quantitative analysis of nitration of individual Tyr residues at a high coverage of Tyr-containing sequences (92%). Our data do not confirm the role of Tyr613 nitration in the control of enzymatic function. Furthermore, we show here that the enzymatic activity of Ph-b does not directly correlate with the protein nitration levels, and that the modification of Cys and, potentially, other amino acid residues can better rationalize Ph-b inactivation by peroxynitrite.
Keywords: Glycogen phosphorylase b, Enzymatic activity, Tyrosine nitration, Cysteine oxidation, Peroxynitrite, Mass spectrometry, Solvent accessible surface area
There is increasing evidence that reactive nitrogen species nitrate specific tyrosine residues in proteins, and that the formation of 3-nitrotyrosine (3-NT) causes functional changes in targeted proteins [1–4]. Biological nitration of protein tyrosine residues has been demonstrated in various diseases and biological aging [5–13]. Protein nitration appears to be a rather selective process since neither all tyrosine residues in proteins nor all proteins in a given proteome get nitrated both in vivo [6,14,15] and in vitro [15–18]. In addition, modifications of different Tyr residues may not be evenly important for protein function. Several sources of Tyr nitration in vivo have been established involving reactions of peroxynitrite and/or nitrogen dioxide, or nitrite catalyzed by peroxidases [19]. Regardless of the chemical mechanism, tyrosine nitration appears to be a complex process depending on the individual reactivity of Tyr residues in a protein, environment (pH, concentrations of reagents, solvent accessibility and diffusion coefficients), tissue specific protection, protein repair and turnover mechanisms. The knowledge of sequence location and the yields of respective Tyr nitration is therefore very important, both mechanistically and physiologically, to understand the role of specific protein damage and to design potential treatments of ensuing disorders. However, to date most protein nitration targets in vivo have been identified by anti-3-NT antibodies only, and, with a few exceptions [6,15,20–24], a sequence-specific analysis of 3-NT on individual proteins in vivo has not been done. Peptide mass mapping by liquid chromatography-mass spectrometry (LC-MS) has become a key approach in the qualitative and quantitative characterization of protein post-translational modifications. The development of quantitative approaches suitable for virtually complete sequence-specific characterization of “problematic” proteins is, therefore, a necessary step towards understanding a role of protein oxidative post-translational modifications, such as Tyr nitration, in the functional alteration of proteins. Earlier, we found that nanoHPLC-nanoESI-MS/MS analysis of in-gel digests obtained by 1-D SDS-PAGE can be successfully used for the characterization of 3-NT accumulation on even large membrane proteins, such as SERCA [27,28], and this technique is applied here for a comprehensive analysis of phosphorylase b (Ph-b).
Ph-b is a ubiquitous protein, which represents ca. 5% of total soluble protein in muscle tissue. Ph-b (gene name: PYGM) belongs to the glycogen phosphorylase family, which catalyzes the formation of glucose-1-phosphate from the cellular carbohydrate storage form, glycogen:
The activity of Ph-b is controlled both allosterically (through the non-covalent binding of metabolites) and by covalent modification [29–30]. AMP allosterically activates, whereas ATP, ADP, and glucose-6-phosphate allosterically inhibit Ph-b. Phosphorylation of Ser14 (SwissProt and NCBI protein databases refer to this residue as Ser15, which includes the original Met removed in the mature protein) converts Ph-b (a homodimer, T-form) to phosphorylase a (enzymatically more active, tetrameric R-form). High-resolution crystal structures of the protein in different conformational T and R states are now available [31–32] allowing for computational analysis of structure-activity relationships in the protein. Defects in the PYGM gene cause glycogen storage disease type 5 (GSD5), also known as McArdle disease, which is a metabolic disorder resulting in myopathy characterized by exercise intolerance, cramps, muscle weakness and recurrent myoglobinuria. The structure and enzymatic activity of Ph-b are extremely sensitive to the modification of even a single amino acid residue: at least 18 missense mutations of a single amino acid residue of human muscle Ph-b throughout the gene sequence have been identified, which cause functional deficiency of skeletal muscle [33–34]. The Ph-b sequence is highly conserved in mammals sharing 98% homology in the human, mouse and rabbit protein. Generally, Ph-b is an excellent model to study the selectivity of protein tyrosine nitration and represents a good challenge to test new methodology. The enzyme of 97 kDa contains 36 tyrosine residues (out of a total 842 amino acids), which can be differentially mapped to assess the selectivity of tyrosine nitration. The unusually high fraction of protein Tyr residues (4.3%) may be a reason for the susceptibility of the protein to nitration observed in vivo [15].
The aim of the current work was the sequence-specific characterization of Tyr nitration by peroxynitrite in rabbit muscle Ph-b, and a potential correlation to enzymatic activity. The motivation for the current study originated from two findings. First, Ph-b from rat skeletal muscle suffers an age-associated loss of function, which is accompanied (but not necessarily caused) by age-associated, sequence-specific accumulation of 3-NT on the protein [15–24]. Immunochemical evidence for Ph-b nitration in vivo was also reported by Kuo and others [26]. Second, in a recent study [17] the nitration of a critical residue, Tyr613, located in the allosteric inhibitor site of the enzyme, has been suggested as a mechanism of Ph-b inhibition by peroxynitrite, based on ligand binding and MS analysis. However, this important conclusion was apparently made from qualitative MS analysis using MALDI-TOF peptide fingerprinting only, at relatively low sequence coverage (<50%). Furthermore, the experimental conditions in the cited work did not account for potential changes of peroxynitrite reactions in the presence of physiological concentrations of CO2/HCO3− [19].
Therefore, we have optimized the analysis of in-gel Ph-b digests by high performance liquid chromatography-electro spray ionization-tandem mass spectrometry (HPLC-ESIMS/MS), focusing on a quantitative analysis of individual Tyr residue nitration at a high coverage of Tyr-containing sequences (92%). Our data do not confirm a role of Tyr613 nitration in the control of enzymatic function. Furthermore, we show here that the enzymatic activity of Ph-b does not directly correlate with the protein nitration levels, and that the modification of Cys and, potentially, other amino acid residues can better rationalize Ph-b inactivation by peroxynitrite.
Materials and methods
Chemicals
TPCK-treated sequence grade trypsin was from Promega (Madison, WI). Rabbit muscle phosphorylase b, bovine serum albumin (BSA), DTT, SDS, urea, NADP, glycogen from rabbit liver, adenosine-5’-monophosphate, glucose-6-phosphate dehydrogenase and sodium iodoacetate were purchased from Sigma (St. Louis, MO). Phosphoglucomutase was obtained from Boeringer Mannheim (Indianapolis, IN). Pre-cast Novex® tris-glycine-SDS gels, molecular weight standard Mark12, running and sample buffers were from Invitrogen (Carlsbad, CA). All other chemicals of highest commercially available grade were obtained from Fisher (Pittsburgh, PA, USA).
Reaction with peroxynitrite
Peroxynitrite was prepared by the reaction of ozone with cooled aqueous sodium azide as described previously [35], aliquoted and kept at −70°C and pH 12 until use. The concentration of stock peroxynitrite (ca. 50 mM) was determined by absorbance (ε302=1,670 M−1cm−1 [19]), and all dilutions were made in 0.1 % NaOH (pH 12). Ph-b was reconstituted in a buffer containing 1 mM DTPA and either 25 mM Na2HPO4 and 25 mM NaHCO3, or 50 mM Na2HPO4 alone (pH 7.4). A small volume of stock peroxynitrite was quickly added while vortexing to get desired final concentrations of peroxynitrite (bolus addition of peroxynitrite). For control experiments, peroxynitrite was added to the respective buffer 5 min prior to mixing with the protein solution (reverse-order-of-addition experiment). We did not observe any differences in nitration, electrophoretic mobility, and HPLC elution profile between control (not treated with peroxynitrite) and the reverse-order-of-addition samples.
Assay of phosphorylase b activity
Phosphorylase b activity was assayed at room temperature with an assay described earlier [15]. The test mixture contained 0.1 mM EDTA, 3 mM MgCl2, 0.3 mg/ml NADP, 0.2 mg/ml glycogen, 30 µM adenosine-5’-monophosphate, 0.7 U/ml phosphoglucomutase, 3 U/ml glucose-6-phosphate dehydrogenase in 0.05 M potassium phosphate (pH 6.8). After recording the blank rate for 2 min, the reaction was initiated by the addition of phosphorylase b solution, and the absorbance at 340 nm was monitored for 8 min. The activity was determined as ΔA340/min and calibrated through a standard commercial enzyme with known activity.
Gel electrophoresis
For reduction and alkylation of protein Cys residues, samples were incubated for 1 h with 6 M urea and 2 mM DTT at 37°C followed by the addition of 5 mM sodium iodoacetate and incubation for an additional 1 h at room temperature under exclusion of light. Aliquots of Ph-b containing 20 µg protein were mixed with an equal volume of tris-glycine-SDS sample buffer, heated for 2 min in a boiling water bath, and loaded onto 1.5-mm thick 10-well Novex 4–12% Tris-glycine gradient gels (Invitrogen, Carlsbad, CA). After running gel electrophoresis at 200 V for 90 min, the gels were stained with 0.2% Coomassie R250 in 7.5% acetic acid/30% methanol/62.5% H2O for 2 h, followed by destaining in 7.5% acetic acid/ 40% methanol/52.5% H2O until the bands were visible and the background was clear.
In-gel protein digestion
Protein bands were excised from the gel, cut into pieces of about 1 mm3 size, and processed as described elsewhere [27]. In brief, gel slices were washed twice for 45 min at 37°C in 0.5 ml of 200 mM NH4HCO3/50% (v/v) acetonitrile with agitation. For protein alkylation, the gel slices were incubated with 2 mM DTT in 200 mM NH4HCO3 buffer (pH 8.1) for 30 min at 50°C, followed by the reaction with 5 mM iodoacetic acid for 30 min at room temperature. The solvent was discarded; the gel slices were additionally washed for 1 h in 0.5 ml of 200 mM NH4HCO3/50% acetonitrile with agitation, and shrunk in pure acetonitrile for 15 min. After removal of acetonitrile and drying under vacuum, the samples were re-swollen with a buffer containing 40 mM NH4HCO3, 10% acetonitrile and sequencing-grade modified trypsin at a ca. 1:10 molar ratio of trypsin to the protein (i.e., 2 µg trypsin to 20 µg protein in a band). The volume of buffer was ca. 1.5-times that of the excised gel band. After the absorption of trypsin, additional buffer (30–50 µL) was used to cover the gel pieces to keep them wet during overnight digestion (16–18 h) at 37°C. For the extraction of peptides from the gel, octyl-β-glucopyranoside (OBG) was added to a final concentration of 0.1% (w/v), and gels were additionally incubated in a sonication bath for 30 min. The supernatant of the in-gel digest obtained after short centrifugation (ca. 60 µL volume) was collected and mixed with a second extraction from the gel with 60 µL of 60% acetonitrile in 0.2% aqueous trifluoroacetic acid (TFA) containing 0.1% OBG. The resulting peptide solution was analyzed by mass spectrometry.
MALDI-TOF MS
For MALDI-TOF MS analysis, either desalted (using C18 ZipTips, Millipore Co., Bedford, MA) or non-treated samples were directly applied onto the stainless steel sample plate with 2 µl of 50% (v/v) acetonitrile/0.1% TFA saturated with α-cyano-4-hydroxycinnamic acid. Mass spectra were acquired in the mass range between 500 and 3000 a.m.u. on a Voyager-DE STR mass spectrometer (PerSeptive Biosystems, Framingham, MA) equipped with a 337 nm nitrogen laser. The instrument was operated in the positive reflector mode with the following parameters: accelerating voltage: 20,000 V; grid voltage: 70–75%; mirror voltage ratio: 1.12; guide wire: 0.02%; extraction delay time: 150 nsec. For internal mass calibration we used trypsin autoproteolysis peaks (M+H+ of 842.50 and 2211.10). Detected masses were matched against protein sequences in the SWISS-PROT database using the ProFound program. The search parameters included a maximal number of 2 missing cleavage sites, carboxymethylation of Cys, nitration of Tyr, and oxidation of Met.
Capillary HPLC-ESI-Q-TOF MS analysis of tryptic peptides and proteins
Peptide separation was performed on an Ultra Plus II Micro LC System (Micro Tech Scientific, Sunnyvale, CA), equipped with a Zorbax C18SB (300Å, 5 cm length and 0.32 mm i.d.) column using a flow rate of 10 µl/min. Peptides were eluted with non-linear gradients of solvent A and solvent B, containing 0.08 % (v/v) formic acid and 1% or 99% (v/v) MeOH in water, respectively. The gradient was held at 20% B for 5 min., then ramped to 95% B within 25 min., and finally held for 5 min. at 95% B. Peptides were directly eluted into a Q-Tof™-2 mass spectrometer (Micromass Ltd, Manchester, UK), which was operated for maximum resolution with all lenses optimized on the MH22+ ion from a standard cyclic peptide, gramicidin S. The cone voltage was 30 eV, and argon gas was admitted to the collision cell at collision energy of 5 eV. Mass spectra were acquired with the time of flight analyzer at 11364 Hz pusher frequency covering a mass range between 400 and 3000 a.m.u., and accumulating data for 5 seconds per cycle. Time-to-mass calibration was made with CsI cluster ions acquired under the same conditions. An exhaustive search for peaks corresponding to nitrated peptides and their native analogues was performed with the MassLynx 3.5 software (Micromass Ltd, Manchester, UK), assuming that up to 2 cleavage sites could be missed during tryptic digestion and that Met sulfoxide could be formed in addition to 3-NT. The retention times of the ions obtained during the first HPLC-MS run were used for programming collision-induced dissociation (CID) experiments on peptides of interest.
For the analysis of non-digested proteins, a Vydac C4 column (1-mm ID × 50 mm length) was used. 20 µL of Ph-b (1.5 mg/ml) were injected and eluted into the source of the Q-Tof™-2 mass spectrometer by a gradient of solvent B (equilibrated at 20%B, than ramped to 95%B from 1 to 25 min., followed by washing with 95%B for additional 5 min.). Multiply charged ion series (from +30 to +70) were integrated and deconvoluted to convert data to a spectrum of a singly charged protein ion using the MassLinx 3.1 software.
Capillary HPLC-ESI-Q-TOF MS/MS analysis
CID spectra were acquired by setting the MS1 quadrupole to transmit a precursor mass window of ±1.5 a.m.u. As a collision gas, Ar was admitted at a density that attenuates the beam to 20%, which corresponds to 16 psi on the supply regulator. The collision energy was varied from 20 to 35 eV to obtain a distribution of fragments from low to high mass. Spectra were acquired for 2–5 min at 5 s cycles. Analysis of the MS/MS spectra was based on a search for the major sequence-indicating ions resulting from the cleavage of the parent ion at specific locations relative to the peptide bond: N-terminal (b) and C-terminal (y”) fragments.
Capillary HPLC-FT-ICR-CID analysis
Samples were introduced into an LTQ-FT hybrid linear quadrupole ion trap Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer (ThermoFinnigan, Bremen, Germany) via capillary liquid chromatography, analogous to published procedure [36]. Raw experimental files were processed using the BioWorks 2.0 software followed by peptide identification using Sequest and Mascot and X!Tandem database-searching programs. The peptide assignments obtained were validated using a statistical method of the Scaffold 1.7 software from Proteome Software Inc. (Portland, OR; http://www.proteomesoftware.com).
Determination of solvent accessible surface area (SASA)
For determination of relative solvent accessibility of Tyr residues in the protein we used the NACCESS 2.1.1 program, which can be downloaded from the website http://www.bioinf.manchester.ac.uk/naccess. This program calculates the solvent accessible area of a molecule from a PDB (Protein Data Bank) format file. The atomic and residue accessibilities are calculated as the atomic accessible area when a probe (typically of a radius of a water molecule, i.e., 1.4 Å) is rolled around the Van der Waal's surface of a macromolecule, and the path traced out by its center represents the accessible surface [37].
Results
Nitration of Ph-b by peroxynitrite
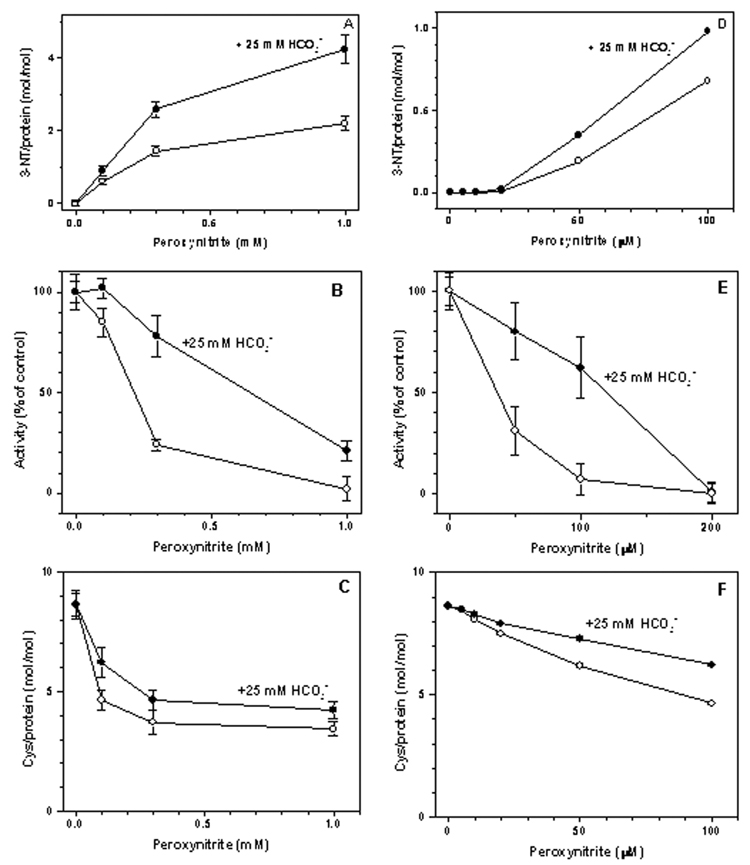
Bolus addition of peroxynitrite to Ph-b solutions resulted in the yellow staining indicating the formation of protein 3-NT, with a characteristic absorbance at 430 nm when measured at pH>9 (ε430=4,400 M−1cm−1 [19]). Fig.1A shows peroxynitrite concentration dependencies of 3-NT formation on Ph-b in the absence and presence of a physiological concentration of sodium bicarbonate. In the presence of HCO3−, 3-NT formation was ca. 2-fold more efficient, yielding ca. 4.3 mol 3-NT/mol Ph-b at a concentration of added peroxynitrite of 1 mM. We further used these conditions for peroxynitrite treatment of Ph-b for tryptic mapping of protein 3-NT residues by MS. Peroxynitrite concentration dependencies of Ph-b nitration in the absence and presence of bicarbonate displayed a “sigmoidal” shape showing negligible yields of 3-NT at low molar ratios of peroxynitrite to protein (<1) (Fig. 1D), followed by a linear increase, and leveling off of 3-NT yields at a high excess of peroxynitrite over protein (Fig. 1A).
Figure 1.
Peroxynitrite concentration dependencies of 3-NT formation (A,D), Ph-b activity (B,E), and of DTNB-reactive protein Cys residues (C,F). Rabbit muscle Ph-b (20 µM, except for 2 uM in E) was treated with peroxynitrite in the absence (open circles) or in the presence of 25 mM sodium bicarbonate (solid circles) in 25 mM sodium phosphate buffer containing 1 mM DTPA, pH 7.4. The Ph-b activity in controls (26 U/mg protein) was set equal 100%.
Peroxynitrite inhibition of Ph-b activity
Bolus addition of peroxynitrite to a Ph-b solution at pH 7.4 resulted in the loss of enzymatic activity in a concentration-dependent manner (Fig. 1B). Addition of 1 mM peroxynitrite to 20 µM Ph-b in the absence of bicarbonate led to nearly complete inactivation of the enzyme. Lower concentrations of Ph-b (2 µM) were more sensitive to peroxynitrite: here, a complete inhibition was observed for ca.100 µM peroxynitrite (Fig. 1E), in accord with results published earlier [17]. Surprisingly, bicarbonate afforded protection against the inactivation of Ph-b by peroxynitrite in spite of increasing 3-NT formation (compare Fig.1A, 1B, 1D, and 1E). In contrast, bicarbonate attenuated the loss of DTNB-reactive Cys residues (Fig. 1C and 1F). After the exposure to 1 mM peroxynitrite, the molar content of Cys residues decreased from 8.63±0.56 to 3.43±0.30 and 4.24±0.35 in the absence and in the presence of bicarbonate, respectively. The addition of 1 mM DTT in 10 min after peroxynitrite treatment did not significantly reverse loss of either activity or molar content of DTNB-reactive Cys residues (data are not shown).
Preparation of Ph-b in-gel digests for MS analysis
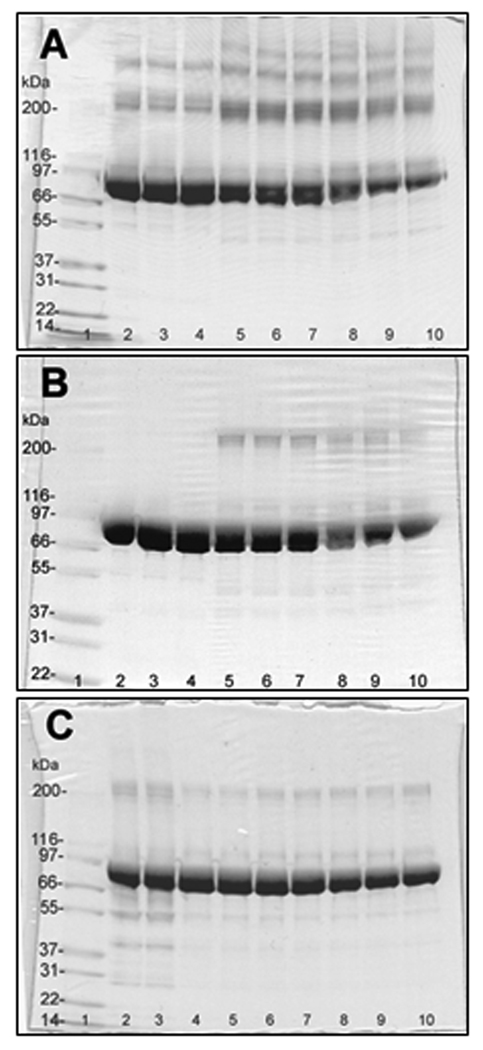
Treatment of Ph-b solutions with peroxynitrite induced slow-developing protein aggregation and precipitation. SDS-PAGE analysis followed by Coomassie Blue R250 staining indicated a loss of protein in the 97 kDa (monomer) band and the appearance of a series of additional higher molecular weight bands (Fig.2A). This effect could be attenuated by alkylation of the protein (Fig. 2B) suggesting that the formation of intermolecular disulfide bridges contributes to the aggregation of the protein. Dityrosine based cross-linking was ruled out based on our previous studies where we failed to detect an increase in the specific fluorescence of dityrosine (data not shown). We did not further focus on the mechanisms of aggregation. However, we note that reductive alkylation of Cys residues (i) may change protein structure, and (ii) does not completely protect from protein aggregation. More importantly, incubation in SDS-PAGE buffer and gel separation of the protein immediately after reaction with peroxynitrite prevents the loss of the monomeric protein even under non-reducing conditions (Fig.2C). Within the gel band the protein is available for trypsin digestion and subsequent peptide analysis by HPLC-MS and MS/MS with high sequence coverage (shown below).
Figure 2.
SDS-PAGE of rabbit muscle phosphorylase b (20 µg/lane) treated with peroxynitrite in the presence of 25 mM NaHCO3 at pH 7.4 (non-reducing conditions). A and B: lanes 2–4 – none; 5–7 – 0.3 mM; 8–10 – 1 mM peroxynitrite. In C: lanes 2 and 3 – reverse-order of peroxynitrite addition (1 mM) and control; lanes 4 and 5 – 0.1 mM; lanes 6 and 7 – 0.3 mM; lanes 8 and 9 – 1 mM peroxynitrite; lane 10 – 1 mM peroxynitrite addition in the absence of NaHCO3 in 25 mM sodium phosphate, pH 7.4. Lane 1 contained molecular weight standard Mark12 (3 µg of each protein). Samples were loaded either 2 hours (A and B) or immediately (C) after treatment with peroxynitrite. In B, samples were reduced with 2 mM DTT at 37°C for 30 min followed by alkylation with iodoacetic acid for 30 min at room temperature in the dark.
A low yield of peptides after in-gel digestion is the main problem for the characterization of particularly hydrophobic and protease-resistant proteins or protein domains. Common ionic detergents, such as SDS, suppress the MS detection of peptides and proteins and should be generally washed out before the analysis [39–40]. Application of nonionic surfactants, e.g., octyl-β-glucopyranoside (OBG), allows improving the recovery of peptides by MALDI-MS analysis [40–41]. However, it has been reported that at relatively high concentrations (1%) they may also have a negative impact on ESI-MS analysis (direct infusion) due to suppression of peptide detection by multiply charged surfactant cluster ions [42]. In our experiments, we found that the addition of 0.1% OBG during extraction of tryptic Ph-b peptides after in-gel digestion significantly enhances the yield of tryptic peptides and protein sequence coverage by HPLC-ESI-MS/MS analysis (data not shown). Tryptic peptides were separated from OBG by reverse-phase HPLC; moreover, even in the region of OBG elution we successfully resolved Ph-b peptides in ESI-MS and ESI-MS/MS modes. Therefore, SDS-PAGE separation and in-gel digestion were selected for the characterization of Ph-b nitration by LC-MS, similar to our methodology for SERCA1 [27].
Analysis of tyrosine nitration sites in Ph-b
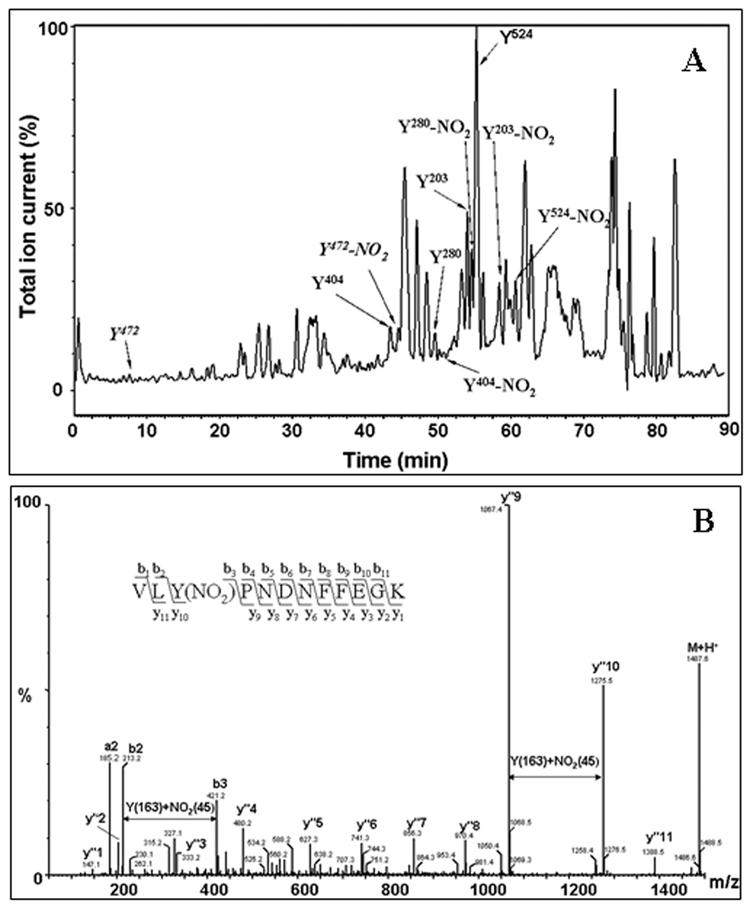
Table 1 summarizes all the MS data on the coverage of tyrosine-containing sequences in Ph- b obtained by capillary HPLC-ESI-Q-TOF MS analysis. The only Tyr residues not covered by this specific mode of analysis are those at positions 113, 553, and 613 (here and further we will use sequence positions for mature protein not including initial Met residue). Therefore, our coverage of Tyr residues in Ph-b amounts to 33 out of 36, or 92%. Among these peptides, sequences containing 28 Tyr residues (78% from total Tyr) were confirmed by tandem MS analysis and respective HPLC-MS peaks were used for the relative quantitation of sequence-specific Ph-b nitration. Figure 3 illustrates our approach. The mass chromatograms (Fig.3A) were searched for masses corresponding to both native Tyr-containing peptides and their respective nitropeptides with a gain of +45 atomic mass units (AMU). Tandem MS analysis was employed to confirm that the selected peptide ions contained indeed 3-NT, illustrated in Fig.3B. All the nitrated peptide peaks were shifted to longer retention times compared to the native peptides in the mass chromatogram (by ca. 3–5 min, see Fig.3A) reflecting an increased interaction of 3-NT peptides with the stationary phase. We calculated specific Tyr nitration yields through the ratios of mass chromatographic peak area of a specific nitropeptide to the sum of the peak areas of the native and the nitropeptide.
Table 1.
Coverage of Tyr-containing peptide sequences in rabbit muscle phosphorylase b by capillary HPLC-ESI-Q-TOF MS analysis. Peptides containing 33 out of total 36 Tyr residues (92% Tyr-containing sequence coverage) were resolved in the MS1 mode, and sequence of peptides containing 28 (78%) Tyr residues were confirmed by MS/MS. N.f. – not found.
| Tyr # | Start | End | M+H+ | M+2H+ | M+3H+ | Sequence | Rt, min | Intensity | m/z exp. | MS/MS |
|---|---|---|---|---|---|---|---|---|---|---|
| 51,52 | 50 | 60 | 1355.67 | 678.34 | 452.56 | (R)DYYFALAHTVR(D) | 10.5–11 | 47 | 678.37 | + |
| 74,75 | 70 | 77 | 1096.51 | 548.76 | 366.17 | (R)TQQHYYEK(D) | n.f. | |||
| 74,75 | 67 | 77 | 1551.77 | 776.39 | 517.93 | (R)WIRTQQHYYEK(D) | 9.25–10 | 204 | 776.42 | |
| 83,84,90 | 82 | 93 | 1554.77 | 777.89 | 518.93 | (R)IYYLSLEFYMGR(T) | 16.5–17.5 | 191 | 777.93 | + |
| 113 | 94 | 138 | 4897.23 | 2449.12 | 1633.08 | (R)TLQNTMVNLALENACDEATYQLGLD MEELEEIEEDAGLGNGGLGR(L) | n.f. | |||
| 155,157 | 139 | 160 | 2335.15 | 1168.08 | 779.05 | (R)LAACFLDSMATLGLAAYGYGIR(Y) | 21–22.5 | 115 | 779.08 | |
| 161 | 161 | 169 | 1145.56 | 573.29 | 382.53 | (R)YEFGIFNQK(I) | 10.5–11.5 | 4241 | 573.30 | + |
| 185 | 185 | 191 | 893.42 | 447.21 | 298.48 | (R)YGNPWEK(A) | 7.5–8.25 | 363 | 447.24 | + |
| 203 | 192 | 205 | 1689.89 | 845.45 | 563.97 | (K)ARPEFTLPVHFYGR(V) | 11–12.0 | 1118 | 563.99 | + |
| 226,233 | 215 | 234 | 2307.15 | 1154.08 | 769.72 | (K)WVDTQVVLAMPYDTPVPGYR(N) | 15.5–16.5 | 4155 | 769.73 | + |
| 262 | 256 | 269 | 1566.79 | 783.90 | 522.94 | (K)DFNVGGYIQAVLDR(N) | 19.5–21 | 2352 | 783.92 | + |
| 280 | 278 | 289 | 1442.70 | 721.85 | 481.57 | (R)VLYPNDNFFEGK(E) | 10.75–11.5 | 6364 | 721.87 | + |
| 297 | 295 | 309 | 1765.95 | 883.48 | 589.32 | (K)QEYFVVAATLQDIIR(R) | 18.5–20 | 937 | 883.52 | + |
| 374 | 371 | 386 | 1875.89 | 938.45 | 625.97 | (K)TCAYTNHTVLPEALER(W) | 9.75–10.25 | 3317 | 625.98 | + |
| 404 | 399 | 409 | 1426.78 | 713.89 | 476.27 | (R)HLQIIYEINQR(F) | 9.6–10.2 | 1218 | 713.92 | + |
| 472 | 470 | 478 | 1177.55 | 589.28 | 393.19 | (K)DFYELEPHK(F) | 8.5–9.5 | 1604 | 589.31 | + |
| 511 | 507 | 519 | 1550.77 | 775.89 | 517.60 | (R)IGEEYISDLDQLR(K) | 12–12.5 | 3089 | 775.92 | + |
| 524 | 521 | 532 | 1440.74 | 720.87 | 480.92 | (K)LLSYVDDEAFIR(D) | 11.5–12.5 | 5229 | 720.89 | + |
| 548 | 545 | 551 | 869.45 | 435.23 | 290.49 | (K)FAAYLER(E) | 8–8.5 | 335 | 435.25 | + |
| 553 | 552 | 554 | 439.22 | 220.11 | 147.08 | (R)EYK(V) | n.f. | |||
| 573 | 570 | 574 | 689.36 | 345.19 | 230.46 | (R)IHEYK(R) | 7.5–8 | 247 | 689.40 | |
| 587 | 576 | 589 | 1757.94 | 879.47 | 586.65 | (R)QLLNCLHVITLYNR(I) | 15–15.8 | 192 | 586.69 | + |
| 613 | 609 | 617 | 945.46 | 473.23 | 315.83 | (K)AAPGYHMAK(M) | n.f. | |||
| 648 | 642 | 649 | 1053.57 | 527.29 | 351.86 | (R)VIFLENYR(V) | 10–10.5 | 6006 | 527.31 | + |
| 726,731,732 | 725 | 734 | 1278.54 | 639.77 | 426.85 | (R)GYNAQEYYDR(I) | 7.6–8.2 | 290 | 639.80 | + |
| 777,780 | 773 | 782 | 1262.59 | 631.80 | 421.54 | (K)VFADYEEYVK(C) | 10–11.0 | 6658 | 631.82 | + |
| 791 | 787 | 792 | 680.40 | 340.70 | 227.47 | (R)VSALYK(N) | 6.6–8 | 158 | 680.43 | + |
| 820 | 816 | 822 | 822.45 | 411.73 | 274.82 | (R)TIAQYAR(E) | 9.5–10.2 | 353 | 822.45 | + |
Figure 3.
Sequence-specific identification of tyrosine nitration in rabbit muscle phosphorylase b (15 µM) incubated with 1 mM peroxynitrite in vitro. A – Total ion current chromatogram showing retention times for selected native and respective nitropeptides. B - Identification of nitrated Tyr residues by tandem MS for a representative 3-NT containing peptide, VLY280(NO2)PNDNFFEGK. The insert illustrates the use of a ladder of sequence-indicating fragments from N- (b) and C- (y”) termini for the identification of the peptide sequence.
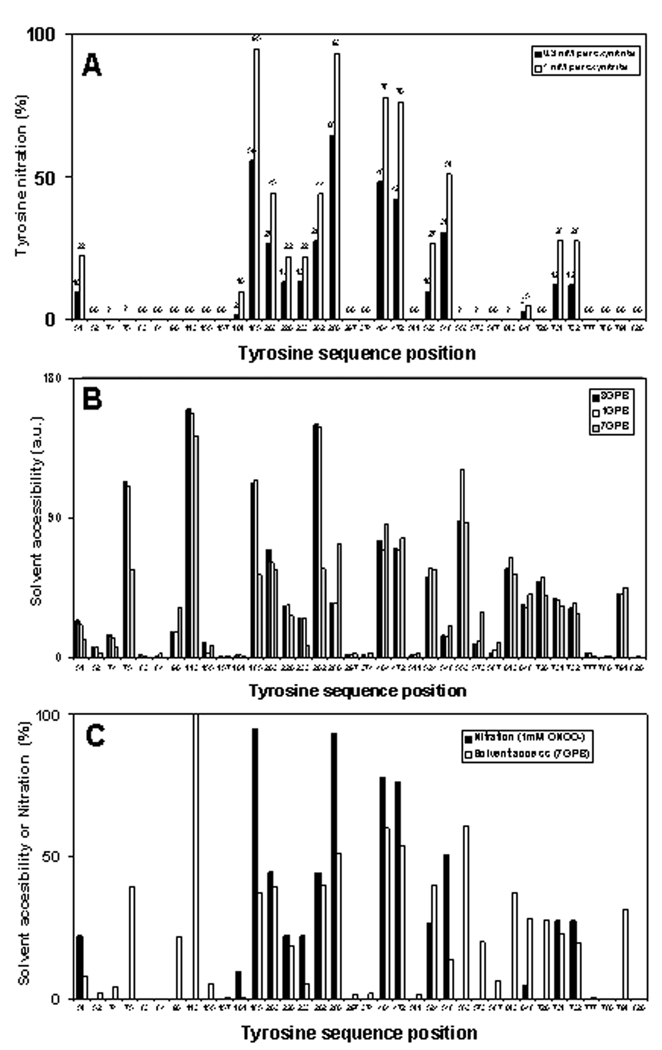
A total of 15 Tyr residues were detected in the nitrated form after treatment of the protein with peroxynitrite (Fig.4A). Generally the quality of the tandem MS spectra allowed unambiguous identification of mononitrated peptides only; in the case of peptides containing Tyr227,228 and Tyr727,732,733 we could not quantify nitration of the adjacent individual Tyr residues separately, and, thus, assumed that they are nitrated evenly. The yields of nitration for different Tyr residues were obviously not random. Relative nitration ranged from 0% for 16 Tyr residues to 99% for Tyr280. Neither native Ph-b nor reverse-order-of-peroxynitrite-addition control samples showed significant (more than 0.5%) nitration of individual Tyr residues. Taking into account that we did not detect multiple nitrations for peptides containing two or three Tyr residues we can assume that our conditions chosen for the treatment of Ph-b by peroxynitrite are appropriate for a sequence-specific characterization of Tyr sensitivity to in vitro nitration. Importantly, the figures of total molar 3-NT content in Ph-b measured by UV absorbance (Fig.1A) and obtained as a sum of molar fractions for individual Tyr residues (Fig.4A) are comparable: 4.24±0.38 vs. 6.46 and 2.58 vs. 3.72 for 1 and 0.3 mM peroxynitrite, respectively. The apparent overestimation of nitration levels for individual Tyr residues by MS analysis suggests that the assumption of equal LC-MS recovery and yields for native and respective nitropeptides may not always be accurate. However, this systematic error does not seriously affect the results of comparative analysis of sequence-specific Tyr nitration.
Figure 4.
A - Sequence specific nitration of different tyrosine residues in rabbit Ph-b after in vitro treatment with 0.3 (open columns) and 1 mM peroxynitrite (closed columns) in the presence of bicarbonate. For calculation of the relative nitration of each tyrosine-containing peptide by HPLC- ESI-MS, a mass chromatographic peak area for a nitropeptide was divided by a sum of chromatographic peak areas for both native and nitrated peptides. Masses and retention times for respective peptides were confirmed by tandem MS analysis in a separate run. Zero values are shown for tyrosine residues from peptides with confirmed native peptide sequences but lacking respective nitrated peptide sequences by tandem MS. A question mark in the panel A indicates that the sequence position of Tyr residues was not resolved by HPLC-ESI-tandem MS analysis in the native state.
B – NACCESS data for SASA values of respective Tyr residues obtained for different 3-D structures of rabbit muscle Ph-b: 1GPB – T-state without AMP (2 Å resolution), 8GPB – T-state with AMP (2.2 Å resolution), and 7GPB – R-state with AMP (2.8 Å resolution).
C – Overlay of relative nitration (experimental data at 1 mM peroxynitrite) and SASA values for the R-state of Ph-b (pdb: 7GPB).
Correlation between surface accessibility and nitration yields of individual tyrosine residues
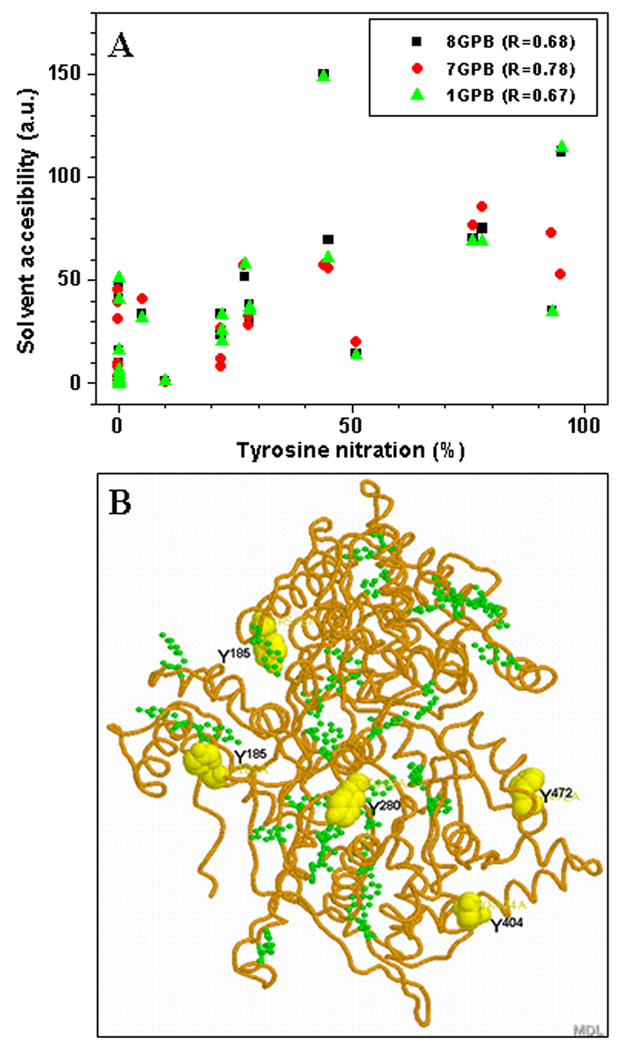
The SASA of Tyr residues was estimated using the NACCESS 2.1.1 program for 3 different PDB files of rabbit muscle Ph-b representing different structural states of the protein: here, 1GPB and 8 GPB represent the less active T-form (without allosteric activators), and 7GPB represents the active R-form (with either phosphorylated Ser14 or in the presence of the allosteric activator, AMP) [31–33]. Our experimental conditions fit to the T-state of Ph-b. However, even the exposure of Ph-b to peroxynitrite in the presence of 1 mM AMP did not significantly affect peroxynitrite concentration dependencies of Ph-b inactivation (provided that the enzymatic activity was assayed in the presence of the same AMP concentration), in accord with the data of Dairou and co-workers [17]. Plots for the relative Tyr residue SASA values for these three crystal structures of Ph-b are displayed in Fig.4B, which show that SASA values greatly vary between different Tyr residues. However, for most Tyr residues there is little difference between the Ph-b structures, except for Tyr in positions 75, 185, 262, 280, and 553. Fig.4C represents a comparison of Tyr nitration yields (1 mM peroxynitrite) and SASA values for the T-form of Ph-b (1GPB). This comparison demonstrates little correlation specifically for two regions in the N- and C-terminal domains of the protein. Quantitative correlation factors for all three structures of Ph-b are shown in Fig.5A. Low R-values (0.67, 0.68, and 0.78 for 1GPB, 8GPB, and 7GPB, respectively) confirm the lack of a direct correlation between solvent accessibility and Tyr nitration yields by peroxynitrite in vitro.
Figure 5.
A – Correlation analysis of Tyr nitration vs. solvent accessibility surface area (SASA) for different PDB structures of rabbit muscle phosphorylase b. The respective correlation coefficients are shown in the insert. B – Localization of the peroxynitrite-reactive Tyr residues: yellow colored “spacefill” residues (Tyr185, Tyr280, Tyr404, Tyr472, and Tyr548) exhibit >50% nitration level after reaction of 1 mM peroxynitrite with 15 µM Ph-b, while green “ball-and-stick” Tyr residues show lesser nitration levels or lack of nitration by peroxynitrite.
MALDI-TOF and capillary HPLC-FT-ICR-CID analysis of Tyr613-containing sequences
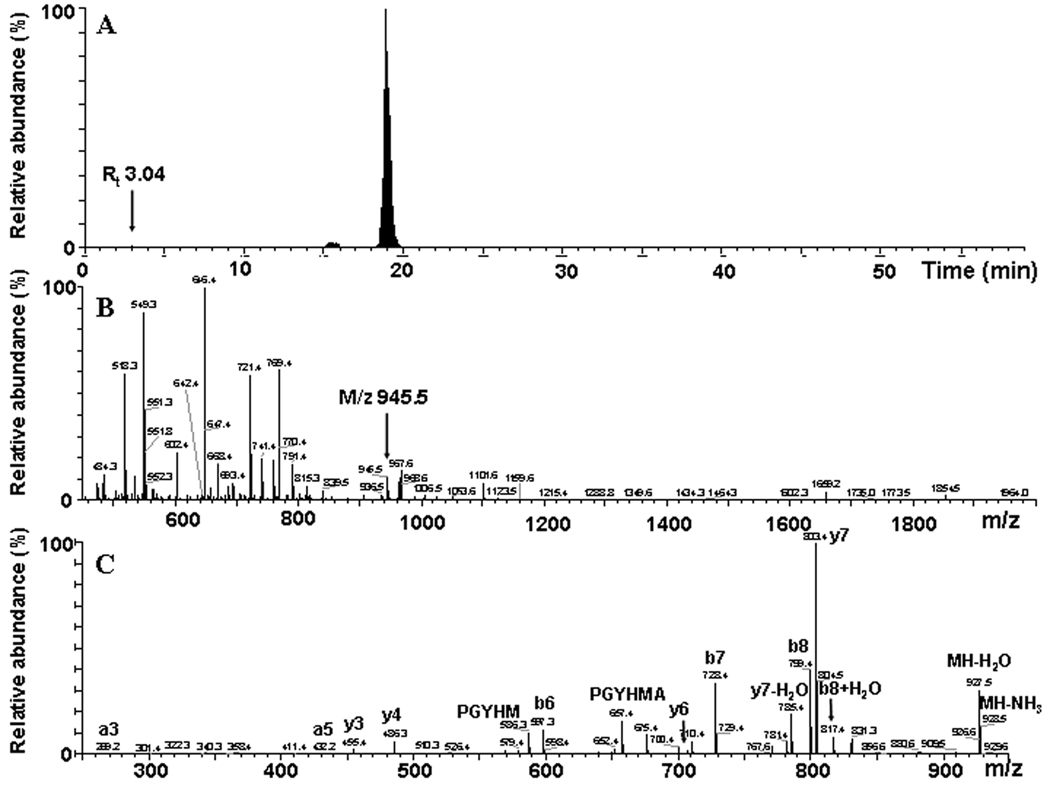
So far, the lack of identification of even native peptides containing Tyr613 by our capillary HPLC-ESI-Q-TOF MS method did not allow any conclusion about its putative role in the inactivation of Ph-b by peroxynitrite [17]. We need to consider at least three potential reasons for the lack of Tyr613 containing sequences in our data: poor recovery either from the gels after trypsin digestion or from the HPLC column prior to MS analysis, and low efficiency of peptide ionization and detection by MS. Dairou et al. [17] detected the nitration of Tyr613 on the peptide F597VVPRTVMIGGKAAPGY613(NO2)HMAK617 (M+H+ = 2274.7 AMU) by MALDI-TOF analysis. We, therefore, attempted to monitor the nitration of Tyr613 through two complimentary though not quantitative MS approaches. Our MALDI-TOF analysis of in-gel Ph-b digests revealed neither masses of the native peptides containing Tyr613, i.e. 945.5 (AAGY613HMAK; Table 1) and 2229.7 (for the tryptic peptide 597–617), nor masses for the respective modified peptides (Tyr nitration and Met oxidation were considered as potential modifications) in Ph-b samples exposed to peroxynitrite (spectra not presented). We then introduced in-solution tryptic digests of higher loads of Ph-b (1 mg/ml protein) into a more sensitive mass spectrometer (LTQ-FT-ICR) and detected the native peptide AAGY613HMAK (Fig. 6). However, the relative abundance of this peptide ion is very low, as demonstrated by mass chromatogram and representative mass spectrum (Fig.6A and 6B, respectively) for m/z 945.5, which nevertheless can be identified by tandem MS analysis (Fig. 6C). However, we did not detect any peptide containing 3-NT in position 613, while multiple peptides containing 3-NT at positions 84, 161, 185, 203, 226, 233, 262, 297, 404, 511, 524, 548, 648, 726, 777, 780, 791, and 820 were identified in the same sample. Hence, we conclude that nitration yields of Tyr613 are below the detection limit, and likely not as significant as originally proposed.
Figure 6.
Identification of the peptide containing Tyr613, AAPGYHMAK, by capillary HPLCFT-ICR-CID analysis. A – A selected mass chromatogram for m/z 945.5; B – mass spectrum integrated over the time window 2.5–3.5 min.; and C – tandem MS spectrum for the singly charged ion of m/z 945.5. The positions for this peptide ion in the mass chromatogram (A) and in the spectrum (B) are indicated by arrows.
Quantitative MS analysis of non-digested Ph-b
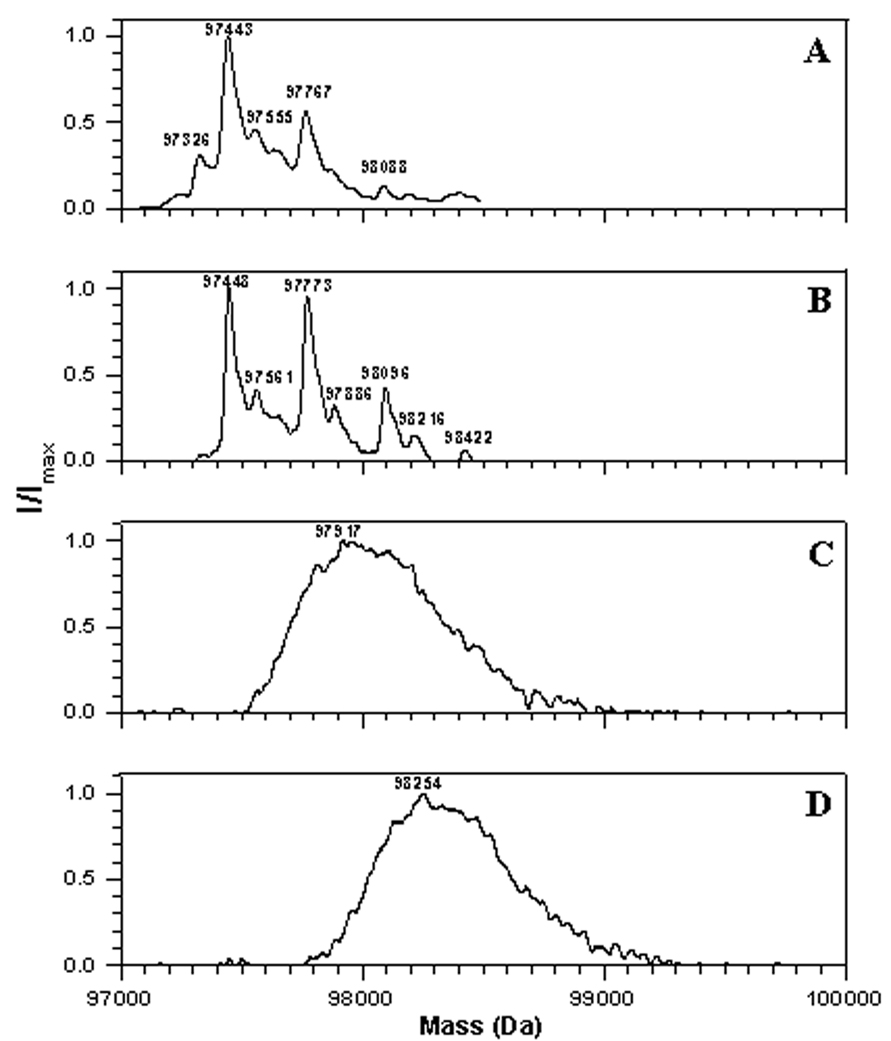
Mass spectra obtained for native Ph-b show that the preparation contains predominantly one isoform with an average mass of 97,443±10 AMU (Fig. 7A), which corresponds well to the predicted average mass of 97,431 AMU obtained from the published sequence (average mass of N-acetylated protein monomer with 9 free Cys thiol groups plus mass of pyridoxal phosphate Schiff base formed with Lys681 of 231 AMU). However, the commercial preparation of rabbit muscle phosphorylase b contains a minor isoform (about 15% of total protein) with a mass increase of ca. 323±2 AMU, which most probably represents a lactose adduct with one of the ε-NH2 groups of Lys (mass gain of 324 AMU) [43–44]. Such a modification would not be surprising since lactose is used as a main excipient in the dry preparation of Ph-b for protein stabilization; moreover, storage of Ph-b solution even at low temperatures is accompanied by an elevation of the fractions of the monolactosylated isoform and by the formation of multiply lactosylated protein fractions (Fig. 7B). Treatment of freshly prepared Ph-b solutions with peroxynitrite (in the presence of physiological concentrations of bicarbonate) produces a significant broadening of the mass spectra with a peroxynitrite concentration-dependent shift of the maximum to 97,917 and 98,254 AMU for 0.3 and 1 mM peroxynitrite, respectively (Fig.7, C and D). The changes in mass spectra are attributed to peroxynitrite-mediated protein modification since the respective reverse order of peroxynitrite addition controls did not significantly change the mass spectra (not shown). These data show that, in addition to Tyr nitration and Cys oxidation peroxynitrite can generate multiple, and different, modifications on the protein, producing a broad spectrum of oxidized and nitrated isoforms.
Figure 7.
LC-MS analysis of non-digested rabbit muscle phosphorylase b freshly dissolved (A), after 12 h storage in solution (B), and treated by either (C) 0.3 or (D) 1 mM peroxynitrite. Spectra were deconvoluted to convert series of multiply charged protein ions to a spectrum of single M+H+ protein ion.
Discussion
Differential sensitivity of tyrosine residues to nitration by peroxynitrite
The data presented here demonstrate that HPLC-MS and HPLC-MS/MS analysis of in-gel protein digests are instrumental for targeted characterization of large proteins with multiple Tyr nitration sites, such as Ph-b. In the proposed approach, capillary HPLC-ESI-QTOF MS quantitation of native and modified peptides in Ph-b provided a high sequence coverage and reliability of peptide identification. Though absolute quantitation of all potential 3-NT-containing peptides is barely possible, requiring multiple stable isotope labeled peptide standards even for a single protein, relative quantitation can be achieved to address a specific question: to compare the sensitivity of individual Tyr residues in Ph-b to nitration by peroxynitrite in order to identify Tyr residues, which may be critical for enzyme activity. The importance of such model studies is underlined by the earlier finding that Ph-b purified from muscle of aged rats exhibits a significant loss of specific activity compared to the enzyme isolated from the tissue of young rats; this loss of function was accompanied by the accumulation of 3-NT [15]. In the latter study, nanoHPLC-nanoESI-MS/MS analysis of in-gel digests of Ph-b allowed for the comparison of identified 3-NT-containing sequences between Ph-b from old and young rats, and Ph-b isolated from young rats, which was exposed to peroxynitrite in vitro. Peptides containing 3-NT at positions 113, 161, and 573 were detected in the in vivo “aged” protein, whereas exposure of the purified protein to 1 mM peroxynitrite in vitro yielded 3-NT at positions 51, 52, 113, 185, 203, 262, 280, 404, 473, 731 and 732. Such differences in the Tyr nitration patterns were rationalized by a variety of parameters such as potentially different mechanisms of Tyr nitration, the in vivo abundance of antioxidant systems, protein-ligand and protein-protein interactions, as well as protein repair and turnover mechanisms. Unfortunately, the qualitative MS approach used in that former study could not yield fractions of 3-NT on each affected Tyr residue in order to correlate Tyr nitration with age-dependent loss-of-function. In the present study, the nitrated sequences detected on rabbit Ph-b (3-NT at positions 51, 161, 185, 203, 226, 233, 262, 280, 404, 472, 523, 548, 648, 731, and 732) coincide qualitatively with those identified in the rat protein [15]. The few differences could be explained by differences in the analytical method, and differences in the sequences of rabbit and rat Ph-b. Of note is that in both studies, using different HPLC-ESI-MS techniques, the sequence containing Tyr613 was not resolved. Therefore, we have some concern about identity of the 3-NT-containing peptide FVVPRTVMIGGKAAPGY613(NO2)HMAK detected by MALDI-TOF analysis [17], based on a peptide mass only, without tandem MS sequence confirmation. In addition, the application of laser-induced ionization for the analysis of nitropeptides may generate artifacts due to the photochemical instability of 3-NT residues [45–47]. In fact, the application of more sensitive capillary HPLC- FT-ICR-CID analysis of in-solution Ph-b digests in our studies along with increased protein loads led to the identification of the native Tyr613-containing tryptic peptide, AAGYHMAK. Our data show that the relative abundance of the peptide ion in the chromatogram (Fig.6) is very low, which explains the lack of identification of these sequences by other MS methods. Importantly, we did not detect any nitrated peptides containing Tyr613 even under conditions where peroxynitrite exposure caused a 90% inactivation of Ph-b, and the formation of multiple 3-NT residues detected at least for 19 other sequence positions. These data suggest that Tyr613 may not be as significant as originally concluded [17]. At last, Ph-b inactivation by peroxynitrite may be the result of modification of additional amino acid residues (see below).
Correlation with SASA values shows that 3-NT yields are not a simple function of solvent accessibitiy of Tyr residues (Fig.4C and 5A). Most probably, individual reactivities of Tyr residues towards peroxynitrite or nitrosoperoxycarbonate derived radicals implicated in 3-NT formation [19] are determined by the respective microenvironment (hydrophobicity, surface charge depending on pK of neighboring residues, etc.).
Significance of protein tyrosine nitration in peroxynitrite inactivation of Ph-b
Qualitative mass spectrometry, including HPLC-MS/MS identification of nitrated peptides, provides very important information on protein sequence-specific 3-NT formation. However, such analysis must be followed by a quantitative analysis of the modification yields in order to estimate its potential significance for protein structure and function. Nevertheless, even the estimation of molar amounts of 3-NT in the protein, and a high correlation of nitration yields with protein specific activity cannot be conclusive. The reason for this is the potential co-existence of other oxidative modifications on the nitrated protein. It is well-known that peroxynitrite can generate different oxidative modifications on multiple additional oxidation-sensitive amino acid residues, such as Cys and Met [19]. Dairou et al. ruled out a potential role of Cys modification by peroxynitrite in the inactivation of Ph-b based on the lack of effect of a Cys alkylating reagent, N-ethylmaleimide, on enzyme activity [17]; however, here we show that peroxynitrite exposure caused the loss of ca. 5 DTNB-reactive Cys residues (out of a total of 9) per protein monomer under conditions, where Ph-b was nearly completely inactivated by 1 mM peroxynitrite (Fig. 1C). Although the effect of Cys oxidation is sequence- and modification-specific, this value is quite sufficient for structural and functional changes of the protein. Importantly, in the presence of CO2/HCO3− the average number of Cys residues in Ph-b lost upon peroxynitrite modification decreases to ca. 4 (Fig. 1C), which correlates with the decrease in Ph-b inactivation (Fig. 1A). In contrast, the presence of CO2/HCO3− increases the yield of 3-NT almost 2-fold, from 2.2 to 4.24 mol 3-NT/mol protein (Fig. 1A). This negative correlation between 3-NT yields and Phb activity may be considered as additional argument against the critical role of Tyr613 nitration in the inactivation of Ph-b by peroxynitrite. In general, our data do not support the existence of a single critical residue (“redox switch”), the oxidative modification of which leads to the loss of Ph-b activity. Rather, we suggest that the modification of the most reactive Cys and Tyr residues does not result in protein inactivation. The comparison of peroxynitrite concentration dependencies in the presence of physiological relevant concentration of bicarbonate shows that the loss of ca. 4 Cys and nitration of ca. 2.5 Tyr residues after exposure of 20 µM protein to 300 µM peroxynitrite resulted in a ca. 20% loss of Ph-b activity (Fig. 1, A–C). Even if there is a residue that needs to be completely modified for turning off protein function, it is obviously not the most oxidation sensitive one. It is hard to imagine how physiologically relevant oxidants can provide a selective modification of such “critical” residues except for a situation where other oxidation-sensitive residues are shielded by formation of special protein complexes or by low molecular weight ligands. At least for a single protein in vitro, the inactivation of Ph-b by peroxynitrite seems to result from a gradual accumulation of oxidative modifications of different amino acid residues. Although Cys residues show greater potential for oxidative Ph-b modification, as evidenced by a higher sensitivity to peroxynitrite (compare panels D and F in Fig.1) and by the effect of bicarbonate (positive correlation between loss of Cys and Ph-b activity), they also may not be exclusively implicated in the peroxynitrite inactivation of the protein.
Further analysis is needed to reveal, which specific modifications of specific amino acid residues may be responsible for the observed effects. We need to underline that the MS analysis of non-digested protein demonstrates a series of products, which may not necessarily be rationalized by the modification of Cys and Tyr only (Fig. 7A, 7C, and 7D). For example, in addition to an average of 4.24 mol 3-NT/mol protein at 1 mM peroxynitrite (average mass gain of ca. 190 AMU) there is a mass gain of 620 AMU, which cannot be rationalized by the oxidation of 5 Cys residues to a maximal oxidation state, sulfonic acid (mass gain of 240 AMU). Obviously, peroxynitrite modification of Ph-b may also target other sensitive residues, e.g., Met, His, or Trp (21, 22, and 12 residues per Ph-b monomer, respectively). These modifications can also potentially contribute to the overall change in the protein activity.
ACKNOWLEDGEMENTS
This study was supported by NIH grants PO1AG12993, AG23551 and AG25350. We also thank Dr. Krzysztof Kuczera for permitting us to use NACCESS program on the computational resource provided by the Department of Molecular Biosciences, University of Kansas.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Beckman JS. Chem. Res. Toxicol. 1996;9:836–844. doi: 10.1021/tx9501445. [DOI] [PubMed] [Google Scholar]
- 2.Ischiropoulos H, Al-Mehdi AB. FEBS Lett. 1995;364:279–282. doi: 10.1016/0014-5793(95)00307-u. [DOI] [PubMed] [Google Scholar]
- 3.Greenacre SAB, Ischiropoulos H. Free Rad. Res. 2001;34:541–581. doi: 10.1080/10715760100300471. [DOI] [PubMed] [Google Scholar]
- 4.Ischiropoulos H. Biochem. Biophys. Res.Commun. 2003;305:776–783. doi: 10.1016/s0006-291x(03)00814-3. [DOI] [PubMed] [Google Scholar]
- 5.Beckman JS, Ze YZ, Anderson PG, Chen J, Accavity MA, Tarpey MM, White CR. Biol. Chem. Hoppe-Seyler. 1994;375:81–88. doi: 10.1515/bchm3.1994.375.2.81. [DOI] [PubMed] [Google Scholar]
- 6.Viner RI, Ferrington DA, Williams TD, Bigelow DJ, Schöneich C. Biochem. J. 1999;340:657–669. [PMC free article] [PubMed] [Google Scholar]
- 7.Giasson BI, Duda JE, Murray IV, Chen Q, Souza JM, Hurtig HI, Ischiropoulos H, Trojanowski JQ, Lee VM. Science. 2000;290:985–989. doi: 10.1126/science.290.5493.985. [DOI] [PubMed] [Google Scholar]
- 8.Turko IV, Murad F. Pharmacol. Rev. 2002;54:619–634. doi: 10.1124/pr.54.4.619. [DOI] [PubMed] [Google Scholar]
- 9.Haqqani AS, Kelly JF, Birnboim HC. J. Biol. Chem. 2002;277:3614–3621. doi: 10.1074/jbc.M105730200. [DOI] [PubMed] [Google Scholar]
- 10.Drew B, Leeuwenburgh C. Ann. N.Y. Acad. Sci. 2002;959:66–81. doi: 10.1111/j.1749-6632.2002.tb02084.x. [DOI] [PubMed] [Google Scholar]
- 11.Ischiropoulos H, Beckman JS. J. Clin. Invest. 2003;111:163–169. doi: 10.1172/JCI17638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pacher P, Beckman JS, Liaudet L. Physiol. Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Souza JM, Peluffo G, Radi R. Free Radic. Biol. Med. 2008. pp. 357–366. [DOI] [PubMed]
- 14.Viner RI, Ferrington DA, Hühmer AF, Bigelow DJ, Schöneich C. FEBS Lett. 1996;379:286–290. doi: 10.1016/0014-5793(95)01530-2. [DOI] [PubMed] [Google Scholar]
- 15.Sharov VS, Galeva NA, Kanski J, Williams TD, Schöneich C. Exp. Gerontol. 2006;41:407–416. doi: 10.1016/j.exger.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 16.Souza JM, Daikhin E, Yudkoff M, Raman CS, Ischiropoulos H. Arch. Biochem. Biophys. 1999;371:169–178. doi: 10.1006/abbi.1999.1480. [DOI] [PubMed] [Google Scholar]
- 17.Dairou J, Pluvinage B, Noiran J, Petit E, Vinh J, Haddad I, Mary J, Dupret JM, Rodrigues-Lima F. J. Mol. Biol. 2007;372:1009–1021. doi: 10.1016/j.jmb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 18.Blanchard-Fillion B, Souza JM, Friel T, Jiang GC, Vrana K, Sharov VS, Barron L, Schoneich C, Quijano C, Alvarez B, Radi R, Przedborski S, Fernando GS, Horwitz J, Ischiropoulos H. J. Biol. Chem. 2001;76:46017–46023. doi: 10.1074/jbc.M105564200. [DOI] [PubMed] [Google Scholar]
- 19.Alvarez B, Radi R. Amino Acids. 2003;25:295–311. doi: 10.1007/s00726-003-0018-8. [DOI] [PubMed] [Google Scholar]
- 20.Aslan M, Ryan TM, Townes TM, Coward L, Kirk MC, Barnes S, Alexander CB, Rosenfeld SS, Freeman BA. J. Biol. Chem. 2003;278:4194–4204. doi: 10.1074/jbc.M208916200. [DOI] [PubMed] [Google Scholar]
- 21.Xu S, Ying J, Jiang B, Guo W, Sharov VS, Lazar H, Menzoian J, Bigelow D, Schöneich C, Cohen R. Am. J. Physiol. Heart Circ. Physiol. 2006;290:H2220–H2227. doi: 10.1152/ajpheart.01293.2005. [DOI] [PubMed] [Google Scholar]
- 22.Kanski J, Behring A, Pelling J, Schöneich C. Am. J. Physiol. Heart Circ. Physiol. 2005;288:H371–H381. doi: 10.1152/ajpheart.01030.2003. [DOI] [PubMed] [Google Scholar]
- 23.Sacksteder CA, Qian W-J, Knyushko TV, Wang H, Chin MH, Lacan G, Melega WP, Camp DG, Smith RD, Smith DJ, Squier TC, Bigelow DJ. Biochemistry. 2006;45:8009–8022. doi: 10.1021/bi060474w. [DOI] [PubMed] [Google Scholar]
- 24.Kanski J, Hong SJ, Schöneich C. J. Biol. Chem. 2005;280:24261–24266. doi: 10.1074/jbc.M501773200. [DOI] [PubMed] [Google Scholar]
- 25.Hong SJ, Gokulrangan G, Schöneich C. Exp. Gerontol. 2007;42:639–651. doi: 10.1016/j.exger.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuo WN, Kreahling JM, Shanbhag VP, Shanbhag PP, Mewar M. Mol. Cell Biochem. 2000;214:121–129. doi: 10.1023/a:1007118300731. [DOI] [PubMed] [Google Scholar]
- 27.Sharov VS, Galeva NA, Knyushko TV, Bigelow DJ, Williams TD, Schöneich C. Anal. Biochem. 2002;308:328–335. doi: 10.1016/s0003-2697(02)00261-0. [DOI] [PubMed] [Google Scholar]
- 28.Knyushko TV, Sharov VS, Williams TD, Schöneich C, Bigelow DJ. Biochemistry. 2005;44:13071–13081. doi: 10.1021/bi051226n. [DOI] [PubMed] [Google Scholar]
- 29.Johnson LN. FASEB J. 1992;6:2274–22782. doi: 10.1096/fasebj.6.6.1544539. [DOI] [PubMed] [Google Scholar]
- 30.Kurganov BI, Kornilaev BA, Chebotareva NA, Malikov VP, Orlov VN, Lyubarev AE, Livanova NB. Biochemistry. 2000;39:13144–13152. doi: 10.1021/bi000975w. [DOI] [PubMed] [Google Scholar]
- 31.Oikonomakos NG, Zographos SE, Skamnaki VT, Archontis G. Bioorg. Med. Chem. 2002;10:1313–1319. doi: 10.1016/s0968-0896(01)00394-7. [DOI] [PubMed] [Google Scholar]
- 32.Oikonomakos NG, Chrysina ED, Kosmopoulou MN, Leonidas DD. Biochim. Biophys. Acta. 2003;1647:325–332. doi: 10.1016/s1570-9639(03)00085-2. [DOI] [PubMed] [Google Scholar]
- 33.DiMauro S, Andreu AL, Bruno C, Hadjigeorgiou GM. Curr. Mol. Med. 2002;2:189–196. doi: 10.2174/1566524024605770. [DOI] [PubMed] [Google Scholar]
- 34.Martin MA, Rubio JC, Buchbinder J, Fernandez-Hojas R, del Hoyo P, Teijeira S, Gamez J, Navarro C, Fernandez JM, Cabello A, Campos Y, Cervera C, Culebras JM, Andreu AL, Fletterick R, Arenas J. Ann. Neurol. 2001;50:574–581. [PubMed] [Google Scholar]
- 35.Pryor WA, Cueto R, Jin X, Koppenol WH, Ngu-Schwemlein M, Squadrito Gl, Uppu PL, Uppu RM. Free Radic. Biol. Med. 1995;18:75–83. doi: 10.1016/0891-5849(94)00105-s. [DOI] [PubMed] [Google Scholar]
- 36.Ikehata K, Duzhak TG, Galeva NA, Ji T, Koen YM, Hanzlik RP. Chem. Res. Toxicol. 2008;21:1432–1442. doi: 10.1021/tx800093k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee B, Richards FM. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 38.Ogorzalek Loo RR, Dales N, Andrews PC. Protein Sci. 1994;3:1975–1983. doi: 10.1002/pro.5560031109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li F, Dong M, Miller LJ, Naylor S. Rapid. Commun. Mass Spectrom. 1999;13:464–465. doi: 10.1002/(SICI)1097-0231(19990315)13:5<464::AID-RCM486>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 40.Barnidge DR, Dratz EA, Sunner J, Jesaitis AJ. Protein Science. 1997;6:816–824. doi: 10.1002/pro.5560060408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Montfort BA, Canas B, Duurkens R, Godovac-Zimmermann J, Robillard GT. J. Mass Spectrom. 2002;37:322–330. doi: 10.1002/jms.288. [DOI] [PubMed] [Google Scholar]
- 42.Ishihama Y, Katayama H, Asakawa N. Anal. Biochem. 2000;287:45–54. doi: 10.1006/abio.2000.4836. [DOI] [PubMed] [Google Scholar]
- 43.Leonil J, Molle D, Fauquant J, Maubois JL, Pearce RJ, Bouhallab S. J. Dairy Sci. 1997;80:2270–2281. doi: 10.3168/jds.S0022-0302(97)76176-9. [DOI] [PubMed] [Google Scholar]
- 44.Scaloni A, Perillo V, Franco P, Fedele E, Froio R, Ferrara L, Bergamo P. Biochim. Biophys. Acta. 2002;1598:30–39. doi: 10.1016/s0167-4838(02)00290-x. [DOI] [PubMed] [Google Scholar]
- 45.Sheeley SA, Rubakhin SS, Sweedler JV. Anal. Bioanal. Chem. 2005;382:22–27. doi: 10.1007/s00216-005-3145-0. [DOI] [PubMed] [Google Scholar]
- 46.Nauser T, Koppenol WH, Pelling J, Schöneich C. Chem. Res. Toxicol. 2004;17:1227–1235. doi: 10.1021/tx049862u. [DOI] [PubMed] [Google Scholar]
- 47.Schöneich C, Sharov VS. Free Radic. Biol. Med. 2006;41:1507–1520. doi: 10.1016/j.freeradbiomed.2006.08.013. [DOI] [PubMed] [Google Scholar]