Abstract
Influenza A virus is an important human pathogen causing significant morbidity and mortality every year and threatening the human population with epidemics and pandemics. Therefore, it is important to understand the biology of this virus to develop strategies to control its pathogenicity. Here we demonstrate that live influenza A virus infection causes accumulation of autophagosomes by blocking their fusion with lysosomes. Matrix protein 2 is sufficient and necessary for this inhibition of autophagosome degradation. Macroautophagy inhibition compromises cell survival of influenza virus infected cells, but does not influence viral replication. We propose that influenza A virus, which also encodes pro-apoptotic proteins, is able to determine the death of its host cell by inducing apoptosis and blocking macroautophagy.
Keywords: influenza, macroautophagy, CD4+ T cells, Atg8/LC3, matrix protein 1
INTRODUCTION
Influenza virus is a segmented negative-sense RNA virus. The eight RNA segments of influenza A viruses encode 11 proteins (Palese and Shaw, 2007). The three largest segments are dedicated to produce each of the three RNA polymerase components, and one of them encodes an additional polypeptide, polymerase basic protein 1- frame 2 (PB1-F2), which promotes cell death (Chen et al., 2001). Two of the segments encode the surface glycoproteins hemagglutinin (HA) and neuraminidase (NA), involved in sialic acid adhesion and fusion in endosomes. One segment encodes the nucleoprotein (NP) involved in viral RNA packaging and transport, and one the two matrix proteins (M1 and M2). M2 possesses proton channel activity that allows virion acidification for efficient uncoating after fusion in endosomes. On top of these mostly structural proteins encoding RNA segments, the smallest segment encodes the non-structural proteins NS1 and NS2. NS2 or NEP supports the nuclear export of viral RNA packaged with NP. NS1 compromises the interferon (IFN) response to viral infection by multiple mechanisms, including binding to double-stranded RNAs and preventing their recognition by the innate immune system. Although many aspects of influenza virus pathogenesis have been investigated, a more detailed understanding of the virus-host interaction, especially infection-induced changes in the biology of the host cell, could reveal critical points of the influenza virus life cycle that could be targeted to cripple virus replication and diminish virus production. This is especially important, because the virus causes significant morbidity and mortality even outside pandemics. Influenza virus infection is associated with 20,000 deaths in the USA in a typical year, and causes, in addition, significant losses in the tens of billions of US dollars to the economy due to debilitating infections (Yewdell and Garcia-Sastre, 2002). Even more dramatically, the virus causes periodic pandemics with a higher death toll. Therefore, it is important to characterize additional control mechanisms and the virus’ countermeasures against them.
One such control mechanism could be macroautophagy, and indeed many pathogens have been found to interfere with this catabolic process (Schmid and Münz, 2007). Autophagic and proteasomal degradation are the two main catabolic processes for proteins in eukaryotic cells. While the proteasome discards primarily short-lived soluble ubiquitinated proteins (Ciechanover, 2005), autophagy is thought to degrade cellular organelles and protein aggregates of primarly long-lived proteins (Mizushima and Klionsky, 2007). Autophagy consists of at least three pathways microautophagy, macroautophagy and chaperone-mediated autophagy. Of those macroautophagy is best characterized and has been implicated in both innate and adaptive immunity (Schmid and Münz, 2007). During macroautophagy, an isolation membrane forms around its substrate, to form an autophagic vesicle, surrounded by two membranes. This autophagosome then fuses with late endosomes and lysosomes to degrade the inner autophagosome membrane and the autophagic cargo. More than 30 essential autophagy genes (atg) have been identified in yeast for this process (Mizushima and Klionsky, 2007). While the function of most of these is still unknown, two ubiquitin-like systems have been characterized as essential for autophagosome generation (Ohsumi, 2001). In one of them Atg12 is coupled to Atg5, which then in complex with Atg16L1 decorates the outer membrane of the isolation membrane. Upon autophagosome completion this complex is recycled. Several components of this ubiquitin-like system have been targeted to inhibit the process. Conditional knock-out mice have been generated for the E1-like enzyme Atg7, which activates Atg12, and for the Atg12 acceptor Atg5 (Komatsu et al., 2006)(Hara et al., 2006). In addition, we and others have used siRNA mediated silencing of Atg12 to inhibit macroautophagy (Paludan et al., 2005)(Jackson et al., 2005). The Atg12/Atg5/Atg16L1 complex is essential for autophagosome formation and might influence the curvature of the forming autophagosome as well as guide coupling of Atg8/LC3, the second ubiquitin-like molecule involved in macroautophagy (Hanada et al., 2007)(Fujita et al., 2008). This second ubiquitin-like system couples Atg8/LC3 to phosphatidylethanolamine at the outer and inner autophagosomal membrane. Atg8/LC3 stays with the completed autophagosome and is in part degraded with the inner vesicle membrane by lysosomal hydrolysis. Therefore, GFP tagged Atg8/LC3 can be used as a specific marker of autophagosomes and the turnover of the lipidated Atg8/LC3-II can be used as a measure of the macroautophagy rate (Klionsky et al., 2008). These ubiquitin-like conjugation reactions are initiated by class III phosphatidylinositol-3 (PI-3) kinase complexes, which include the Atg6/Beclin-1 protein. Interestingly, Atg6/Beclin-1 containing complexes that contain Atg14L support autophagosome formation, while others that contain UVRAG facilitate the fusion between autophagosomes and lysosomes (Matsunaga et al., 2009; Zhong et al., 2009)(Itakura et al., 2008).
This knowledge of the molecular mechanisms of autophagosome generation allowed us to analyze macroautophagy regulation by influenza virus infection. We found that influenza A virus infection arrests autophagosome degradation in large perinuclear structures. We could identify influenza A matrix protein 2 (M2) as sufficient and required for this phenotype, compromising macroautophagy, which is required for the survival of the infected host cells. Therefore, we propose that influenza A virus has at least two mechanisms to regulate cell death of its host cell: PB1-F2 inducing apoptosis and M2 inhibiting anti-apoptotic macroautophagy.
RESULTS
Autophagosomes accumulate after influenza virus infection
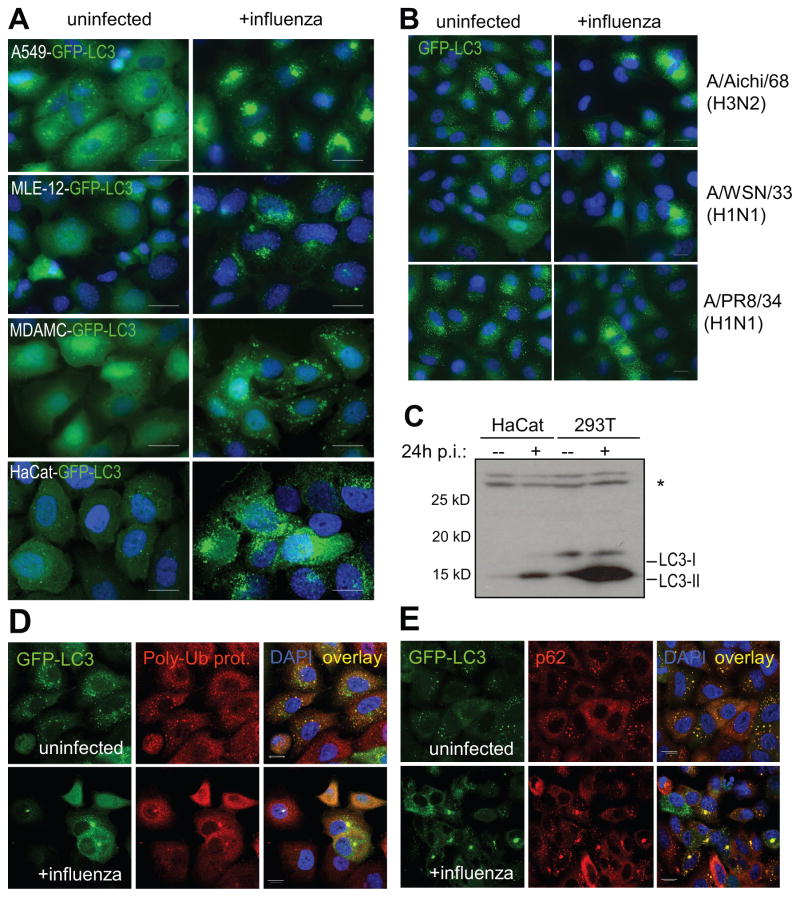
In order to investigate whether the cellular macroautophagy level is altered in response to influenza A virus infection, we infected stably GFP-LC3-transfected human epithelial cell lines with influenza virus strain A/Aichi/68 (H3N2). Cells were infected at a low multiplicity of infection (MOI between 0.6 or 0.8), which typically resulted in an infection rate of 50–70% (data not shown). At these MOIs, cell viability was maintained at 90% 24 hours post-infection, whereas 48 hours post-infection, up to 50% of apoptotic cells could be detected (data not shown). We therefore chose to analyze cells 24 hours post-infection. In comparison to uninfected cells, influenza-infected cells showed a strong increase in GFP-LC3+ autophagosomes (Figure 1A). Autophagosome accumulation was observed in A549 human lung epithelial cells, MLE-12 mouse lung epithelial cells, MDAMC human breast carcinoma cells, and HaCat human keratinocyte cells (Figure 1A). The increase in autophagosomes in A549 human lung epithelial cells occurred after infection with different strains of influenza virus [A/Aichi/68 (H3N2), A/WSN/33 (H1N1) and A/PR8/34 (H1N1)] (Figure 1B). The increased autophagosome number could also be observed by Western blot at the level of endogenous lipidated Atg8/LC3 (Figure 1C). When lysates of uninfected and infected cells were analyzed by anti-Atg8/LC3 immunoblot, the lipidated, and therefore faster migrating Atg8/LC3-II form was strongly increased in influenza-infected epithelial cell lines (Figure 1C). In addition to Atg8/LC3 as an autophagosome marker, we also analyzed accumulation of previously described autophagosome cargo (Klionsky et al., 2008). Both polyubiquitinated protein aggregates and p62/sequestosome 1, previously described macroautophagy substrates (Komatsu et al., 2006)(Hara et al., 2006)(Bjorkoy et al., 2005)(Pankiv et al., 2007), accumulated in autophagosomes upon influenza infection (Figure 1D and E). These data demonstrate that autophagosomes accumulate after epithelial infection with influenza A virus, suggesting that macroautophagy is a process that is regulated during influenza virus infection.
Figure 1. Classical autophagosomes accumulation after influenza A virus infection.
(A) Stably GFP-Atg8/LC3 transfected epithelial cells lines A549, MLE-12, MDAMC and HaCat were infected with influenza A/Aichi/68 virus and were analyzed for GFP-Atg8/LC3 positive autophagosome accumulation 24h later by fluorescence microscopy. DAPI was used to stain nuclear DNA. Scale bar: 30 μm. (B) GFP-Atg8/LC3 transfected A549 human lung epithelial cells were infected with different influenza A virus strains (A/Aichi/68, A/WSN/33 and A/PR8/34) and analyzed after 24h. One of three experiments is shown. Scale bar: 30 μm. (C) The human epithelial cell lines HaCat and 293T were infected with influenza A/Aichi/68 virus at a MOI of 0.1. 24h post-infection, lysates of uninfected and infected cells were analyzed by Western blot analysis for Atg8/LC3. Unconjugated (LC3-I) and lipidated Atg8/LC3 (LC3-II) can be distinguished by there apparent molecular weights of 18kD and 16kD, respectively. The asterisk (*) marks an unspecific band, detected by our Atg8/LC3 antiserum. One of three experiments is shown. 24h after influenza A virus infection co-localization of GFP-ATG8/LC3 and the classical macroautophagy substrates poly-ubiquitinated proteins (D) and p62/sequestosome 1 (E) was analyzed by immune fluorescence microscopy. Nuclear DNA was stained with DAPI. Scale bar: 20 μm. One of three experiments is shown.
Live influenza virus infection is required for autophagosome accumulation
To investigate how long after infection the autophagosome increase occurs, we prepared lysates at various time points post-infection and analyzed them by anti- Atg8/LC3 Western blot. Atg8/LC3-II was first detectably increased 10 hours after infection of lung epithelial cells and gradually accumulated to higher levels at later time points (Supplemental Figure 1A). Thus, the autophagosome accumulation was not a rapid response to influenza virus infection, suggesting that viral or cellular factors might have to be produced first.
To see whether cells have to be productively infected or whether uptake of inactivated viral particles and perhaps recognition of pathogen associate molecular patterns (PAMPs) within these is sufficient to mediate autophagosome accumulation, we exposed cells to live or heat-inactivated influenza A virus and analyzed their lipidated Atg8/LC3-II levels by Western blot analysis (Supplemental Figure 1B) and autophagosome content by GFP-Atg8/LC3 fluorescence microscopy (Supplemental Figure 1C). Only live influenza virus was able to elicit an increase of autophagosomes as indicated by GFP-Atg8/LC3-II, endogenous Atg8/LC3-II and GFP-Atg8/LC3 vesicle accumulation, whereas heat-inactivated virus did not cause any change in Atg8/LC3-II levels or GFP- Atg8/LC3 distribution (Supplemental Figure 1B and C). Thus, contact with the components or endocytosis of inactivated virus particles is not sufficient to elicit autophagosome accumulation.
To visualize more directly that productive infection is necessary for macroautophagy regulation, we stained uninfected and infected cells with M1-specific antibodies, to see if the autophagosome accumulation would only occur in cells that produce viral protein. Indeed, autophagosome accumulation and formation of large perinuclear autophagosomes was only detectable in cells that expressed influenza A M1, whereas M1-negative cells did not show these changes in intracellular GFP-Atg8/LC3 distribution (Supplemental Figure 1D). This indicates that autophagosome accumulation occurs only in directly and productively infected cells and is not mediated by a soluble factor secreted in response to influenza virus infection. In line with these observations, we could rule out that type I IFNs were responsible for the observed effect, because blocking of the IFN receptor did not abrogate the influenza-mediated autophagosome accumulation (data not shown). Therefore, productive influenza A infection causes autophagosome accumulation.
Influenza virus infection blocks autophagosome degradation
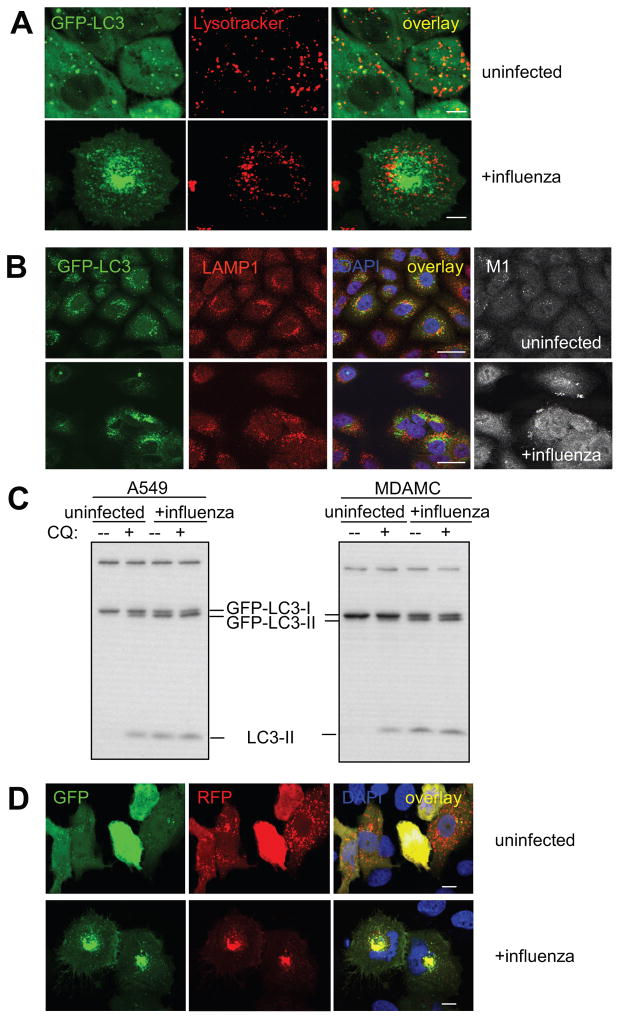
Since autophagosomes are transient vesicles that deliver their cargo within minutes for lysosomal hydrolysis, their accumulation could result from increased formation or decreased degradation (Klionsky et al., 2008). To investigate which of these two mechanisms was responsible for autophagosome accumulation upon influenza virus infection and if fusion of autophagosome with lysosomes was intact in infected cells, we labelled acidic compartments with red fluorescent lysotracker and performed live cell imaging (Figure 2A). In uninfected cells, about 25% of GFP-Atg8/LC3+ vesicles overlapped with lysotracker staining, indicating that a fraction of autophagosomes had fused with lysosomes and had become acidified. In contrast, in influenza-infected cells, almost no overlap between autophagosomes and lysotracker staining was observed, especially the large perinuclear autophagosomes were devoid of lysotracker staining (Figure 2A). This suggests that after influenza virus infection, autophagosomes do not fuse with acidic compartments, even so acidified lysosomes were present at similar numbers in infected cells. In order to rule out that autophagosomes fuse with lysosomes, but are not efficiently acidified in infected cells, we investigated GFP-Atg8/LC3 colocalization with the lysosome-associated membrane protein 1 (LAMP1) (Figure 2B). While GFP- Atg8/LC3 partially overlapped with LAMP1 in uninfected lung epithelial cells, this autophagosome marker did not co-localize with LAMP1 in influenza virus infected cells. Especially, the large GFP-Atg8/LC3 positive perinuclear vesicles were LAMP1 negative. These data suggested that autophagosomes do not efficiently fuse with lysosomes in influenza infected cells.
Figure 2. Autophagosomes do not fuse with acidified proteolytic lysosomes in influenza infected cells.
(A) Live cell imaging was performed to analyze co-localization of lysotracker stained acidified vesicles and GFP-Atg8/LC3 positive autophagosomes in uninfected and influenza A virus infected cells. Scale bar: 15 μm. Representative still images of two independent experiments are shown. (B) Furthermore, fusion of autophagosomes with lysosomes was analyzed as co-localization of the autophagosomes marker GFP-Atg8/LC3 with the lysosome marker LAMP1. Nuclear DNA was stained with DAPI and infected cells with matrix protein 1 (M1) specific antibodies. Scale bar: 30 μm. One of three experiments is shown. (C) GFP-Atg8/LC3 transfected A549 human lung epithelial and MDAMC human breast carcinoma cells were infected with influenza A virus at a MOI of 0.4 for 24h. Then the indicated samples were treated for 6h with the lysosomal inhibitor chloroquine (CQ), and lipidated endogenous Atg8/LC3-II and lipidated transfected GFP-Atg8/LC3-II content was analyzed by Atg8/LC3 specific Western blot analysis. One of three experiments is shown. (D) The tandem reporter construct mRFP-GFP-Atg8/LC3 was transiently transfected into uninfected or influenza A virus infected A549 human lung epithelial cells. GFP (sensitive to acidification and lysosomal degradation) and RFP fluorescence (insensitive to acidification and lysosomal degradation) of the reporter construct were analyzed by fluorescence microscopy. DAPI was used to stain nuclear DNA. Scale bar: 20 μm. One of three experiments is shown.
In order to confirm this block of autophagosome fusion with lysosomes in influenza virus infected cells, we analyzed the turnover of autophagosomes. For this purpose, influenza virus-infected and mock-infected cells, were treated, 24h post-infection, with the lysosomal proteolysis inhibitor chloroquine (CQ) for 6 hours. In uninfected cells, lipidated Atg8/LC3-II and GFP-Atg8/LC3-II accumulated upon CQ treatment, indicating that autophagosomes are turned over by lysosomal proteolysis (Figure 2C). In contrast, in influenza-infected cells, no further accumulation of Atg8/LC3-II and GFP-Atg8/LC3-II could be observed in response to CQ treatment, demonstrating that autophagosomes were not turned over at a significant rate by lysosomal proteolysis in infected cells. As a second indication that lysosomal turnover of autophagosomes is defective in influenza virus infected cells, we made use of a tandem reporter construct mRFP-GFP-Atg8/LC3 (Kimura et al., 2007). The GFP moiety of this tandem autophagosome marker is sensitive to lysosomal proteolysis and quenching in acidic pH, while the mRFP is not. Therefore, the green fluorescent component of the composite yellow fluorescence for this mRFP-GFP-LC3 reporter is lost upon autophagosome fusion with lysosomes. This fluorescence change from yellow to red can be used to visualize lysosomal proteolysis and localization in acidified compartments of macroautophagy targeted GFP. In uninfected lung epithelial cells few yellow autophagosomes, but a high number of mRFP positive autolysosomes could be detected after transient transfection of mRFP-GFP-Atg8/LC3 (Figure 2D). In contrast, influenza virus infection led to the accumulation of mRFP and GFP double positive vesicles, especially in the perinuclear region, suggesting impaired autophagosome fusion with lysosomes. These data suggest that influenza virus infection inhibits fusion of autophagosomes with acidified LAMP1+ lysosomes and thereby prevents degradation of macroautophagy substrates.
Influenza virus infected cells accumulate small autophagosomes with high mobility and immobile large perinuclear autophagosomes
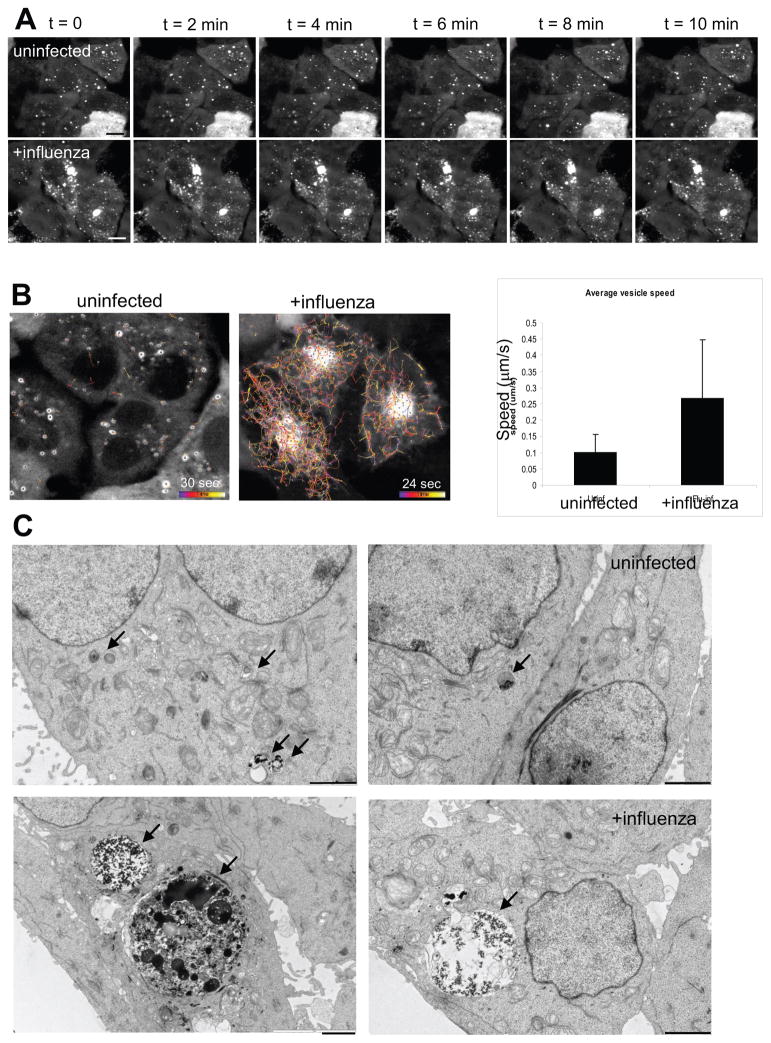
In order to investigate if the block of autophagosome fusion with lysosomes in influenza virus infected cells could result from impaired autophagosome mobility, we observed GFP-Atg8/LC3 transfected A549 lung epithelial cells by live cell imaging. Most influenza-infected cells contained numerous small autophagosomes in addition to one large GFP-LC3+ perinuclear vesicle of ≥5 μm diameter, whereas uninfected cells contained a smaller number of evenly distributed, small to medium size autophagosomes (Figure 3A). The larger autophagosomes (≥1μm) in both uninfected and infected cells moved only slowly, and especially the very large perinuclear autophagosome, which accumulated upon influenza virus infection, appeared almost immobile when followed over a period of 10 min (Figure 3A). In uninfected cells, smaller autophagosomes (< 1μm) moved slightly more rapidly than large autophagosomes and therefore could be seen to move slowly when their tracks were recorded over a period of 30 seconds (average speed = 0.1 μm/s, Figure 3B left). Strikingly, small autophagosomes in influenza-infected cells moved rapidly throughout the cytosol (Figure 3B and Supplemental Movie 1). The movement seemed to be random and non-directional, since vesicles moved towards and away from the central perinuclear region containing immobile large autophagosomes. The average speed of autophagosomes in infected cells was about 0.3 μm/s, and, therefore, significantly elevated compared to autophagosomes in uninfected cells. These findings suggest that the block in autophagosome fusion with lysosomes is not due to impaired autophagosome mobility.
Figure 3. Mobility and ultrastructure of autophagosomes in influenza A virus infected cells.
(A) Live cell imaging of GFP-Atg8/LC3 transfected A549 human lung epithelial cells was performed to analyze autophagosome mobility. Still images of uninfected and influenza A virus infected cells taken at the indicated timepoints of live cell imaging document the immobility of the accumulated large perinuclear autophagosomes. (B) Autphagosome tracking in uninfected and influenza A infected cells. Left: Vesicle tracks are displayed as color coded tracks, indicating the distance traveled by one vesicle during the indicated observation period (t=0 in blue, t=endpoint in white). Right: Average speed of autophagosomes was determined in three independent fields and summarized. One of three experiments is shown. Error bars indicate standard deviation. (C) Electron micrographs of uninfected and influenza A virus infected A549 human lung epithelial cells. Small autophagic structures (black arrows, upper row) were detected in uninfected cells, whereas large autophagic structures (black arrows, lower row) were visible in infected cells. Scale bar: 2 μm. One of three experiments is shown.
In addition to this kinetic description of the autophagosome populations in influenza virus infected cells, we analyzed the ultrastructural features of the large perinuclear GFP- Atg8/LC3+ vesicles. We analyzed influenza virus-infected human lung epithelial cells by electron microscopy in order to distinguish if these large GFP-Atg8/LC3-positive structures are autophagosomes of very large size or clusters of small autophagosomes. This also allowed us to look at the autophagosome content/composition at the ultrastructural level. In uninfected cells, a few small (0.5 – 1 μm) autophagosomes or autolysosomes containing internal lipid vesicles and electron dense material could be observed (Figure 3C, top row, black arrows). In contrast, 24 hours after influenza infection, most cells contained extremely large autophagosomes (up to 7 μm) that sometimes even reached the size of the nucleus (Figure 3C, bottom row, black arrows). The giant autophagosomes contained electron-dense, amorphous material, and did not seem to contain large amounts of other organelles, such as mitochondria or rough ER, which we could readily identify in the cytoplasm (Figure 3C). Our ultrastructural analysis of influenza-infected cells confirms that autophagosome numbers are strongly increased after influenza infection. Furthermore, it suggests that at least some of the large GFP-LC3-positive structures observed in fluorescence microscopy represent one unusually large autophagosome that might originate from the fusion of smaller autophagosomes.
Matrix protein 2 protein of influenza A virus is sufficient to block autophagosome degradation
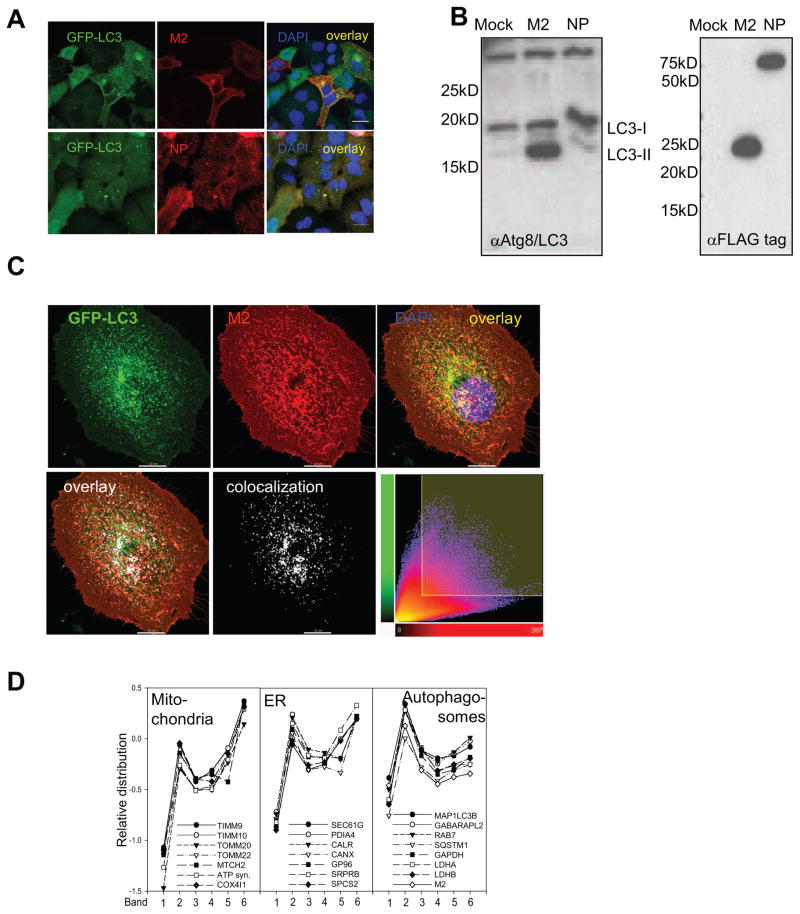
In order to investigate how influenza A virus causes this dramatic change in autophagic activity of infected cells, we transiently transfected GFP-Atg8/LC3 positive A549 cells with expression plasmids encoding for the influenza A virus proteins PB2, NP, NS1, M1, M2 and HA. 24 hours post transfection, we observed that only M2 transfected cells showed a strong accumulation of GFP-Atg8/LC3 vesicles by immunofluorescence microscopy analysis (Figure 4A), while the introduction of the other influenza A virus proteins did not result in autophagosome accumulation. The increased autophagosome number upon M2 expression could also be observed by Western blot analysis as an accumulation of lipidated Atg8/LC3-II (Figure 4B). In order to compare transfection levels of influenza A proteins, we used flag-tagged constructs (Figure 4B). Although the flag-tag can interfere with protein function, it did not cause autophagosome accumulation when coupled to NP and allowed autophagosome stabilization by M2. M2 mediated autophagosome accumulation was also observed in different human and mouse cell types, 293T human kidney epithelial cells, MDAMC human breast carcinoma cells, mouse embryonic fibroblasts and human melanoma cell lines (data not shown). In order to confirm that M2 expression also recapitulated the block of autophagosome fusion with lysosomes as observed during live influenza virus infection, we investigated autophagosome acidification and GFP degradation/quenching of the mRFP-GFP-Atg8/LC3 reporter construct in M2 transfected cells. Indeed no overlay between autophagosomes and lysotracker staining was observed in M2 transfected A549 cells (data not shown). Moreover co-expression of M2 and mRFP-GFP-LC3 tandem construct led to the accumulation of mRFP and GFP double positive vesicles, especially in the perinuclear region, suggesting impaired autophagosome fusion with lysosomes (data not shown). This observation was not the consequence of a general defect in the fusion with lysosomes, because we observed that fusion of fluorescent sepharose beads containing phagosomes with lysosomes was intact in influenza A virus infected and in M2 transfected cells (data not shown). These data indicate that M2 transfection recapitulates the macroautophagy block observed after influenza A virus infection.
Figure 4. Influenza A virus matrix protein 2 (M2) blocks autophagosome fusion with lysosomes.
(A) GFP-Atg8/LC3 transfected A549 human lung epithelial cells were transfected with expression plasmids encoding either FLAG tagged M2 or NP of influenza A/WSN/33 virus. 24 hours post transfection GFP-ATG8/LC3 positive autophagosome accumulation was analyzed by fluorescence microscopy. Influenza M2 specific antibodies were used for M2 detection, and an anti-Flag antibody for NP detection. Scale bar: 80 μm. One of three experiments is shown. (B) Left: Western Blot analysis of lipidated endogenous Atg8/LC3-II accumulation in A549 cells transfected with M2 or NP. Right: Western blot analysis of M2 and NP expression in transfected cells, using anti-FLAG tag staining. One of three experiments is shown. (C) Colocalization analysis of M2 (red fluorescence) and GFP-LC3 (green fluorescence) in infected A549 cells: Serial optical sections, (0.2 um; 40 sections) were acquired and images were then deconvoluted using Huygens software. Pearson coefficients were calculated using the Imaris 6.3.0 software. White fluorescence represents colocalization. Scale bar: 10μm. One representative of 36 analyzed cells is shown. (D). Mass spectrometric analysis of density gradient fractions (1–6) of influenza A virus infected A549 cells content upon influenza infection in A549 cells. Cell organelle marker protein distribution in these fractions was determined by mass spectrometric sequencing of protein fragments. Influenza A virus M2 distribution follows the distribution of autophagosome marker protein (right panel), but not the distribution pattern of mitochondria and endoplasmic reticulum (ER) proteins. One of four experiments is shown.
During our immune fluorescence microscopy analysis, we noted that M2 staining localized to the cell membrane and to perinuclear autophagosomes (Figure 4A). We therefore performed a detailed colocalization analysis of GFP-LC3 and M2 in infected A549 cells (Figure 4C). We found a significant partial colocalization of these two proteins with a Pearson coefficient of 0.7, as depicted in the scatter plot (4C lower panel) showing the pixel distribution of the two corresponding channels (number of colocalized voxels 63619). This example is representative of 36 cells that were analyzed. Overall a Pearson coefficient of 0.6 was reached, and 42% of autophagosomes contained 12% of M2 in influenza A virus infected cells.
In order to further support this M2 localization to autophagosomes, we performed cell fractionation and density gradient centrifugation to enrich autophagosomes from influenza A virus infected cells. The autophagosome distribution in these density fractions was then determined by characterizing the enrichment of the Atg8 homologues LC3 (MAP1LC3B) and Gate16 (GABARAPL2), of Rab7, involved in autophagosome fusion with lysosomes (Jager et al., 2004), and of the classical macroautophagy substrates p62/sequestosome 1 (SQSTM1) (Komatsu et al., 2007), glyceraldehydephosphate dehydrogenase (GAPDH) (Fengsrud et al., 2000) and lactate dehydrogenase A/B (LDHA/B) (Stromhaug et al., 1998). The distribution of these proteins was determined by peptide fragment analysis via mass spectrometry (Kristensen et al., 2008). This analysis revealed that influenza A virus M2 enrichment in density gradient fractions of infected A549 cells followed the profile of autophagosome markers, but not mitochondrial and endoplasmic reticulum proteins (Figure 4D). These findings suggest that M2 accumulates in regular autophagosomes and prevents their fusion with lysosomes.
Matrix protein 2 is necessary for autophagosome accumulation during influenza virus infection
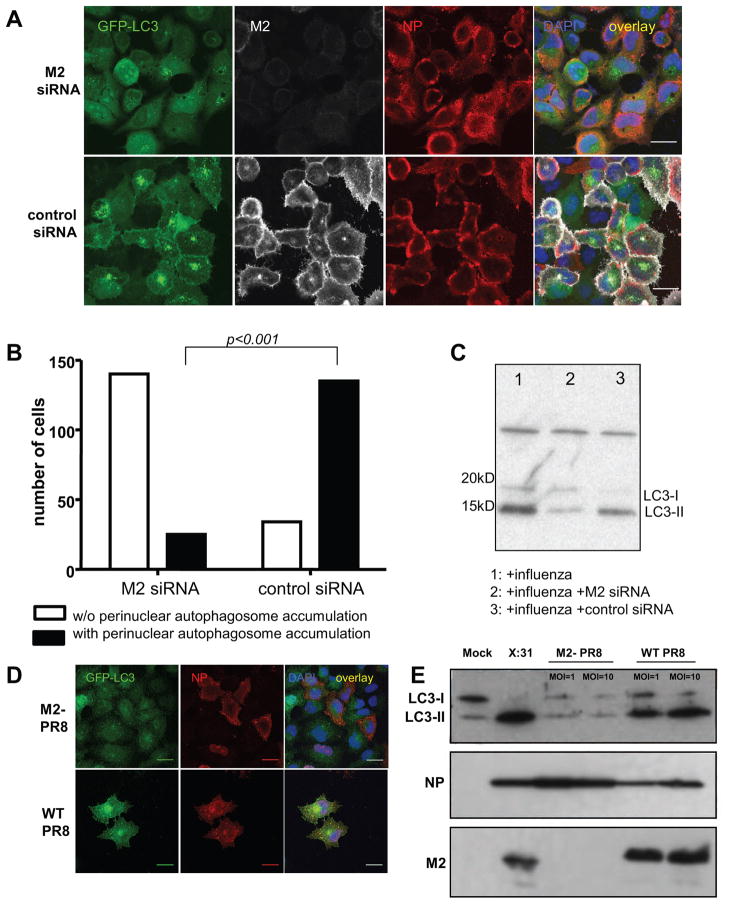
In order to demonstrate that M2 is necessary for the block of autophagosome degradation by influenza A virus infection, we used two strategies. In a first set of experiment we used M2 specific RNA interference, and in a second set of experiment we use a M2 knock out virus.
Silencing of M2 protein expression during influenza A virus infection significantly decreased autophagosomes accumulation in infected cells (Figure 5A). Indeed down-regulation of M2 by RNA interference reverted the phenotype of infected cells from containing perinuclear autophagosome accumulations to homogenous GFP-Atg8/LC3 distribution. This difference was statistically significant (p<0.001) (Figure 5B). Loss of M2 by this treatment did, however, not prevent infection by influenza A virus, because NP expression was similar in control small interfering RNA (siRNA) or M2 specific siRNA treated cells (Figure 5A). This loss-of-function by M2 silencing was also observed in Western blot analysis. Atg8/LC3-II upregulation during influenza A virus infection was significantly decreased upon M2 specific RNA interference (Figure 5C). To further confirm this finding we constructed a M2 knock-out virus on the A/PR8/34 influenza strain background by reverse genetics. This strain background was chosen, since it can only perform single round replications in cell culture, and therefore allows the analysis of cell biological effects of M2 loss without confounding effects due to blocked secondary infections. A549 cells infected with the M2 deficient virus did not show any accumulation of autophagosomes as depicted by the GFP-Atg8/LC3 fluorescence analysis (Figure 5D). We also could not detect any Atg8/LC3-II accumulation by Western blot analysis at 24 hours post infection with two different concentration of M2 knock-out virus virus (MOI=1 and MOI=10) (Figure 5E), while wild-type influenza A/PR8/34 virus, produced with the same reverse genetics approach was capable to arrest autophagosome degradation. This difference was not due to a diminished infectivity of the M2 deficient virus as documented by NP expression (immunofluorescence and western blot analysis Figure 5D and 5E). These data suggest that M2 protein is sufficient and necessary to inhibit fusion of autophagosomes with the lysosomal compartment and thereby prevents degradation of macroautophagy substrates during influenza A virus infection.
Figure 5. Loss of influenza A virus matrix protein 2 (M2) expression during infection prevents autophagosome accumulation.
(A) GFP-Atg8/LC3 transfected A549 cells were transfected with M2 specific siRNA or a control siRNA, and analyzed 24 hours after infection by fluorescence microscopy. In order to monitor influenza A virus infection and M2 expression, co-staining for NP and M2 was performed. Scale bar: 30 μm. One of three experiments is shown. (B) Summary of perinuclear autophagosomes accumulation in influenza A virus infected cells with and without M2 specific RNA silencing of three independent experiments. In each experiment at least 150 cells were analyzed per condition. Statistical analysis was performed by applying Pearson’s chi-squared test. (C) Western blot analysis of lipidated Atg8/LC3-II accumulation in influenza infected A549 cells transfected with M2 siRNA or control siRNA. One of three experiments is shown. (D) GFP-Atg8/LC3 transfected A549 cells at 24 hours post infection with M2 deficient M2- PR8) or wild-type influenza A virus (WT PR8). Infection was visualized by staining for NP. Scale bar: 100 μm. (E) Western blot analysis of Atg8/LC3-II, M2 and NP expression in influenza A virus infected A549 cells using the two recombinant influenza A viruses (M2- PR8 and WT PR8) at MOIs of 1 and 10, and influenza A/X:31 virus as a control. One of five experiments is shown.
The proton channel activity of matrix protein 2 is not involved in blocking autophagosome degradation
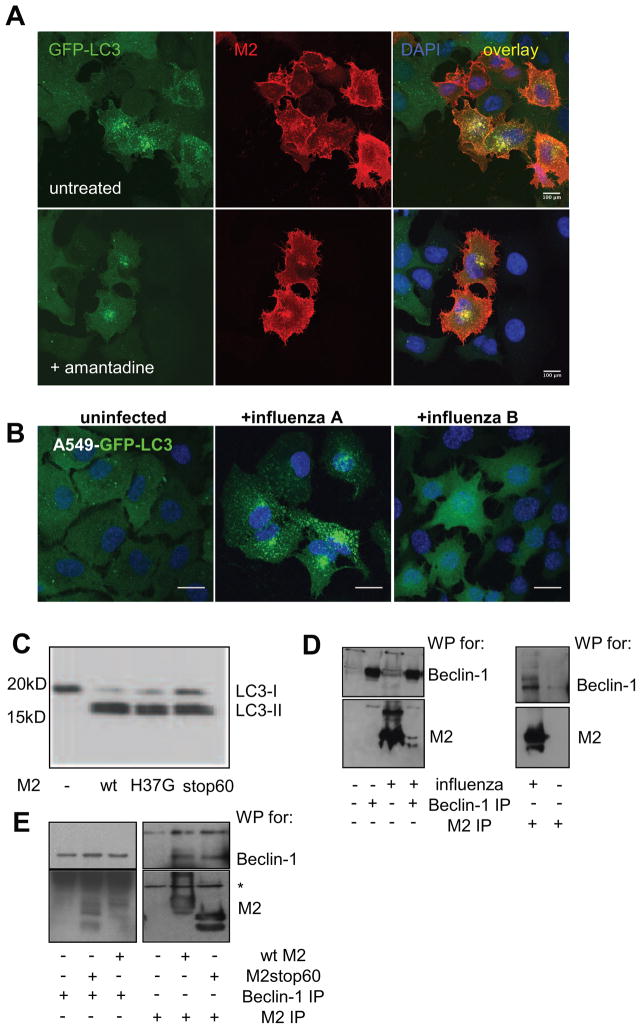
The major function of M2 is its activity as selective proton channel during viral entry. Therefore, we hypothesized that M2 localization in the autophagosomal membranes could prevent the acidification of these vesicles and thereby inhibit their efficient fusion with lysosomes (Yamamoto et al., 1998). We first used the antiviral drug amantadine hydrochloride (AMA), an amino-adamantane, known to inhibit the proton channel activity of M2 (Hay et al., 1985). This inhibitor was unable to prevent autophagosome accumulation in M2 transfected or influenza A virus infected cells (Figure 6A). In order to confirm that viral proton channel activity alone was not sufficient for autophagosome accumulation we tested influenza B virus infection. Influenza B virus’ proton channel BM2 facilitates influenza B virus acidification during its cellular entry (Mould et al., 2003). The homology between M2 of influenza A virus and the BM2 protein of influenza B virus is, however, restricted to the HXXXW motif of the membrane spanning residues, known to be critical for the proton channel activity (Ma et al., 2008). Surprisingly, influenza B infection did not result in autophagosomes accumulation (Figure 6B). Moreover, mutating the His37 of M2 to glycine (M2H37G), which abolishes proton channel activity (Stouffer et al., 2008), did not decrease lipidated Atg8/LC3-II accumulation in M2 transfected cells (Figure 6C). These data suggest that M2’s proton channel activity is not involved in blocking autophagosome fusion with lysosomes.
Figure 6. The proton channel function of M2 is not involved in autophagosomes accumulation.
(A) GFP-Atg8/LC3 transfected A549 human lung epithelial cells were infected with influenza A virus and incubated one hour later with and without amantadine at a concentration of 50μM. Cells were stained with M2 antibody and analyzed for GFP-Atg8/LC3 positive autophagosome accumulation by fluorescence microscopy. Nuclear DNA was stained with DAPI. Scale bar: 100 μm. Representative images of one out of three experiments are shown. (B) GFP-Atg8/LC3 transfected A549 cells were infected with influenza A or B and analyzed by fluorescence microscopy for autophagosome accumulation. DAPI was used to stain nuclear DNA. Scale bar: 100 μm. One of three experiments is shown. (C) A549 cells were transiently transfected with M2 wild type and M2 mutants (M2 H37G and M2 stop60) encoding plasmids. 24h post transfection Atg8/LC3-II accumulation was analyzed by Western blot analysis. One of three experiments is shown. (D) A549 cells were infected with X:31 virus, 24 hour later cell lysates from infected and non infected cells were immunoprecipitated using Atg6/Beclin-1 specific (left) or M2 specific antibodies (right). Western blotting was performed for Atg6/Beclin-1 and influenza A virus M2 detection. One of 3 experiments is shown. (E) A549 cells were transfected with full-length (wt) or the 60 N-terminal amino acids of M2 (stop60). Transfected or non-transfected cells were immunoprecipitated using Atg6/Beclin-1 or M2 specific antibodies (Atg6/Beclin-1 IP: left panel, M2 IP: right panel). Western blotting was performed for Atg6/Beclin-1 (top) and M2 (bottom) detection. * indicates immunoglobulin heavy chain of the antibodies used for immunoprecipitation. One of three experiments is shown.
We next investigated if the cytoplasmic domain of M2 was involved in autophagosome accumulation we constructed a truncated form of M2 encoding only for the first 60 amino acids (M21–60). M21–60 transfection was still able to cause accumulation of lipidated Atg8/LC3-II (Figure 6C). We conclude from these data that the C-terminal cytoplasmic domain of M2 is not involved in autophagosome accumulation during influenza A virus infection.
Finally, in order to get a first insight into how M2 blocks autophagosome fusion with lysosomes, we analyzed if it could interact with the molecular machinery of macroautophagy. Therefore, we performed immunoprecipitations of M2 to identify associated Atg proteins. We identified Atg6/Beclin-1 in M2 immunoprecipitates from influenza A virus infected A549 cells. Similarly, M2 co-immunoprecipitated with Atg6/Beclin-1 from infected cells (Figure 6D). Furthermore, the N-terminal 60 amino acids of M2 were able to interact with Beclin-1 (Figure 6E). M21–60 and full-length M2 were co-immunoprecipitated with Atg6/Beclin-1 to a similar degree. These results give a first indication that M21–60 might interfere with the Atg6/Beclin-1 and UVRAG containing PI-3 kinase complex, which has been recently described to facilitate autophagosome fusion with lysosomes (Matsunaga et al., 2009; Zhong et al., 2009)(Itakura et al., 2008).
Thus M2 of influenza A virus blocks autolysosome formation with its N-terminal 60 amino acids, independently of its proton channel function, possibly through interfering with the Atg6/Beclin-1 and UVRAG containing PI-3 kinase complex.
Macroautophagy deficient cells are more susceptible to apoptosis upon influenza infection
In order to understand the functional relevance of macroautophagy inhibition upon influenza A virus infection, we used wild-type and macroautophagy deficient mouse embryonic fibroblasts (MEFs). Loss of macroautophagy was achieved by knockout of the essential macroautophagy gene atg5 (Kuma et al., 2004). Although wild-type and Atg5−/− MEFs were similarly susceptible to apoptosis as measured by Annexin V and 7-AAD staining in flow cytometry, influenza A virus infection induced significantly more cell death in macroautophagy deficient MEFs across a broad range of different infectious doses (Figure 7A). Moreover, this susceptibility to apoptosis of Atg5−/− MEFs was observed at several timepoints (6, 12 and 24h) after influenza A virus infection (Figure 7B). Across the different infectious doses and infection timepoints influenza A infected Atg5−/− MEFs consistently contained up to 4 fold more apoptotic and secondary necrotic cells than infected wild-type MEFs (paired t test p=0.005). The difference in apoptosis was most pronounced 12h after infection and at an infectious viral dose of 120 HAU/106 cells. Thus loss of autophagosome formation compromises the survival of influenza infected cells. Furthermore, loss of the M2 block of autophagosome degradation resulted in increased survival of M2 knock-out influenza A virus infected cells (Figure 7C and D). The cell death was analysed 24 hours after the infection of A549 lung epithelial cells with M2 deficient and wild-type virus. Therefore, allowing completion of macroautophagy by lifting the M2 mediated block of autophagosome fusion with lysosomes enhances survival of influenza infected cells. These data suggest that macroautophagy serves as a survival pathway in influenza infected cells, since its deficiency leads to enhanced apoptosis, and restoring completion of macroautophagy leads to decreased apoptosis.
Figure 7. Macroautophagy inhibition enhances cell death after influenza A virus infection.
(A) Wild-type (Atg5+/+) and macroautophagy deficient (ATG5−/−) MEFs were infected with the indicated doses of influenza A virus (hemagglutinin units [HAU] per 106 cells), and analyzed 24 hours later by flow cytometry. Staining for M2, 7-AAD and annexin V was performed. Cells were gated on M2 positive cells, and apoptosis (annexin V positive) and secondary necrosis (annexin V and 7-AAD positive) cells were quantified. The percentage of double positive cells (annexin V and 7-AAD positive) is significantly higher in ATG5−/−cells (paired t test p=0.005). One of five experiments is shown. (B) Cell death induction was analyzed as in (A), but with one constant influenza A virus infection dose (1200HAU/106 cells) at the indicated time points. One of three experiments is shown. (C). A549 cells were infected with the M2 deficient or the wild-type recombinant virus at an MOI of 1 and cell death was analyzed at 24 hours post infection. Cells were gated on infected cells, (anti-H1N1 FITC) and apoptosis (annexin V positive) and secondary necrosis (annexin V and 7-AAD positive) cells were quantified. (D) Composite data on five independent experiments comparing apoptosis (Annexin V+) and secondary necrosis (7AAD+ Annexin V+) after M2 deficient (M2- PR8) and wild-type (WT PR8) influenza A virus infection of A549 cells.
Macroautophagy deficiency enhances viral protein release, but not viral replication
In order to investigate if this enhanced cell death of macroautophagy deficient cells after influenza A virus infection, affects virus release, we performed HA specific ELISA assays with the supernatants of infected Atg5+/+ and Atg5−/− MEFs. We detected increased viral protein release from infected macroautophagy-deficient cells (Supplemental Figure 2A). Four times more HA was released from infected Atg5 deficient MEFs than from infected wild-type cells. Furthermore, the viral protein content was higher in infected macroautophagy-competent than in macroautophagy-deficient cells (Supplemental Figure 2B). Similarly, more viral RNA was retained in Atg5 positive than negative cells after influenza A virus infection (7.94 fold ± 2.68; p=0.0005). This values were normalized to GAPDH (Supplemental Figure 2C left), These data suggest that macroautophagy competent cells retain more viral protein and RNA within influenza A virus infected cells.
However, we did not detect higher viral titers in the supernatants of macroautophagy competent cells after influenza A virus infection (Supplemental Figure 2D). In these experiments, influenza A virus titers were determined by MDCK plaque assay. No significant differences were detected in virus infectivity from supernatant collected from Atg5+/+ and Atg5−/− MEFs. Overall these data suggest that macroautophagy competent cells accumulate more viral RNA and viral proteins than macroautophagy deficient cells, but this does not result in higher infectious virus release.
DISCUSSION
Our study characterizes inhibition of macroautophagy by influenza A virus at the check-point of autophagosome fusion with lysosomes. We identify M2 as the viral protein mediating this block in autophagosome degradation, and document that macroautophagy down-modulation leads to enhanced virus induced cell death of infected cells and elevated viral antigen release. These findings establish M2 as the first viral inhibitor of autophagosome fusion with lysosomes.
However, influenza A virus is not the first pathogen that has been described to interfere with macroautophagy. Indeed, as early as 1965 Sam Dales and colleagues documented that poliovirus infection leads to the accumulation of double-membrane coated vesicles (Dales et al., 1965). Later, it was reported that the formation of these membrane compartment depends on some components of the macroautophagic machinery and is required for efficient viral replication (Jackson et al., 2005). Similarly, hepatitis C virus (HCV) infection leads to the accumulation of Atg8/LC3 positive vesicles, while not affecting the turn-over of classical macroautophagy substrates (Sir et al., 2008). The accumulating vesicles seem to be required for HCV replication. However, even so autophagic replication compartments of pathogens seem to be stabilized and not degraded within minutes as regular autophagosomes, an individual protein blocking autophagosome fusion with lysosomes has not been identified to date (Münz, 2009). In contrast to this second check-point of macroautophagic flux, inhibition of autophagosome formation, as a first check-point during macroautophagy, has been reported and characterized in more detail for several pathogens. Herpesviruses have proven a rich source of inhibitors of macroautophagy. Members of the γ-herpesviridae, namely Kaposi Sarcoma associated herpesvirus (KSHV) and mouse herpesvirus 68 (MHV-68), have been demonstrated to encode Bcl-2 homologues that inhibit autophagosome initiation via their binding to Atg6/Beclin-1, a component of the class III PI-3 kinase complexes that are required for macroautophagy (Ku et al., 2008) (Pattingre et al., 2005)(Sinha et al., 2008). Similarly, the α-herpesvirus herpes simplex encodes ICP34.5, which also contains an Atg6/Beclin-1 interacting domain, which inhibits macroautophagy (Orvedahl et al., 2007). Interestingly, HSV1 with an ICP34.5 protein deficient in this domain is attenuated in its neurovirulence in mice. Consistent with this role of macroautophagy for resistance to HSV infection in vivo is that Sindbis virus, transgenic for Atg6/Beclin-1 and thereby enhancing macroautophagy upon infection, is attenuated in mice (Liang et al., 1998). While our study was being prepared for publication, influenza A virus was also reported to affect autophagosome formation to enhance its replication (Zhou et al., 2009). While we cannot exclude that influenza A virus might up-regulate macroautophagy via innate immune recognition by PAMP receptors in certain cell types (Delgado et al., 2008), we could only identify one viral protein, namely M2, that blocks autophagosome degradation and this was also the main mechanism of autophagosome accumulation in human lung epithelial cells after infection with a variety of influenza A virus strains. Interestingly, M2 might also target Atg6/Beclin-1 for this function, possibly interfering with the Atg6/Beclin-1 and UVRAG containing PI-3 kinase complex, which was recently characterized to support autophagosome fusion with lysosomes (Matsunaga et al., 2009; Zhong et al., 2009)(Itakura et al., 2008). Therefore, pathogens target macroautophagy at two check-points, during autophagosome formation and autophagosome fusion with lysosomes, and we have characterized the first viral protein blocking autophagosome degradation.
Apart from mediating immunity, as has been demonstrated for other pathogens, we demonstrate that macroautophagy can also influence the death of the pathogen’s host cell. In addition to regulation of the pro-survival pathway of macroautophagy by influenza A virus, several viruses influence apoptosis of infected cells (Galluzzi et al., 2008). Although, one would intuitively assume that it is in the virus’ interest to prolong the life of infected cells in order to maximize viral replication, pro- and anti-apoptotic mechanisms are employed by viruses to nearly the same extent (Galluzzi et al., 2008). While more anti-apoptotic proteins have been identified in DNA viruses, like Bcl-2 homologues of herpesviruses, RNA viruses encode more pro-apoptotic factors. Among the most prominent pathogens that trigger apoptosis, are the RNA viruses human immunodeficiency virus (HIV) and influenza. Viral protein R (Vpr) of HIV directly interacts with adenine nucleotide translocator (ANT) and voltage-dependent anion channel (VDAC) in the mitochondrial membrane to facilitate mitochondrial membrane permeabilization and cytochrome C release, triggering apoptosis (Jacotot et al., 2001)(Jacotot et al., 2000)(Muthumani et al., 2002). Apoptosis induction by Vpr is thought to contribute to depletion of HIV’s host cells, primarily CD4+ T cells, and progression to the acquired immunodeficiency syndrome (AIDS). Indeed, a mutation that decreases apoptosis induction by Vpr has been identified in long-term nonprogressors (Lum et al., 2003). Furthermore, influenza A virus encodes the pro-apoptotic protein PB1-F2 (Chen et al., 2001). It inserts into the mitochondrial membrane (Gibbs et al., 2003), causing pores in lipid bilayers (Chanturiya et al., 2004), and also directly interacts with ANT and VDAC (Zamarin et al., 2005). Although it is not entirely clear if PB1-F2 causes mitochondrial membrane permeabilization by regulation of ANT and VDAC, or its direct pore forming capacity, this pro-apoptotic function of influenza A virus is required for pathogenesis in mice (Zamarin et al., 2006). Furthermore, PB1-F2 is in part responsible for the increased pathogenesis of the H5N1 “bird flu” and the 1918 “Spanish flu” (McAuley et al., 2007)(Conenello et al., 2007). Indeed, only a single amino acid mutation was responsible for the increased pathogenicity of PB1-F2 of these highly virulent influenza A strains (Conenello et al., 2007). These studies suggest that RNA viruses like influenza induce apoptosis and thereby regulate the cell death of their host cells.
Even so pro-apoptotic proteins have now been identified from many pathogens and we show in this study that influenza inhibits the prosurvival pathway of macroautophagy, the functional benefit of enhanced host cell death during infection still remains largely unclear. We propose three possible advantages of macroautophagy inhibition for influenza A virus. Firstly, depletion of immune system cells and lung epithelial cells by apoptosis could facilitate influenza A virus infection by limiting virus specific immune responses and compromising the lung barrier function. Within the cells of the immune system, macrophages have been found to be especially sensitive to influenza A virus induced cell death (Bender et al., 1998), and highly pathogenic influenza virus strains were superior in lung lesion induction and dendritic cell depletion early during influenza A virus infection of macaques (Baskin et al., 2009). Secondly, influenza A virus induced apoptosis has been demonstrated to limit proinflammatory cytokine production (Brydon et al., 2003). In good agreement, we find that viral RNA, which can be recognized as a pathogen associated molecular pattern (PAMP) to trigger inflammatory cytokine responses, is more readily retained in macroautophagy competent cells after influenza infection (Supplemental Figure 2). Therefore, induction of apoptotic cell death by influenza A virus in macroautophagy competent cells could limit the immunogenicity of viral infection. Along these lines, it has been suggested that apoptotic bodies are less immunogenic than necrotic cell debris (Sauter et al., 2000). Moreover, ATP production by macroautophagy seems required to expose phosphatidylserine on apoptotic bodies for efficient engulfment by phagocytes, and for the production of lysophosphatidylcholine to attract macrophages (Qu et al., 2007). Thirdly, we demonstrate that macroautophagy competent cells retain more virus derived antigens after influenza infection. The enhanced viral antigen release by macroautophagy deficient cells (Supplemental Figure 2) could increase the antigenicity of influenza virus infection, which might be avoided by trapping of viral antigens in autophagosomes, and preventing their degradation for direct presentation on MHC molecles via M2. Therefore, influenza A virus induced cell death by apoptosis induction and macroautophagy inhibition could eliminate immune and barrier cells that limit viral spreading, and render the death of infected cells as little immunogenic as possible. Since PB1-F2 is mainly expressed during later stages of the infection cycle (Chen et al., 2001) and we observed autophagosome accumulation also only at later timepoints after infection (≥10h), the virus might induce host cell death with low immunogenicity in an ordered fashion after efficient initial replication.
In summary, we have identified the influenza A virus protein M2 as an inhibitor of autophagosome fusion with lysosomes, and this inhibition seems to target with macroautophagy a pathway that prevents apoptosis of infected cells.
EXPERIMENTAL PROCEDURES
Cell lines
The following epithelial cell lines were used: the human keratinocyte cell line HaCat (a gift of Dr. Rajiv Khanna, Brisbane, Australia), the human breast carcinoma cell line MDAMC (a gift from Dr. Irene Joab, Paris, France), the human lung epithelium cell line A549 (a gift from Dr. Thomas Moran, New York, NY), the mouse lung epithelium cell line MLE-12 (a gift from Dr. Arnoud Didierlaurent, London, UK), the canine kidney epithelium cell line MDCK (ATCC, Manassas, VA), the human cervix carcinoma cell line HeLa (ATCC, Manassas, VA); and the wild-type and Atg5 deficient mouse embryonic fibroblasts (a gift from Dr Noboru Mizushima Tokyo, Japan). All epithelial cell lines were routinely cultured in DMEM with 10% fetal calf serum (FCS, Sigma), 2 mM glutamine, 110 μg/ml sodium pyruvate and 2 μg/ml gentamycin, except MLE-12, which was cultured in a 50:50 mix of DMEM:Ham’s F12, supplemented with 2% FCS, 5 μg/ml insulin, 10 μg/ml transferrin, 30 mM sodium selenite, 10 nM hydrocortisone, 10 nM beta-estradiol, 2 mM glutamine and 2 μg/ml gentamycin. Epithelial cell monolayers were detached by one wash in PBS/0.5 mM EDTA followed by incubation in 0.05% trypsin/0.53 mM EDTA (Gibco). Cell lines stably expressing GFP-LC3 were generated by lentiviral infection with MOIs of 10–40.
Antibodies
Primary antibodies
Anti-LC3 antiserum was generated as previously described (Schmid et al., 2007) by immunizing two rabbits with the N-terminal peptide LC31–15 (MPSEKTFKQRRTFEQR) conjugated to KLH carrier protein (Cocalico Biologicals). The rat anti-Flag antibody was a gift from Dr. Cheolho Cheong (New York, USA). The following antibodies specific for influenza proteins were used: two influenza M2 specific monoclonal antibodies, M2-E10, a gift from Dr Tom Moran, (New York, USA), M2-Clone 14C2 (Affinity Bioreagents); for influenza MP1 a specific rabbit antiserum a gift from Dr. Ari Helenius, (Zürich, Switzerland); for influenza NP a specific rabbit antiserum (a gift from Dr Peter Palese, New York, USA), for NS1 a monoclonal antibody (a gift from Dr Peter Palese, New York, USA). For human p62/sequestosome1, a specific guinea pig antiserum (C-terminus specific, American Research Products, Belmont, MA), and monoclonal antibodies against LAMP1 (clone H4A3, Southern Biotech, Birmingham, Alabama), against actin (clone AC-40, Sigma, St. Louis, MO), and against polyubiquitinated proteins (clone FK-1, Cosmo Bio Ltd., Carlsbad, CA) were used. For Atg6/Beclin-1 detection, we used a goat anti-Atg6/Beclin-1 (Santa Cruz, Biotechnology) for immunoprecipitation and a rabbit polyclonal antiserum (Novus Biologicals) for Western-blotting.
Secondary antibodies
For immunofluorescence microscopy we used RhodamineRed-X (RRX)-conjugated, or Cy-3 conjugated donkey anti-mouse from (Jackson ImmunoResearch, West Grove, PA), Alexa fluor-568 conjugated goat anti-rat, Alexa fluor-555 conjugated donkey anti-rabbit, Alexa fluor-647 conjugated rat anti-mouse and Alexa fluor- 647 conjugated goat anti-mouse (all from Invitrogen),
Flow cytometry
Cells were labeled using the following antibodies: For detection of influenza A virus infection anti-H1N1 FITC (ABR) or anti-M2 (ABR) antibodies were used. For apoptosis detection 7-Amino-actinomycin D (7-AAD) (BD) and Annexin V-APC (BD) staining was done using BD Annexin V binding buffer. Acquisition was performed within the hour following the staining, on a BD FACSCanto Flow Cytometer. Analysis was performed using the FlowJo software.
Inhibitors, synthetic peptides and cytokines
Chloroquine and amantidine were purchased from Sigma, St. Louis, MO.
Expression plasmids and lentiviral constructs
The cDNA of human MAP1LC3B sequence (NM_022818) was PCR-amplified from a human B-LCL by RT-PCR with gene specific primers and cloned into the multiple cloning site of the mammalian expression vector pEGFP-C2 (Clontech). The mRFP-GFP-LC3 encoding expression vector was a gift from Dr. Tamotsu Yoshimori, Osaka, Japan. For the expression of individual viral proteins pCAGGS vectors containing influenza A/WSN/33 matrix protein 1 (M1), matrix protein 2 (M2), NP, NS1, PB2 and HA were used. The M2 and NP constructs contain a FLAG tag at their C terminus.
siRNA-mediated gene silencing
For delivery of siRNAs into epithelial cell lines, siRNAs were transfected with lipofectamine 2000 (Invitrogen), using 200 pmol siRNA + 7μl lipofectamine/well in a 6-well format. 18 hours after the transfection, cells were infected and analyzed after 24 hours. M2 knockdown efficiency was analyzed by flow cytometric analysis, using the M2-E10 antibody, and by Western blot analysis using the M2 14-C2 antibody.
The Atg12 specific siRNA was purchased from Dharmacon composed of Atg12 sense 5′-GUGGGCAGUAGAGCGAACAUdT-3′ and Atg12 antisense 5′-UCAUGUAGUAGCAAGUUGAUdT-3′. The M2 specific siRNA was purchased from Invitrogen (stealth modified): M2 sense 5′-UUUCUGAUAGGCGUUUCGACCUCGG-3′ and M2 antisense 5′-CCGAGGUCGAAACGCCUAUCAGAAA-3′.
Influenza A virus infection
For most experiments, influenza A virus strain Aichi X:31, A/Aichi/68 (H3N2) (purified virus, purchased from Charles River Laboratories) was used. Where indicated, Influenza B/Lee/40 (from Charles River Laboratories), influenza A/PR8/34 and A/PR8/34DNS1 (H1N1) (Garcia-Sastre et al., 1998), or A/WSN/33 and A/WSN/33 PB1F2 (H1N1) (Zamarin et al., 2005) were used. Before infection, cells were washed three times in RPMI-1640 to remove FCS and then were incubated with influenza virus in a small volume of RPMI-1640 for 1 h at 37°C (MOI = 0.1–2). After one hour, cells were washed once in PBS, than incubated in culture medium (DMEM + 10% FCS + glutamine + gentamicin) for the indicated amount of time, usually 24 hours. For heat-inactivation, virus was incubated in a 56°C water bath for 30 min and then was added to cells as described above.
M2 deficient recombinant influenza A/PR8/34 virus generation
A recombinant influenza virus that expresses only the extracellular domain (aa 1 to 25) of the M2 was generated according to methods described elsewhere (McCown and Pekosz, 2005). The A/WSN/33 M segment was mutated such that codons 27 and 28 of the M2 open reading frame were mutated to stop codons by PCR mutagenesis (Supplementary Figure 3A). PCR reactions included one of two primer pairs (each primer sequence is relative to the virus cRNA): 1) [forward primer Ambi-A] 5′-GATCGCTCTTCTGGGAGCAAAAGCAGG-3′ and [reverse ΔM2 primer] 5′-GGTGCAGATGCAACGATTCAAGTGATTGATAGGTCATTGCAGCAAA-3′; or ii) [forward ΔM2 primer] 5′-AGATGCAACGATTCAAGTGATTGATAGGTCATTGCAGCAAATATCATTGG-3′ and [reverse primer NC-SAP] 5′-CCTTGTTTCTACTAATAGAAGAGCGATG-3′. The forward ΔM2 primer and reverse ΔM2 primer retained the BsrDI restriction enzyme site which facilitated cloning of the full-length mutated M segment. The resulting WSN M segment only expresses the extracellular domain (aa 1–26) of the M2 protein. Recombinant A/Puerto Rico/8/1934 (A/PR8/34) possessing the wild-type WSN M segment (PR8:WSN M) or the ΔM2 WSN M segment (PR8:WSN M ΔM2) was rescued from plasmid transfections as described elsewhere (Fodor et al., 1999)(Quinlivan et al., 2005). Sequencing of the rescued PR8:WSN M ΔM2 virus confirmed the retention of the two stop codons in the M segment (Supplementary Figure 3B). Plaque assays indicated that while the PR8:WSN M virus formed plaques on wild-type or M2 complementing MDCK cells, the PR8:WSN M ΔM2 virus could not form plaques on non-complementing MDCK cells (Supplementary Figure 3C).
Immunocytochemistry and confocal microscopy
Epithelial cells were grown on microscopy cover glasses in 24 well plates. Cells were fixed in 3% paraformaldehyde in PBS for 15 min and permeabilized in 0.1% Triton X-100 in PBS for 3 min. Primary and secondary antibodies were applied in PBS 1X 0.1% saponin + 10% of serum from the species of the secondary antibody for 45–60 min, followed by three 5 min-washes in PBS 1X.
Finally, cells were stained with DAPI nucleic acid stain (Invitrogen-Molecular Probes) for 1 min and cover glasses were mounted onto microscope slides using Aqua Polymount (Polysciences) or Prolong Gold antifade reagent (Invitrogen-Molecular Probes). All steps were carried out at room temperature. Cells were analyzed either on an Olympus wide-field microscope with a 60x or 100x/1.4 N.A. oil immersion lens and pictures were processed with the Image J or the Metamorph Software (Universal Imaging Corporation) or on an inverted LSM 510 laser scanning confocal microscope (Zeiss Axiovert 200) with a 63 or 100x/1.4 N.A. oil immersion lens using a pinhole diameter of 1 Airy unit. Pictures were taken with the LSM 510 confocal software (Zeiss). For colocalization analysis, serial optical sections were acquired, and images were then deconvoluted using Huygens software. Pearson coefficients were calculated using the Imaris 6.3.0 or Image J software.
Live cell imaging
Cells were grown on 35 mm culture dishes with No. 1.5 coverglass inserts (MatTek Corporation) and infected with influenza virus as described above. 24 hours post-infection, medium was replaced by CO2-independent Medium 199 supplemented with 10% FCS, 2 mM glutamine and 25 mM Hepes (Gibco) prewarmed to 37°C. For staining of acidic compartments, 50 nM Lysotracker Red (Molecular Probes) was added to the medium. After 30 min, medium was replaced with fresh supplemented Medium 199 and cells were analyzed on an inverted Zeiss Axiovert 200 microscope equipped with an UltraView spinning disk confocal head (Perkin-Elmer) and a 37°C environmental chamber, using a 63x/1.4 N.A. oil immersion lens. Pictures were taken at various intervals with a Hamamatsu Orca ER cooled CCD camera using Metamorph Software (Universal Imaging Corporation).
Electron microscopy
Human lung epithelium A549 cells with and without influenza infection were fixed for 1h at RT with 4% paraformaldehyde (PFA, Electron Microscopy Sciences) in 0.25 M Hepes, pH 7.4, followed by overnight fixation at 4°C in 8% PFA/Hepes. Cells were embedded in 5% gelatin in PBS, small pieces of gelatin pellets were infiltrated overnight at 4°C with 2.3 M sucrose in PBS, mounted onto cryospecimen pins and frozen in liquid nitrogen. Ultrathin sections (80 nm) were cut using a Leica ultracut ultramicrotome with an FCS cryoattachment at −108°C and collected on formvar- and carbon-coated nickel grids using a 1:1 mixture of 2% methyl cellulose (25 centipoises; Sigma) and 2.3 M sucrose in PBS. Sections were infiltrated for 10 min on ice with a mixture of 1.8% methylcellulose and 0.5% uranyl acetate (Electron Microscopy Sciences), washed 3× in 0.5% uranyl acetate/1.8% methylcellulose and air-dried. Samples were analyzed in a Tecnai 12 Biotwin (FEI) microscope and pictures were taken using Kodak 4489 film.
Lysate preparation, SDS-PAGE and immunoblotting
Cells were lysed in ice cold lysis buffer (50 mM Tris-HCl pH 8.0, 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40 with Complete protease inhibitor cocktail from Roche) for 10 min on ice (about 106 cells/200μl). Whole cells and cell debris were pelleted by low speed centrifugation (400 g, 3 min) and cleared supernatants were transferred to a new tube. Protein concentration was determined by BCA protein assay (Pierce). Samples were boiled for 5 min in the presence of 4x SDS-PAGE-loading buffer (250 mM Tris-HCl pH 6.8, 40% glycerol, 8% SDS, 0.57 M β-mercaptoethanol, 0.12% bromophenol blue). Equal amounts of protein were run on 12% SDS-PAGE gels and transferred onto a PVDF membrane (Hybond-P, Amersham Biosciences). Primary antibodies were visualized with HRP-conjugated goat anti-rabbit or anti-mouse IgG and the ECL plus detection system (Amersham Biosciences).
Immunoprecipitation
Cells were lysed in isotonic TBS (150mM Nacl, 50mM Tris-HCl, pH=8) plus 1% NP40 and 1% Triton X-100 as well as complete protease inhibitors (Roche). Lysates were pre-cleared for 1 hour and supernatants were immunoprecipitated overnight at 4C with Proteine-G sepharose beads (GE Healthcare) coupled with the antibody of interest. Immunocomplexes were washed 4 times and subjected to western-blotting.
Hemagglutinin ELISA assay
An enzyme immunossay kit specific for Influenza A virus by GenWay (San Diego, CA, USA) was used to determine viral protein release in culture supernatants of infected cells.
Quantification of viral RNA
Cellular RNA was extracted using a Qiagen RNeasy Mini kit and viral RNA from the supernatants was isolated using a Qiagen viral RNA isolation kit. First strand cDNA was synthesized using Improm-II reverse transcription (RT) kit (Promega, Dübendorf, Switzerland). RT product was diluted 500 fold and used as a template for quantitative PCR (qPCR) on a BioRad MyIQ detection system with Platinum Quantitative PCR SuperMix-UDG (Invitrogen). The following specific forward (F) and reverse (R) primer pairs (all Eurofins MWG, Ebersberg, Germany) were used:
GAPDH-F: 5′-ATGGGGAAGGTGSSGGTCG-′;
GAPDH-R 5′-GGGTCATTGATGGCAACAATATC-3′;
WSN-M-F: 5′-TAGCCAGCACTACAGCTAAG-3′;
WSN-M-R: 5′-GGCCTGACTAGCAATATCCA-3′;
WSN-HA-F 5′-TAACCTGCTCGAAGACAGAC-3′;
WSN-HA-R: 5′-AGAGCCATCCGGTGATGTTA-3′;
WSN-NA-F 5′-TTGGTCAGCAAGTGCATGTC-3′;
WSN-NA-R: 5′-ACAGCCACTGCTCCATCATC-3′.
Relative concentration of viral RNA in the cells was determined by analysis of cycle threshold values (CT), normalizing the mean CT for WSN-M, WSN-HA and WSN-NA to the expression of the product of the housekeeping gene GAPDH. Viral RNA content in the supernatants of Atg5−/− and ATG5+/+ Mefs was determined by comparison of the mean CT for WSN-M, WSN-HA and WSN-NA.
Influenza virus titration
Viral titers were determined from supernatants of Atg5+/+ and Atg5−/− cells infected with the influenza A/WSN/33 virus. Briefly, aliquots of 1 ml of serial 10-fold dilutions of infectious supernatants collected at 24 hours post-infection, were inoculated into Madin-Darby canine kidney (MDCK) cells in 6-well plates. After 1 hour of incubation at 37 C, each well was overlaid with 2 ml of agar medium. 48 hours later, the number of plaques in each well was determined.
Statistics
Statistical analyses were performed with the paired two-tailed Student t-test, the Mann-Whitney U test or the Pearson’s chi-squared test as indicated. The p value of significant differences is reported. Plotted data represent mean plus standard deviation (SD), unless otherwise stated.
Supplementary Material
Supplemental Figure 1: Autophagosome accumulation occurs only in directly and productively influenza A virus infected cells. (A) A549 human lung epithelial cells were infected with influenza A virus and lysates were prepared at the indicated timepoints after infection. Lipidated Atg8/LC3-II content was analyzed by Western blot analysis. One of three experiments is shown. (B) GFP-Atg8/LC3 transfected A549 human and MLE-12 mouse lung epithelial cells were incubated with live or heat-inactivated influenza A virus. Endogenous lipidated Atg8/LC3-II and transfected lipidated GFP-Atg8/LC3-II levels were analyzed by Atg8/LC3 Western blot analysis after 24h. One of three experiments is shown. (C) GFP-Atg8/LC3 transfected MDAMC cells were exposed to live or heat-inactivated influenza A virus. GFP-Atg8/LC3 positive autophagosomes were visualized by fluorescence microscopy 24h later. Nuclear DNA was stained with DAPI. Scale bar: 80 μm. One of three experiments is shown. (D) GFP-Atg8/LC3 transfected MDAMC cells were infected with influenza A virus and matrix protein 1 (M1) expression was analyzed by immune fluorescence microscopy. Nuclear DNA was stained in addition with DAPI. Scale bar: 80 μm. One of three experiments is shown.
Supplemental Figure 2: Macroautophagy deficiency does not affect viral production. (A) HA units ELISA: culture supernatants from Atg5−/− and Atg5+/+ MEFs were collected 24 hours after influenza A infection, 6 independent experiments were analysed (paired t test p=0.010). (B) Western blot analysis of viral protein content (M2 and NP) in both cell types (C) Relative concentration of viral RNA in the cells was determined by analysis of cycle threshold values (CT), normalizing the mean CT for WSN-M, WSN-HA and WSN-NA to the expression of the product of the housekeeping gene GAPDH. (left graph). Viral RNA content in the supernatants of Atg5−/− and wt Mefs was determined by comparison of the mean CT for WSN-M, WSN-HA and WSN-NA (right graph). 3 independants experiments were analyzed. (D) Viral titer: Viral titer of culture supernatants from Atg5−/− and Atg5+/+ MEFs collected 24 hours post infection. Three independent experiments are shown (paired t test p=0.7 ns).
Supplemental Movie 1: Mobility of autophagosomes in influenza A virus infected cells. Live cell imaging of GFP-Atg8/LC3 transfected A549 human lung epithelial cells was performed to analyze autophagosome mobility.
Acknowledgments
This research is partly supported by grants from the National Cancer Institute (R01CA108609 and R01CA101741) and from the Foundation for the National Institutes of Health (Grand Challenges in Global Health) to CM, and by NIAID grants U54AI57158, U19AI62623 and an NIAID contract to support a Center for Research on Influenza Pathogenesis (CRIP, HHSN266200700010C) to AG-S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baskin CR, Bielefeldt-Ohmann H, Tumpey TM, Sabourin PJ, Long JP, Garcia-Sastre A, Tolnay AE, Albrecht R, Pyles JA, Olson PH, et al. Early and sustained innate immune response defines pathology and death in nonhuman primates infected by highly pathogenic influenza virus. Proc Natl Acad Sci U S A. 2009;106:3455–3460. doi: 10.1073/pnas.0813234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A, Albert M, Reddy A, Feldman M, Sauter B, Kaplan G, Hellman W, Bhardwaj N. The distinctive features of influenza virus infection of dendritic cells. Immunobiology. 1998;198:552–567. doi: 10.1016/S0171-2985(98)80078-8. [DOI] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brydon EW, Smith H, Sweet C. Influenza A virus-induced apoptosis in bronchiolar epithelial (NCI-H292) cells limits pro-inflammatory cytokine release. J Gen Virol. 2003;84:2389–2400. doi: 10.1099/vir.0.18913-0. [DOI] [PubMed] [Google Scholar]
- Chanturiya AN, Basanez G, Schubert U, Henklein P, Yewdell JW, Zimmerberg J. PB1-F2, an influenza A virus-encoded proapoptotic mitochondrial protein, creates variably sized pores in planar lipid membranes. J Virol. 2004;78:6304–6312. doi: 10.1128/JVI.78.12.6304-6312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O’Neill R, Schickli J, Palese P, et al. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med. 2001;7:1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Cell Death Differ. 2005;12:1178–1190. doi: 10.1038/sj.cdd.4401692. [DOI] [PubMed] [Google Scholar]
- Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 2007;3:1414–1421. doi: 10.1371/journal.ppat.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dales S, Eggers HJ, Tamm I, Palade GE. Electron Microscopic Study of the Formation of Poliovirus. Virology. 1965;26:379–389. doi: 10.1016/0042-6822(65)90001-2. [DOI] [PubMed] [Google Scholar]
- Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. Embo J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fengsrud M, Raiborg C, Berg TO, Stromhaug PE, Ueno T, Erichsen ES, Seglen PO. Autophagosome-associated variant isoforms of cytosolic enzymes. Biochem J. 2000;352(Pt 3):773–781. [PMC free article] [PubMed] [Google Scholar]
- Fodor E, Devenish L, Engelhardt OG, Palese P, Brownlee GG, Garcia-Sastre A. Rescue of influenza A virus from recombinant DNA. J Virol. 1999;73:9679–9682. doi: 10.1128/jvi.73.11.9679-9682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L Complex Specifies the Site of LC3 Lipidation for Membrane Biogenesis in Autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Brenner C, Morselli E, Touat Z, Kroemer G. Viral control of mitochondrial apoptosis. PLoS Pathog. 2008;4:e1000018. doi: 10.1371/journal.ppat.1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- Gibbs JS, Malide D, Hornung F, Bennink JR, Yewdell JW. The influenza A virus PB1-F2 protein targets the inner mitochondrial membrane via a predicted basic amphipathic helix that disrupts mitochondrial function. J Virol. 2003;77:7214–7224. doi: 10.1128/JVI.77.13.7214-7224.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Hay AJ, Wolstenholme AJ, Skehel JJ, Smith MH. The molecular basis of the specific anti-influenza action of amantadine. Embo J. 1985;4:3021–3024. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson WT, Giddings TH, Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacotot E, Ferri KF, El Hamel C, Brenner C, Druillennec S, Hoebeke J, Rustin P, Metivier D, Lenoir C, Geuskens M, et al. Control of mitochondrial membrane permeabilization by adenine nucleotide translocator interacting with HIV-1 viral protein rR and Bcl-2. J Exp Med. 2001;193:509–519. doi: 10.1084/jem.193.4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacotot E, Ravagnan L, Loeffler M, Ferri KF, Vieira HL, Zamzami N, Costantini P, Druillennec S, Hoebeke J, Briand JP, et al. The HIV-1 viral protein R induces apoptosis via a direct effect on the mitochondrial permeability transition pore. J Exp Med. 2000;191:33–46. doi: 10.1084/jem.191.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen EL. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the Autophagosome Maturation Process by a Novel Reporter Protein, Tandem Fluorescent-Tagged LC3. Autophagy. 2007;3:452–60. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Kristensen AR, Schandorff S, Hoyer-Hansen M, Nielsen MO, Jaattela M, Dengjel J, Andersen JS. Ordered organelle degradation during starvation-induced autophagy. Mol Cell Proteomics. 2008;7:2419–2428. doi: 10.1074/mcp.M800184-MCP200. [DOI] [PubMed] [Google Scholar]
- Ku B, Woo JS, Liang C, Lee KH, Hong HS, E X, Kim KS, Jung JU, Oh BH. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68. PLoS Pathog. 2008;4:e25. doi: 10.1371/journal.ppat.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum JJ, Cohen OJ, Nie Z, Weaver JG, Gomez TS, Yao XJ, Lynch D, Pilon AA, Hawley N, Kim JE, et al. Vpr R77Q is associated with long-term nonprogressive HIV infection and impaired induction of apoptosis. J Clin Invest. 2003;111:1547–1554. doi: 10.1172/JCI16233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Soto CS, Ohigashi Y, Taylor A, Bournas V, Glawe B, Udo MK, Degrado WF, Lamb RA, Pinto LH. Identification of the pore-lining residues of the BM2 ion channel protein of influenza B virus. J Biol Chem. 2008;283:15921–15931. doi: 10.1074/jbc.M710302200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- McAuley JL, Hornung F, Boyd KL, Smith AM, McKeon R, Bennink J, Yewdell JW, McCullers JA. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe. 2007;2:240–249. doi: 10.1016/j.chom.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCown MF, Pekosz A. The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. J Virol. 2005;79:3595–3605. doi: 10.1128/JVI.79.6.3595-3605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Klionsky DJ. Protein Turnover Via Autophagy: Implications for Metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- Mould JA, Paterson RG, Takeda M, Ohigashi Y, Venkataraman P, Lamb RA, Pinto LH. Influenza B virus BM2 protein has ion channel activity that conducts protons across membranes. Dev Cell. 2003;5:175–184. doi: 10.1016/s1534-5807(03)00190-4. [DOI] [PubMed] [Google Scholar]
- Münz C. Enhancing immunity through autophagy. Annu Rev Immunol. 2009;27:423–429. doi: 10.1146/annurev.immunol.021908.132537. [DOI] [PubMed] [Google Scholar]
- Muthumani K, Hwang DS, Desai BM, Zhang D, Dayes N, Green DR, Weiner DB. HIV-1 Vpr induces apoptosis through caspase 9 in T cells and peripheral blood mononuclear cells. J Biol Chem. 2002;277:37820–37831. doi: 10.1074/jbc.M205313200. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib D, Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host & Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Palese P, Shaw ML. Orthomyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Lippincott, Williams & Wilkins; 2007. pp. 1647–1689. [Google Scholar]
- Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Münz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, Gilpin C, Levine B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- Quinlivan M, Zamarin D, Garcia-Sastre A, Cullinane A, Chambers T, Palese P. Attenuation of equine influenza viruses through truncations of the NS1 protein. J Virol. 2005;79:8431–8439. doi: 10.1128/JVI.79.13.8431-8439.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter B, Albert ML, Francisco L, Larsson M, Somersan S, Bhardwaj N. Consequences of cell death: exposure to necrotic tumor cells, but not primary tissue cells or apoptotic cells, induces the maturation of immunostimulatory dendritic cells. J Exp Med. 2000;191:423–434. doi: 10.1084/jem.191.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid D, Münz C. Innate and adaptive immunity through autophagy. Immunity. 2007;26:11–21. doi: 10.1016/j.immuni.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid D, Pypaert M, Münz C. MHC class II antigen loading compartments continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Colbert CL, Becker N, Wei Y, Levine B. Molecular basis of the regulation of Beclin 1-dependent autophagy by the gamma-herpesvirus 68 Bcl-2 homolog M11. Autophagy. 2008;4:989–997. doi: 10.4161/auto.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir D, Chen WL, Choi J, Wakita T, Yen TS, Ou JH. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, Soto CS, Tereshko V, Nanda V, Stayrook S, DeGrado WF. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451:596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromhaug PE, Berg TO, Fengsrud M, Seglen PO. Purification and characterization of autophagosomes from rat hepatocytes. Biochem J. 1998;335(Pt 2):217–224. doi: 10.1042/bj3350217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- Yewdell J, Garcia-Sastre A. Influenza virus still surprises. Curr Opin Microbiol. 2002;5:414–418. doi: 10.1016/s1369-5274(02)00346-6. [DOI] [PubMed] [Google Scholar]
- Zamarin D, Garcia-Sastre A, Xiao X, Wang R, Palese P. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog. 2005;1:e4. doi: 10.1371/journal.ppat.0010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamarin D, Ortigoza MB, Palese P. Influenza A virus PB1-F2 protein contributes to viral pathogenesis in mice. J Virol. 2006;80:7976–7983. doi: 10.1128/JVI.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Jiang X, Liu D, Fan Z, Hu X, Yan J, Wang M, Gao GF. Autophagy is involved in influenza A virus replication. Autophagy. 2009;5:321–328. doi: 10.4161/auto.5.3.7406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Autophagosome accumulation occurs only in directly and productively influenza A virus infected cells. (A) A549 human lung epithelial cells were infected with influenza A virus and lysates were prepared at the indicated timepoints after infection. Lipidated Atg8/LC3-II content was analyzed by Western blot analysis. One of three experiments is shown. (B) GFP-Atg8/LC3 transfected A549 human and MLE-12 mouse lung epithelial cells were incubated with live or heat-inactivated influenza A virus. Endogenous lipidated Atg8/LC3-II and transfected lipidated GFP-Atg8/LC3-II levels were analyzed by Atg8/LC3 Western blot analysis after 24h. One of three experiments is shown. (C) GFP-Atg8/LC3 transfected MDAMC cells were exposed to live or heat-inactivated influenza A virus. GFP-Atg8/LC3 positive autophagosomes were visualized by fluorescence microscopy 24h later. Nuclear DNA was stained with DAPI. Scale bar: 80 μm. One of three experiments is shown. (D) GFP-Atg8/LC3 transfected MDAMC cells were infected with influenza A virus and matrix protein 1 (M1) expression was analyzed by immune fluorescence microscopy. Nuclear DNA was stained in addition with DAPI. Scale bar: 80 μm. One of three experiments is shown.
Supplemental Figure 2: Macroautophagy deficiency does not affect viral production. (A) HA units ELISA: culture supernatants from Atg5−/− and Atg5+/+ MEFs were collected 24 hours after influenza A infection, 6 independent experiments were analysed (paired t test p=0.010). (B) Western blot analysis of viral protein content (M2 and NP) in both cell types (C) Relative concentration of viral RNA in the cells was determined by analysis of cycle threshold values (CT), normalizing the mean CT for WSN-M, WSN-HA and WSN-NA to the expression of the product of the housekeeping gene GAPDH. (left graph). Viral RNA content in the supernatants of Atg5−/− and wt Mefs was determined by comparison of the mean CT for WSN-M, WSN-HA and WSN-NA (right graph). 3 independants experiments were analyzed. (D) Viral titer: Viral titer of culture supernatants from Atg5−/− and Atg5+/+ MEFs collected 24 hours post infection. Three independent experiments are shown (paired t test p=0.7 ns).
Supplemental Movie 1: Mobility of autophagosomes in influenza A virus infected cells. Live cell imaging of GFP-Atg8/LC3 transfected A549 human lung epithelial cells was performed to analyze autophagosome mobility.