Abstract
Proline-, glutamic acid–, leucine-rich protein 1 (PELP1), a novel nuclear receptor coactivator, and its expression is deregulated in hormone-dependent cancers, including those of the breast, endometrium, and ovary. PELP1 interacts with estrogen receptor and modulates its genomic and nongenomic functions. In this study, we examined whether PELP1 functions as an oncogene. The overexpression of PELP1 in fibroblasts and epithelial model cells resulted in cellular transformation. PELP1 also enhanced the transformation potential of c-Src kinase in focus formation assays, and PELP1 overexpression potentiated estradiol-mediated cell migratory potential and anchorage-independent growth. Using PELP1-small interfering RNA, we provided evidence that endogenous PELP1 plays an essential role in E2-mediated anchorage-independent growth, cell migration, and cytoskeletal changes. When compared with control vector transfectants, breast cancer cells stably overexpressing PELP1 showed a rapid tumor growth in xenograft studies. Immunohistochemical analysis of PELP1 expression using a tumor progression array of 252 breast carcinomas and normal breast tissue specimens revealed that PELP1 expression is deregulated to a greater degree in higher grade node-positive invasive tumors than in normal breast tissue or ductal carcinoma in situ. Our data suggest that PELP1 is a potential oncogene, that its expression is deregulated during cancer progression, and that PELP1 may play a role in oncogenesis.
Introduction
The human estrogen receptor (ER) is a ligand-dependent transcription factor (1) that has been implicated in the progression of breast cancer (2). ER requires both a ligand and interactions with other proteins, such as coregulators, to achieve optimal transcriptional activation of target genes (3). Emerging evidence suggests that in addition to exerting its well-studied nuclear functions, the ER also participates in cytoplasmic and membrane-mediated signaling events through the stimulation of the Src kinase, mitogen-activated protein kinase (MAPK), and protein kinase B (AKT) pathways (4, 5). However, the significance of ER nongenomic signaling in tumorigenesis in vivo is not completely understood.
ER signaling seems to be complex and seems to involve multiple coregulatory proteins as well as cross-talk with a number of cellular pathways (1, 6, 7). Multiprotein complexes containing ER, coactivators, and transcriptional regulators assemble in response to hormone binding and activate the transcription of hormonal target genes (8). Recent evidence suggests that several ER-coregulatory proteins are differentially expressed in tumors (9). Although much is known about the molecular basis of interaction between ER and coregulators, very little is known about the pathologic role of ER coregulators and the mechanisms by which they influence the progression of cancer.
Advances in molecular biology techniques have identified several novel proteins as being ER coregulators (10). One of those ER coregulators, proline-, glutamic acid–, leucine-rich protein 1 [PELP1; ref. 11; also known as modulator of the nongenomic actions of the ER (MNAR); ref. 12], is unique because it plays an important role both in the genomic (13) and nongenomic actions of the ER (14, 15). PELP1 promotes E2-mediated cell proliferation by sensitizing cells to G1-S progression via its interactions with the pRb pathway (16). Recent evidence also suggests that PELP1 couples ER to several signaling axis such as Src-MAPK, phosphoinositide-3-kinase (PI3K)–AKT, and epidermal growth factor receptor (EGFR)–signal transducers and activators of transcription 3 (STAT3) (15, 17), and that PELP1 expression is deregulated in human breast cancer (15, 18). Although these studies suggested that PELP1 can modulate ER functions via multiple mechanisms, whether PELP1 has tumorigenic potential is not known.
In this study, we used in vitro and in vivo mouse xenograft models to investigate the tumorigenic potential of PELP1. Our results indicated that PELP1 deregulation promotes the transformation of fibroblasts and epithelial model cells and enhances the hormone-independent growth of breast cancer cells in xenograft studies. In addition, PELP1 expression was deregulated in higher grade invasive breast tumors. These results define a new role for the nuclear receptor coregulator PELP1 as a potential oncogene.
Materials and Methods
Cell cultures and reagents
MCF-7 human breast cancer cells were maintained in DMEM-F12 (1:1) supplemented with 5% DCC FCS. NIH3T3 mouse fibroblasts obtained from the American Type Culture Collection (ATCC) were maintained in DMEM supplemented with 10% bovine calf serum. RK3E cells were obtained from ATCC and were maintained in DMEM supplemented with 5% DCC FCS. Generation and characterization of MCF-7 clones overexpressing PELP1 (clone 20, clone 13, pool) were earlier described (16). Antibodies against vinculin and the steroid hormone 17β-estradiol were purchased from Sigma. Anti–T7-epitope antibody was purchased from Novagen. PELP1/MNAR antibody was purchased from Bethyl Laboratories. Antibodies against phospho-AKT, phospho-MAPK, and phospho-Src were purchased from Cell Signaling. For PELP1 knockdown, siGenome SMARTpool [small interfering RNA (siRNA)] duplexes were purchased from Dharmacon.
Focus formation assays
Fugene-6 (Roche) was used as directed by the manufacturer to transfect NIH3T3 cells or RK3E cells. For each six-well plate, 2 μg of PELP1 expression vector or control vector was transfected. In some assays, PELP1 was cotransfected with c-Src expression plasmid, and in all cases, the expression vector was normalized with the respective empty vectors. After 3 days of transfection, the cells were subcultured at a 1:5 split ratio into 100-mm plates containing growth medium. Two plates were supplemented with 500 μg/mL G418 and were used to correct transfection efficiency, and the other three plates were grown in complete medium. The number of transformed foci was counted in these plates, and the transfection efficiency was corrected by counting the G418 resistance colonies. One of the G418-selected plates was also used to generate pooled clones for use in the soft-agar colony assays. The cells were fed every 2 days, and the foci of the transformed cells were counted after 14 days. The cells were stained with crystal violet, and the foci of the transformed cells were quantified by visual inspection. Three plates per condition were counted, and average of these experiments was used to generate figures. The data are representative of three independent experiments done in triplicate.
Cell migration and immunofluorescence assays
The cell migration was determined using 8 μmol/L pore calorimetric cell migration assay kit (Chemicon) as directed by the manufacturer. For cell migration assay 105 cells/300 μL of serum-free medium were seeded onto the upper chamber, and 0.75 mL of complete growth medium containing 10% fetal bovine serum was added to each well in the lower chamber. To measure estrogen-mediated cell migration potential, cells were serum starved in phenol red–free medium (0% DCC) for 48 h, and 105 cells/300 μL were added into the upper chamber, and 0.75 mL of phenol red–free medium containing 5% DCC serum with or without 10 mmol/L E2 was added to each well in the lower chamber. Indirect immunofluorescence assays were done as described previously (19). For staining of filamentous actin, cells were fixed with paraformaldehyde and then incubated with 0.1 μmol/L of either Alexa 568– or Alexa 488–conjugated phalloidin. Slides were mounted and analyzed using Nikon Eclipse 80i immunofluorescence microscope or by confocal microscopy.
Soft-agar assays
Soft-agar colony-growth assays were done as previously described (15). Briefly, 1 mL of 0.6% Difco agar in DMEM was added to the bottom of the plate. After solidification had occurred, test cells (1 × 104) mixed with 1 mL of 0.36% bactoagar solution in DMEM were layered on top of the 0.6% bactoagar layer. The plates were incubated at 37°C in 5% CO2 for 21 days, and the colonies were counted.
Tumorigenesis assays
In tumorigenesis studies, either MCF-7 pcDNA or MCF7-PELP1 clone 20 model cells (5 × 107 cells) were implanted s. c. into the mammary fat pads of 6- to 7-week-old female nude mice (n = 10) as previously described (19). No E2 pellet was implanted in MCF-7-pcDNA or MCF-7 PELP1–injected mice, and tumors were allowed to grow for 6 weeks. Tumor volumes were measured with a caliper at weekly intervals. After 6 weeks, the mice were sacrificed, and the tumors were removed and processed. To use as a positive control, parental MCF-7 cells were injected into five nude mice into which an E2 pellet had been implanted (Innovative Research of America).
Tissue microarrays
The tissue microarrays (TMA) used in this study were obtained from the Cooperative Breast Cancer Tissue Resource (CBCTR) of the National Cancer Institute (NCI). Those TMAs are designed by NCI statisticians to provide high statistical power and are suitable for use in the investigation of differences in the prevalence of potential markers in three stages of invasive breast cancer: node-negative, node-positive, and metastatic disease. Each TMA consists of 288 cores (0.6 mm) taken from paraffin-embedded specimens that represent a total of 252 breast cancer and normal breast tissue specimens plus 36 controls.
Immunohistochemistry
Paraffin-embedded TMA arrays obtained from the CBCTR were deparaffinized with xylene and were rehydrated with graded ethanol. The TMA array section was boiled for 10 min in 0.01 mol/L citrate buffer and was cooled for 30 min at room temperature to expose antigenic epitopes. The TMA array was blocked with 2% normal goat serum in 1% bovine serum albumin and PBS for 30 min. The TMA array section was then incubated with polyclonal rabbit antihuman-PELP1 antiserum at a dilution of 1:500 and was incubated overnight at room temperature. The PELP1 antibody was developed in our laboratory. It was well characterized and has been used in immunohistochemical research in earlier published studies (11, 15, 20). The TMA sections were washed thrice with 0.05% Tween in PBS for 10 min, incubated with secondary antibody for 1 h, washed thrice with 0.05% Tween in PBS for 10 min, developed with 3,3V-diaminobenzidine-H2O2, and counterstained with Mayer’s hematoxylin. The preparation of negative controls was accomplished by replacing the primary antibody with control rabbit immunoglobulin G or peptide-absorbed PELP1 antibody. The finding that no cells or <10% of the cells were immunoreactive was considered to be a negative result, and the finding that >10% of the cells were immunoreactive was considered a positive result. The sections were scored by three evaluators blinded to the patient’s clinical status. PELP1 staining was done according to the established method (15, 20), and the results were classified as follows: 0, no expression; 1, weak expression; 2, moderate expression; and 3, strong expression (Fig. 5A; Table 1). Two TMAs containing two different cores from each specimen were stained for PELP1, damaged core spots were eliminated from scoring, and the average PELP1-IHC scores were sent to the CBCTR. After receipt of the immunohistochemistry scores, the CBCTR released the clinical data that were then used to correlate PELP1 expression with various clinicopathologic variables, including patient age; cancer stage, grade, and case type; ER; and PR. Associations among variables were analyzed with the χ2 and Spearman’s rank correlation coefficients. All tests were done with SPSS software [Statistical Package for the Social Sciences (SPSS) Inc.].
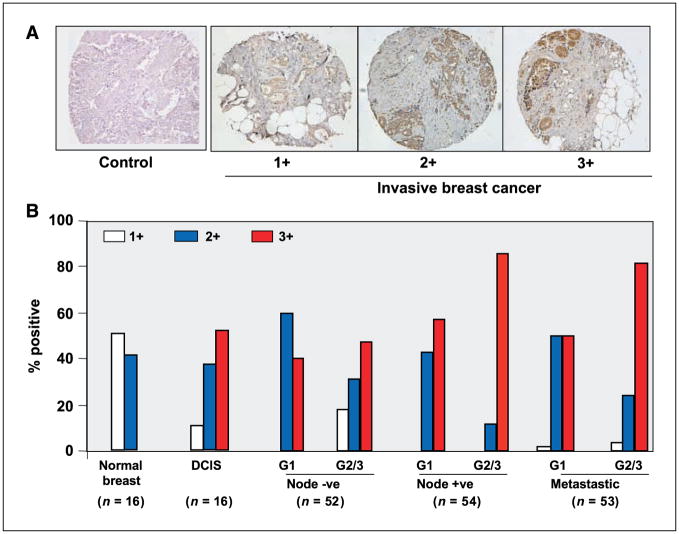
Figure 5.
The expression of PELP1 in node-negative and node-positive breast tumors. A, sections were stained with PELP1 antiserum. Sections were scored according to immunohistochemistry intensity in a range from 0 to 3, in which 0 indicated no expression; 1, low expression; 2, moderate expression; and 3, high expression. A representative sample of each score is shown with the respective PELP1 stain. PELP1 antiserum that was preadsorbed with target peptide was used as a control. B, summary of the staining pattern of the PELP1 expression in normal, DCIS, node-negative, node-positive, and metastatic breast tumors analyzed via immunohistochemical analysis that used PELP1 antibody in a tumor progression array (n = 252). Two arrays containing two different core samples from each tumor were stained, and the average staining intensity was used for data analysis. G1, grade 1; G2, grade 2; G3, grade 3.
Table 1.
Distribution of PELP1 staining and clinical-pathological factors in invasive breast cancer
| Patient number | PELP1 staining |
P | |||
|---|---|---|---|---|---|
| 0–1 | 2 | 3 | |||
| Age | |||||
| <50 | 52 | 4 (7) | 10 (20) | 38 (78) | 0.068 |
| >50 | 107 | 7 (7) | 33 (31) | 67 (62) | |
| Stage | |||||
| T1 (<20 mm) | 88 | 6 (7) | 23 (26) | 59 (67) | 0.983 |
| T2 (20–50 mm) | 71 | 5 (7) | 18 (25) | 48 (68) | |
| Grade | |||||
| 1 | 33 | 0 (0) | 16 (48) | 17 (52) | 0.0005 |
| 2 | 102 | 7 (6) | 24 (24) | 71 (70) | |
| 3 | 24 | 3 (12) | 6 (25) | 15 (63) | |
| Case type | |||||
| Node negative | 52 | 6 (11) | 19 (37) | 27 (52) | 0.003 |
| Node positive | 54 | 2 (4) | 10 (18) | 42 (78) | |
| Metastatic | 53 | 3 (6) | 16 (30) | 34 (64) | |
| ER | |||||
| Positive | 105 | 8 (7) | 32 (31) | 65 (62) | 0.265 |
| Negative (unknown = 4) | 50 | 3 (6) | 11 (22) | 36 (72) | |
| PR | |||||
| Positive | 57 | 8 (7) | 17 (30) | 32 (56) | 0.160 |
| Negative (unknown = 3) | 99 | 3 (3) | 25 (25) | 71 (72) | |
Results
PELP1 promotes the transformation of NIH3T3 mouse fibroblasts
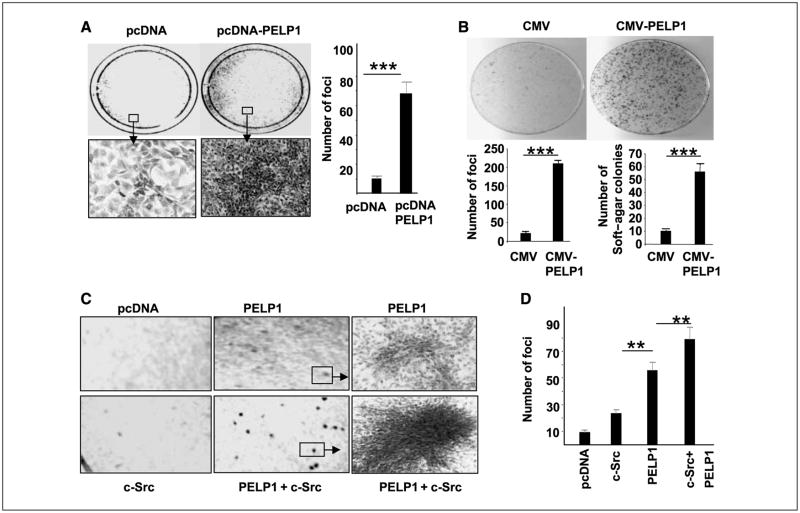
Emerging studies have shown that PELP1 expression is deregulated in several tumors, including those of the breast. To determine significance of that finding, we used NIH3T3 focus formation assays to examine whether PELP1 has transformation potential. pcDNA-PELP1 transfected (but not pcDNA vector-transfected) NIH3T3 cells exhibited potent transforming activity in focus formation assays. Typical cultures of the PELP1-transformed plate contained multiple foci, but pcDNA vector–transformed plates showed a very low number of foci (Fig. 1A). To rule out vector-mediated artifacts, we have repeated focus formation assays using CMV-PELP1 expression vector. CMV-PELP1 (but not CMV vector)–transfected NIH3T3 cells exhibited increased focus formation (Supplementary Fig. S1). To further confirm these findings, we have generated pooled clones of NIH3T3 cells stably expressing either CMV or CMV-PELP1 vector (Fig. 1B). Using these stable cells, we have repeated foci formation (Fig. 1B, top) and soft-agar colony assays (Fig. 1B, bottom). The results from these experiments further confirmed our earlier findings using transient transfection that PELP1 deregulation contributes to transformation of NIH3T3 cells.
Figure 1.
PELP1 transforms NIH3T3 cells. A, pcDNA vector or pcDNA-PELP1 vector was transiently transfected into NIH3T3 cells, and focus formation was counted after 14 d. Bottom, representative morphologic characteristics of a transformed colony induced by PELP1. The number of transformed foci were counted in these plates, and the result was corrected for transfection efficiency by counting G418-resistant colonies (left). B, NIH3T3 cells were cotransfected with either CMV + pNeomycin plasmid or CMV-PELP1 + pNeomycin plasmid. G418 resistant colonies were selected, and transformation potential of pooled clones expressing CMV vector or PELP1 was analyzed for focus formation and soft-agar colony formation. C, NIH3T3 cells were transiently transfected with pcDNA, or c-Src, and/or c-Src + PELP1, and focus formation was recorded after 14 d. A representative portion from each plate is shown. D, quantitation of the focus formation observed in the plates.
Earlier studies showed that PELP1 interacts with and promotes the activation c-Src kinase (12). c-Src is an homologue of Rous sarcoma virus transforming protein (v-Src) and is expressed at low levels in most cell types. c-Src is less active as a protein-tyrosine kinase than v-Src due to tight control of c-Src activity in normal cells and exhibits weak transformation potential. However, cotransfection of c-Src with other nuclear oncogenes (such as c-myc or T antigen) increases c-Src activity and transformation potential (21). To examine whether PELP1 interacts with c-Src in transformation, we cotransfected NIH3T3 cells with PELP1 and then with either c-Src–expressing plasmid or the empty vector. As earlier studies have shown (21), c-Src transformation alone has a minimal effect on focus formation, and the transformed cells in c-Src–induced foci are small. In contrast, PELP1 and c-Src cotransformation stimulated an increase in focus formation substantially greater than that found in vector cells or cells transfected with either c-Src or PELP1 alone (Fig. 1C and D). The foci formed in PELP1 and c-Src cotransfected cells were large and were characterized by the presence of highly refractile, morphologically transformed cells (Fig. 1C). These results suggest that PELP1 has tumorigenic potential and cooperates with proto-oncogenes such as c-Src in cellular transformation.
PELP1 transforms epithelial cells
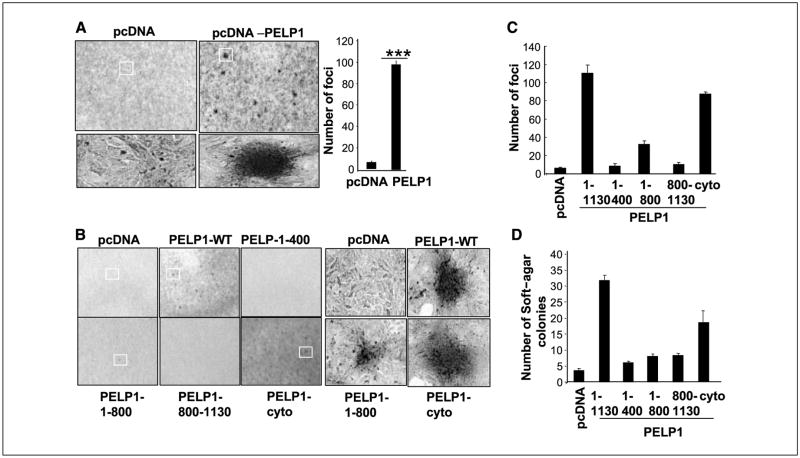
Using the rat kidney epithelial cell line RK3E (22), an E1A-immortalized epithelial cell line derived from neonatal rat kidney, we examined whether PELP1 transforms epithelial cells. RK3E cells exhibit epithelioid morphologic and molecular features, are contact inhibited at confluence, and are polarized. These model cells are susceptible to transformation by several oncogenes (23, 24). In our study, PELP1-transfected (but not vector-transfected) RK3E cells exhibited transformation potential (Fig. 2A). To identify the potential regions of PELP1 that are required for PELP1-mediated transformation, we used several deletions or mutants of PELP1 in our focus formation assays (Fig. 2B). The PELP1 NH2-terminal region (aa 1–400), which contains Src, ER-binding sites, or a COOH-terminal acidic region (aa 800–1,130) characterized by histone-binding and proline-rich regions, did not exhibit any transformation activity. However, PELP1 peptide containing aa 1–800 showed focus formation, suggesting that multiple domains in the PELP1 are required for transformation. Similarly, PELP1-cyto mutant that uniquely localizes in the cytoplasm also showed focus formation, although to a lesser degree than that of PELP1-WT (Fig. 2C). To validate those results, we used a soft-agar colony assay to evaluate the ability of PELP1 mutants to support anchorage-independent growth. We found that vector-transfected RK3E cells and NH2-, COOH-terminal deletions of PELP1 are incapable of promoting anchorage-independent growth, and that PELP1-WT–transfected RK3E cells showed a significant increase in colony formation (Fig. 2D). The PELP1-cyto mutant also showed some potential for anchorage-independent growth. Collectively, these results suggest that transformation potential requires PELP1 aa 1–800, and that PELP1-mediated cytoplasmic signaling may have a role in that transformation.
Figure 2.
PELP1 transforms rat kidney epithelial cells (RK3E). A, focus formation in PELP1-transfected or vector-transfected RK3E cells. Right, quantitation of PELP1-induced focus formation. B, RK3E cells were transfected with various domains of PELP1 or a PELP1 mutant that uniquely localizes in the cytoplasm, and focus formation was counted. Representative morphologic characteristics of PELP1-induced and PELP1-mutant induced foci. C, quantitation of PELP1-induced and PELP1 deletion-induced focus formation. D, pooled RK3E transfectants were plated in soft agar, and the colony formations were counted after 21 d. These experiments were repeated thrice; Columns, average of the results.
PELP1 deregulation promotes migratory potential
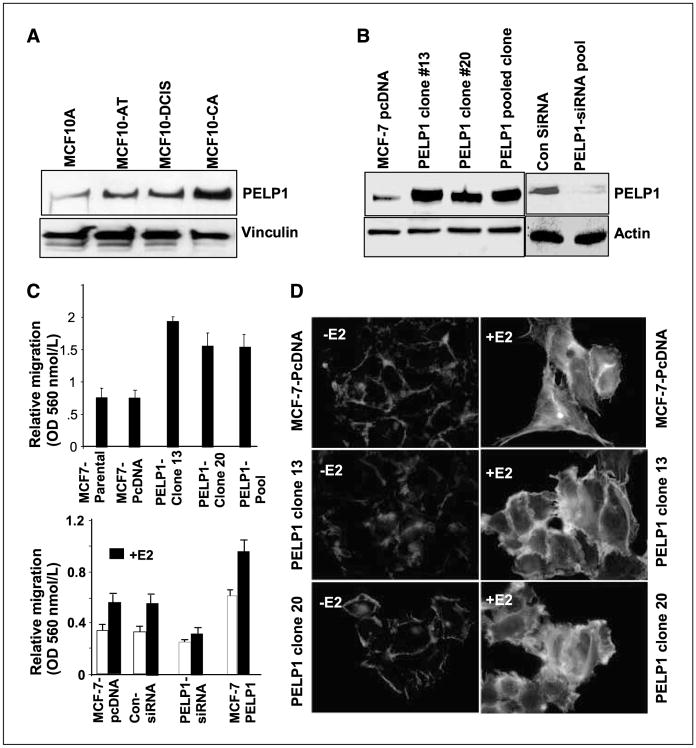
To examine whether the deregulation of PELP1 expression occurs during tumorigenesis, we used lysates from MCF10A model cells developed at Karmanos Cancer Institute to analyze PELP1 expression by Western blot analysis. The MCF10A model contains a spectrum of cell lines that enable the analysis of gene expression during the progression of breast cancer (25, 26). In our study, Western blot results revealed the increased expression of PELP1 as a function of tumorigenesis by showing an increased expression of PELP1 in cells with greater metastatic potential (Fig. 3A). Emerging studies strongly suggest that PELP1 participates in E2-mediated nongenomic actions by promoting ER interactions with c-Src kinase (12). Because many oncogenes (including c-Src) modulate the migratory potential of cancer cells and because PELP1 expression is up-regulated in MCF-10A metastatic variant cells, we used the Boyden chamber assay to investigate whether PELP1 could influence the migration of breast cancer cells. In that evaluation, we used either MCF-7 cells or MCF-7 clones that over-express PELP1 (16). Western analysis revealed that these model cells express PELP1 3- to 4-fold higher than the levels present in MCF-7 cells (Fig. 3B). As a second model, we have used MCF-7 cells in which PELP1 expression is down-regulated by siRNA. The results of Western blot analysis showed that the addition of PELP1-siRNA decreased PELP1 expression levels by ~90%, and that control siRNA transfections did not affect the levels of PELP1 (Fig. 3B). In Boyden chamber assays, PELP1 overexpressing clones exhibited increased cell motility (Fig. 3C, top). Parental MCF-7 cells showed low motility, and E2 further increased the migratory potential of those cells (Fig. 3C, bottom). The overexpression of PELP1 resulted in a significant induction of cell motility upon E2 treatment, and the knockdown of PELP1 expression by siRNA substantially reduced E2-mediated cell motility (Fig. 3C, bottom). These results suggest that PELP1 plays an important role in E2-mediated cell migration. To further understand the mechanism, we used phalloidin staining and immunofluorescence microscopy to investigate whether PELP1 has a role in E2-mediated cytoskeletal changes. We found that E2 treatment significantly increased the formation of filamentous actin structures, including filopodia, membrane ruffles, and stress fibers (Fig. 3D), to a degree greater than that occurring in serum-starved MCF-7 cells. PELP1 overexpression uniquely enhanced ruffles as well as filopodia-like structures (Fig. 3D), both of which enable the attachment and movement of the cytoplasmic components that are responsible for cell migration. The reduction of endogenous PELP1 expression affected E2-mediated ruffle formation while increasing the formation of stress fibers (Supplementary Fig. S2). The overexpression of PELP1 in RK3E cells also caused an increase in filopodia and ruffle formation to a degree greater than that in vector-transfected cells (Supplementary Fig. S3). These results suggest that PELP1 deregulation may have a role in E2-ER–mediated cytoskeletal changes.
Figure 3.
PELP1 promotes cytoskeletal reorganization and motility. A, Western blot analysis of PELP1 expression in lysates from cells derived from the MCF10AT model system. MCF10A, nonmalignant human breast cancer cells; MCF10-AT, weakly tumorigenic; MCF10-DCIS, comedo-type DCIS, highly proliferative, aggressive, and invasive; and MCF10-CA, undifferentiated carcinomas, metastatic. B, total cell lysates from pcDNA- and PELP1-expressing clones (nos. 13, 20, Pool) were analyzed by Western blotting with an anti-PELP1 antibody (left). Actin was used as a loading control. The functionality of PELP1-siRNA was analyzed by Western blotting (right). C, migratory potential of MCF-7 clones overexpressing PELP1 was analyzed by Boyden chamber assay (top). MCF-7 cells stably overexpressing PELP1 and MCF-7 cells transfected with PELP1-siRNA were treated with E2 or were untreated, and the cell migration potential was analyzed by means of the Boyden chamber assay (bottom). D, MCF-7, MCF-7–PELP1 clone nos. 13 and 20 were cultured in a DCC-serum–containing medium, after which they were treated with E2 for 10 min. The status of filamentous actin was visualized by phalloidin staining and was evaluated by fluorescence microscopy.
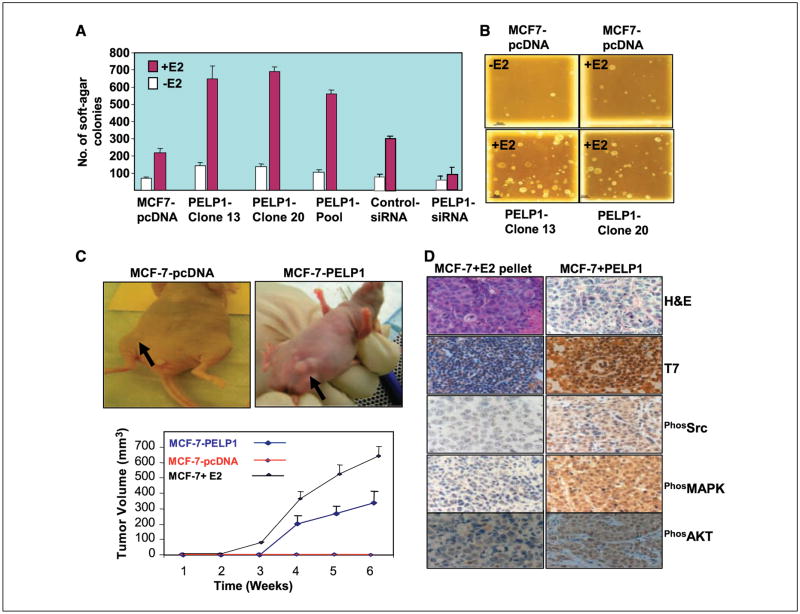
PELP1 promotes tumorigenic potential of MCF-7 cells. We used soft-agar colony formation assays to investigate whether PELP1 promotes the anchorage-independent growth of breast epithelial cells (MCF-7). Parental MCF-7 cells expressing pcDNA showed low or weak potential to form soft-agar colonies (Fig. 4A and B). However, the addition of E2 enhanced the ability of MCF-7–pcDNA cells to form soft-agar colonies. The overexpression of PELP1 in MCF-7 cells substantially increased the ability of those cells to grow in an anchorage-independent manner after E2 stimulation (Fig. 4A and B), and the down-regulation of PELP1 by siRNA substantially reduced the E2-mediated enhancement of soft-agar colony formation (Fig. 4A). Because PELP1 exhibited many characteristics of oncogenes (cellular transformation, anchorage-independent growth, cell motility), we used a nude mouse xenograft model to examine whether PELP1 has tumorigenic potential in vivo. MCF-7 cells stably expressing pcDNA or PELP1 were injected s. c. into the mammary fat pad of the murine subjects. Tumorigenic potential was monitored for 6 weeks in the absence of exogenous estrogen. Under those conditions, no tumors were detected in MCF-7–pcDNA–injected sites. However, 50% of the MCF-7–PELP1–injected sites showed tumors in the absence of exogenous estradiol treatment (Fig. 4C). Immunohistochemistry examination of the tumors revealed that PELP1-induced tumors retained the expression of T7-tagged PELP1 and exhibited excessive activation of Src, MAPK, and AKT to a degree greater than that in tumors obtained from MCF-7 xenografts induced by exogenous E2 pellet implantation (Fig. 4D). These results suggest that PELP1 deregulation has the potential to promote growth of tumor cells in vivo.
Figure 4.
PELP1 deregulation enhances tumorigenic potential of breast cancer cells in vitro and in vivo. A and B, anchorage-independent growth potential of the PELP1-expressing MCF-7 clones or MCF-7 cells were treated with control siRNA or PELP1-specific siRNA was measured by their ability to form colonies on soft agar. A, quantitation of the soft-agar colony formation; B, representative photographs. Results shown are representative of four independent experiments. C, nude mice were injected with 5 × 107 MCF-7 cells that were stably expressing pcDNA or PELP1 into the mammary fat pad, and tumor growth was measured at weekly intervals. No exogenous estrogen was given. As a positive control, MCF-7 cells were injected into the mammary fat pad and xenografts were induced by exogenous E2 pellet implantation. A representative picture of a PELP1-induced tumor in nude mice is shown. D, morphologic characteristics of PELP1-induced tumors and the expression of T7-PELP1, phos-Src, -MAPK, and -AKT in tumors evaluated with H&E staining and anti–phos-Src, anti-MAPK, and anti-AKT antibodies, respectively. E2-pellet–induced tumors of MCF7 cells were used as a positive control.
PELP1 expression is deregulated in node-positive and metastatic tumors
We then used a breast cancer TMA to investigate whether PELP1 expression correlates with any stages of invasive breast cancer (node-negative, node-positive, or metastatic disease). We measured the expression levels of PELP1 by immunohistochemistry, and PELP1 expression was scored as previously described (15, 20, 27). The representative staining for each score range is shown in Fig. 5A. The results of immunohistochemistry examination showed that when compared with node-negative specimens, node-positive and metastatic tumors exhibited increased PELP1 expression (Fig. 5B). Statistical analysis revealed that PELP1 expression was positively correlated with cancer grade and node status. The number of samples with a high level (score 3) of PELP1 staining increased as tumors progressed from grade 1 to grade 2 or 3 (0.005; Table 1). Similarly, node-positive and metastatic tumors exhibited a greater expression of PELP1 than did node-negative tumors (P = 0.003; Table 1). No significant correlation of PELP1 expression with ER, PR, patient age, or tumor stage was observed. These results suggest that PELP1 expression may be altered in higher grade node-positive and metastatic tumors.
Discussion
Mammary tumorigenesis is accelerated by the action of ovarian hormones, and ~70% of malignant breast tumors are ER positive at the time of presentation (28). Emerging evidence suggests that ER signaling is complex and involves multiple coregulatory proteins (1, 3) and cross-talk with a number of cellular pathways (3, 29). PELP1 is a novel ER coactivator, and previous studies have shown that PELP1 up-regulates cyclin D1 expression and couples ER with the oncogenes Src, PI3K, and STAT3 (15–17, 29). In this study, we used fibroblast, epithelial, and xenograft models to provide evidence that the ER coregulator PELP1 has oncogenic potential. Because ER functions depend on the status of the coactivators in a given cell, it is likely that the expression level of PELP1, which has tumorigenic potential, plays an important role in malignant breast tumor development.
Emerging findings suggest that ER-coregulatory proteins have a role in tumorigenesis (10), and a few recent studies have provided direct evidence that supports this concept. In a transgenic murine model, the overexpression of the ER coactivator AIB1 has been shown to promote a high incidence of tumors associated with activation of PI3K-AKT pathway (30). Another study showed that the deregulation of the ER coregulator MTA1 promotes tumorigenesis via up-regulation of the cyclin D1 pathway (31). Our results suggest that PELP1 is another ER coregulator with tumorigenic potential. MCF-7 cells are not tumorigenic in nude mice models in the absence of supplementation of exogenous estrogen. The failure of MCF-7–pcDNA to cause tumors in nude mice is probably related to the low levels of circulating E2 in these mice (32). The promotion of tumorigenesis in a nude mouse xenograft model in the absence of exogenous E2 suggests that PELP1 deregulation may promote the growth of tumors under conditions of low circulating E2. The increased activation of the Src, MAPK, and AKT pathways in PELP1-induced tumors (Fig. 4) also suggests that PELP1-mediated nongenomic signaling contributes to some extent to the tumorigenic potential of PELP1.
Although antiestrogens and selective estrogen receptor modulators (SERM) are effective in curbing the evolution of ER-positive breast tumors into more invasive phenotypes (33), many patients with metastatic breast tumors eventually become resistant to that treatment (34). Growth factor–induced signaling pathways have been shown to modify the ER and its coactivators by phosphorylation, which eventually leads to ligand-independent activation or to differential responses to SERMs (6, 35). Emerging data suggest that coregulators that couple the ER with growth-factor signaling components may play a role in hormonal responsiveness and tumor progression. PELP1 ability to interact with various growth factor signaling components and cytosolic kinases (15, 17), its deregulation in metastatsic tumors (Fig. 5), suggest that PELP1 deregulation in metastatic tumors may contribute to hormonal independence.
The proto-oncogene c-Src is a multifunctional intracellular tyrosine kinase that is implicated in the regulation of a variety of processes, such as cell proliferation, differentiation, and motility (36). Src interacts with multiple cellular factors (e.g., HER2, EGFR, and ER), and breast tumors overexpress Src kinase (37). PELP1 acts as a scaffolding protein that couples ER with c-Src kinase and leads to the activation of the ER-Src–MAPK pathway (12, 14). Mutational analysis of ERα and c-Src mutants has revealed that PELP1 interacts with the c-Src SH3 domain via its NH2-terminal PXXP motif. ER interacts with the Src SH2 domain at phosphotyrosine 537, and MNAR-ER interaction further stabilizes that complex (14). Our results showed that PELP1 enhances the Src transformation potential in focus formation assays, and that PELP1-induced tumors showed increased c-Src kinase activation. Because breast tumors overexpress wild-type c-Src kinase, the deregulation of PELP1 that occurs in breast tumors can contribute to the activation of Src kinase, which leads to the excessive activation of the ER-PELP1–Src signaling pathway.
The ER requires both a ligand and interactions with other proteins, such as coregulators, to achieve the optimal transcriptional activation of target genes (1, 3, 8). In malignant tumors, several ER-coregulatory proteins are differentially expressed, and their dysregulation is thought to influence target gene expression and the eventual development of hormone-resistant cancers (3, 9). The results from our study suggest that PELP1 expression is deregulated in higher grade tumors and in node-positive metastatic tumors. Because PELP1 functions as an adaptor protein that couples ER with several signaling molecules, PELP1 deregulation may enhance ER cross-talk via cellular signaling pathways that lead to hormonal independence. However, the presence of PELP1 in ER-negative tumors also suggests the possibility that PELP1 signaling plays a role in ER-negative cells possibly by promoting nongenomic signaling. Our ongoing studies address the role of PELP1 in ER-negative cells.
The interaction of PELP1 with proteins (Src kinase, PI3K, FHL2) that are involved in cytoskeleton remodeling and the participation of PELP1 in E2-mediated nongenomic signaling pathways (9, 12, 15) also suggest that PELP1 may regulate E2-mediated cell migration and may have a role in metastasis. The results of our study showed that PELP1 overexpression uniquely enhances E2-mediated ruffles and filopodia-like structures, both of which regulate the attachment and movement of cytoplasmic components responsible for cell migration. A reduction in E2-mediated cell motility in cells treated with PELP1 siRNA also supports the theory that PELP1 signaling has a role in E2-mediated cell motility. The increased expression of PELP1 in metastatic model cells and node-positive tumors and its modulation of E2-mediated cytoskeletal changes and cell migration suggest that PELP1 deregulation may have a role in the metastasis of cancer cells.
In summary, the results of our study provide critical evidence that the ER coregulator PELP1 has tumorigenic potential, and that its expression is preferentially up-regulated in higher grade node-positive breast tumors. Future studies of the in vivo mechanism of PELP1 action and profiling the expression of PELP1 in a large number of tumor samples would enable the use of this novel ER-coregulatory protein as a diagnostic marker and as a target for novel therapies.
Supplementary Material
Acknowledgments
Grant support: NIH grant CA095681.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
References
- 1.McDonnell DP, Norris JD. Connections and regulation of the human estrogen receptor. Science. 2002;296:1642–4. doi: 10.1126/science.1071884. [DOI] [PubMed] [Google Scholar]
- 2.Carroll JS, Brown M. Estrogen receptor target gene: an evolving concept. Mol Endocrinol. 2006;20:1707–14. doi: 10.1210/me.2005-0334. [DOI] [PubMed] [Google Scholar]
- 3.Hall JM, McDonnell DP. Coregulators in nuclear estrogen receptor action: from concept to therapeutic targeting. Mol Interv. 2005;5:343–57. doi: 10.1124/mi.5.6.7. [DOI] [PubMed] [Google Scholar]
- 4.Acconcia F, Kumar R. Signaling regulation of genomic and nongenomic functions of estrogen receptors. Cancer Lett. 2006;238:1–14. doi: 10.1016/j.canlet.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 5.Losel R, Wehling M. Nongenomic actions of steroid hormones. Nat Rev Mol Cell Biol. 2003;4:46–56. doi: 10.1038/nrm1009. [DOI] [PubMed] [Google Scholar]
- 6.Schiff R, Massarweh SA, Shou J, et al. Advanced concepts in estrogen receptor biology and breast cancer endocrine resistance: implicated role of growth factor signaling and estrogen receptor coregulators. Cancer Chemother Pharmacol. 2005;561:10–20. doi: 10.1007/s00280-005-0108-2. [DOI] [PubMed] [Google Scholar]
- 7.Smith CL, O’Malley BW. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- 8.McKenna NJ, Lanz RB, O’Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20:321–44. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 9.Gururaj AE, Rayala SK, Vadlamudi RK, Kumar R. Novel mechanisms of resistance to endocrine therapy: genomic and nongenomic considerations. Clin Cancer Res. 2006;12:1001–7s. doi: 10.1158/1078-0432.CCR-05-2110. [DOI] [PubMed] [Google Scholar]
- 10.Barnes CJ, Vadlamudi RK, Kumar R. Novel estrogen receptor coregulators and signaling molecules in human diseases. Cell Mol Life Sci. 2004;61:281–91. doi: 10.1007/s00018-003-3222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vadlamudi RK, Wang RA, Mazumdar A, et al. Molecular cloning and characterization of PELP1, a novel human coregulator of estrogen receptor α. J Biol Chem. 2001;276:38272–9. doi: 10.1074/jbc.M103783200. [DOI] [PubMed] [Google Scholar]
- 12.Wong CW, McNally C, Nickbarg E, Komm BS, Cheskis BJ. Estrogen receptor-interacting protein that modulates its nongenomic activity-crosstalk with Src/Erk phosphorylation cascade. Proc Natl Acad Sci U S A. 2002;99:14783–8. doi: 10.1073/pnas.192569699. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Nair SS, Mishra SK, Yang Z, Balasenthil S, Kumar R, Vadlamudi RK. Potential role of a novel transcriptional coactivator PELP1 in histone H1 displacement in cancer cells. Cancer Res. 2004;64:6416–23. doi: 10.1158/0008-5472.CAN-04-1786. [DOI] [PubMed] [Google Scholar]
- 14.Barletta F, Wong CW, McNally C, Komm BS, Katzenellenbogen B, Cheskis BJ. Characterization of the interactions of estrogen receptor and MNAR in the activation of cSrc. Mol Endocrinol. 2004;18:1096–108. doi: 10.1210/me.2003-0335. [DOI] [PubMed] [Google Scholar]
- 15.Vadlamudi RK, Manavathi B, Balasenthil S, et al. Functional implications of altered subcellular localization of PELP1 in breast cancer cells. Cancer Res. 2005;65:7724–32. doi: 10.1158/0008-5472.CAN-05-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balasenthil S, Vadlamudi RK. Functional interactions between the estrogen receptor coactivator PELP1/MNAR and retinoblastoma protein. J Biol Chem. 2003;278:22119–27. doi: 10.1074/jbc.M212822200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manavathi B, Nair SS, Wang RA, Kumar R, Vadlamudi RK. Proline-, glutamic acid-, and leucine-rich protein-1 is essential in growth factor regulation of signal transducers and activators of transcription 3 activation. Cancer Res. 2005;65:5571–7. doi: 10.1158/0008-5472.CAN-04-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajhans R, Vadlamudi RK. Comprehensive analysis of recent biochemical and biologic findings regarding a newly discovered protein-PELP1/MNAR. Clin Exp Metastasis. 2006;23:1–7. doi: 10.1007/s10585-006-9019-9. [DOI] [PubMed] [Google Scholar]
- 19.Vadlamudi RK, Bagheri-Yarmand R, Yang Z, et al. Dynein light chain 1, a p21-activated kinase 1-interacting substrate, promotes cancerous phenotypes. Cancer Cell. 2004;5:575–85. doi: 10.1016/j.ccr.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 20.Vadlamudi RK, Balasenthil S, Broaddus RR, Gustafsson JA, Kumar R. Deregulation of estrogen receptor coactivator proline-, glutamic acid-, and leucine-rich protein-1/modulator of nongenomic activity of estrogen receptor in human endometrial tumors. J Clin Endocrinol Metab. 2004;89:6130–8. doi: 10.1210/jc.2004-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shalloway D, Johnson PJ, Freed EO, Coulter D, Flood WA., Jr Transformation of NIH3T3 cells by cotransfection with Src and nuclear oncogenes. Mol Cell Biol. 1987;7:3582–90. doi: 10.1128/mcb.7.10.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruppert JM, Vogelstein B, Kinzler KW. The zinc finger protein GLI transforms primary cells in cooperation with adenovirus E1A. Mol Cell Biol. 1991;11:1724–8. doi: 10.1128/mcb.11.3.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foster KW, Ren S, Louro ID, et al. Oncogene expression cloning by retroviral transduction of adenovirus E1A-immortalized rat kidney RK3E cells: transformation of a host with epithelial features by c-MYC and the zinc finger protein GKLF. Cell Growth Differ. 1999;10:423–34. [PubMed] [Google Scholar]
- 24.Hendrix ND, Wu R, Kuick R, Schwartz DR, Fearon ER, Cho KR. Fibroblast growth factor 9 has oncogenic activity and is a downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas. Cancer Res. 2006;66:1354–62. doi: 10.1158/0008-5472.CAN-05-3694. [DOI] [PubMed] [Google Scholar]
- 25.Dawson PJ, Wolman SR, Tait L, Heppner GH, Miller FR. MCF10AT: a model for the evolution of cancer from proliferative breast disease. Am J Pathol. 1996;148:313–9. [PMC free article] [PubMed] [Google Scholar]
- 26.Santner SJ, Dawson PJ, Tait L, et al. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res Treat. 2001;65:101–10. doi: 10.1023/a:1006461422273. [DOI] [PubMed] [Google Scholar]
- 27.3Vadlamudi RK, Balasenthil S, Sahin AA, et al. Novel estrogen receptor coactivator PELP1/MNAR gene and ERβ expression in salivary duct adenocarcinoma: potential therapeutic targets. Hum Pathol. 2005;36:670–5. doi: 10.1016/j.humpath.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002;2:101–12. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- 29.Shou J, Massarweh S, Osborne CK, et al. Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J Natl Cancer Inst. 2004;96:926–35. doi: 10.1093/jnci/djh166. [DOI] [PubMed] [Google Scholar]
- 30.Torres-Arzayus MI, Font de MJ, Yuan J, et al. High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer Cell. 2004;6:263–74. doi: 10.1016/j.ccr.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 31.Bagheri-Yarmand R, Talukder AH, Wang RA, Vadlamudi RK, Kumar R. Metastasis-associated protein 1 deregulation causes inappropriate mammary gland development and tumorigenesis. Development. 2004;131:3469–79. doi: 10.1242/dev.01213. [DOI] [PubMed] [Google Scholar]
- 32.Osborne CK, Hobbs K, Clark GM. Effect of estrogens and antiestrogens on growth of human breast cancer cells in athymic nude mice. Cancer Res. 1985;45:584–90. [PubMed] [Google Scholar]
- 33.Osborne CK, Zhao H, Fuqua SA. Selective estrogen receptor modulators: structure, function, and clinical use. J Clin Oncol. 2000;18:3172–86. doi: 10.1200/JCO.2000.18.17.3172. [DOI] [PubMed] [Google Scholar]
- 34.Clarke R, Leonessa F, Welch JN, Skaar TC. Cellular and molecular pharmacology of antiestrogen action and resistance. Pharmacol Rev. 2001;53:25–71. [PubMed] [Google Scholar]
- 35.Moy B, Goss PE. Estrogen receptor pathway: resistance to endocrine therapy and new therapeutic approaches. Clin Cancer Res. 2006;12:4790–3. doi: 10.1158/1078-0432.CCR-06-1535. [DOI] [PubMed] [Google Scholar]
- 36.Trevino JG, Summy JM, Gallick GE. SRC inhibitors as potential therapeutic agents for human cancers. Mini Rev Med Chem. 2006;6:681–7. doi: 10.2174/138955706777435724. [DOI] [PubMed] [Google Scholar]
- 37.Russello SV, Shore SK. SRC in human carcinogenesis. Front Biosci. 2004;9:139–44. doi: 10.2741/1138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.