Abstract
Defective function of the Sonic Hedgehog (SHH) signaling pathway is the most frequent alteration underlying holoprosencephaly (HPE) or its various clinical microforms. We performed an extensive mutational analysis of the entire human DISP1 gene, required for secretion of all hedgehog ligand(s) and which maps to the HPE 10 locus of human chromosome 1q41, as a HPE candidate gene. Here, we describe two independent families with truncating mutations in human DISP1 that resemble the cardinal craniofacial and neuro-developmental features of a recently described microdeletion syndrome that includes this gene; therefore, we suggest that DISP1 function contributes substantially to both of these signs in humans. While these clinical features are consistent with common HPE microforms, especially those linked to defective signaling by Sonic Hedgehog, we have insufficient evidence so far that functionally abnormal DISP1 alleles will commonly contribute to the more severe features of typical HPE.
Introduction
Holoprosencephaly (HPE) is the most common malformation of the developing brain and face, and its severity is often highly correlated with accompanying craniofacial findings (Muenke and Beachy 2000; Matsunaga and Shiota 1997; Cohen 2006; Ming and Muenke 2002). The latter are often indicative of abnormal midline development manifesting as hypotelorism, hypertelorism, midface hypoplasia, nasal anomalies, midline facial clefting and often extending towards microforms such as simple microcephaly with developmental delay, or solitary single maxillary incisor. While the etiology of HPE is highly heterogeneous and complex (Muenke and Beachy 2000; Cohen 2006), presumably including numerous genes and environmental factors (Ming and Muenke 2002; Krauss 2007), many of the key characteristics of both brain and facial anomalies can be experimentally demonstrated to result from defective function of the hedgehog-signaling pathway in these different structures (Cordero et al. 2004). Previous studies of HPE candidate genes had suggested that family-specific deleterious mutations of HPE genes could be a reliable indicator of the potential role of a gene in the overall pathogenetic process. All mutations thus far identified in components of the human HEDGEHOG pathway have been heterozygous variations in key pathway components including, most commonly, the SHH ligand itself (Roessler et al. 1996; Nanni et al. 1999; Traiffort et al. 2004; Maity et al. 2005), and less frequently the receptor PATCHED (Ming et al. 2002) and the transcription factor GLI2 (Roessler et al. 2003, 2005). Microdeletions have also been reported in HPE genes (Bendavid et al. 2005a, b) and affecting candidate HPE loci, including large microscopically visible deletions of chromosome 1q41q42 involving DISP1 (Shaffer et al. 2007).Therefore, we considered the human DISP1 [originally called DISPA based on its orthologous relationship to murine DispA (Ma et al. 2002)] gene as a strong candidate gene based on its location within the HPE 10 locus and its demonstrated requirement as a positive factor required for the efficient secretion of cholesterol-modified Shh from midline signaling structures. (Burke et al. 1999; Ma et al. 2002; Kawakami et al. 2002; Caspary et al. 2002; Nakano et al. 2004; Tian et al. 2004).
Subjects and methods
Study population
Our study consists of all available 463 unrelated cases from our HPE collection (that span the entire clinical spectrum of severity), 57 additional coded anonymous HPE samples from Rennes, France, as well as 95 unrelated individual normal controls (obtained as anonymous samples from the Coriell Institute for Medical Research that controlled primarily for common variants among the Caucasians of Northern European ancestry typical of the largest segment of our collection). All samples were investigated under an Institutional Review Board approved research protocol in accordance with the National Human Genome Institute, NIH guidelines. Recruitment of affected individuals over the past 17 years has been as inclusive as possible with an increasing fraction of diverse ethnicities (see Sect. “Discussion”).
The procedures for PCR amplification, mutation screening and DNA sequencing
The genomic organization of the human DISP1 gene was characterized by using nucleotide homology searches with the murine DispA gene (Ma et al. 2002) in public databases (BLASTN program: http://www.ncbi.nlm.nih.gov/blast/) and comparing this with the UCSC Bioinformatics Site gene annotation (http://genome.ucsc.edu/). Oligo™4.1 was used to design primers for the seven coding exons as described in Table 2. Amplification of human genomic DNA was performed in a 30-μl reaction volume, using 60–100 ng DNA template, 50 μM each of deoxynucleotide triphosphate, 0.25 μM of each primer, 3 μl of 10× PCR Amplification buffer (Invitrogen), 1.5 μl 10× Enhancer buffer (Invitrogen) and 0.3 μl Taq polymerase. All reactions were performed using a PTC-255 thermocycler (MJ Research, MA). Typical PCR cycling parameters were 95°C for 4 min followed by 30 cycles at 95°C, annealing at 62°C, extension at 72°C for 1 min, and a final extension step of 72°C for 5 min. One half of the PCR product was used for denaturing high-pressure liquid chromatography (dHPLC) analysis (WAVE™, Trangenomic, CA) and the remainder was retained for direct DNA sequencing. Amplicons displaying heterozygous profiles were purified using a high pure PCR purification kit (Roche, IN) and bi-directionally sequenced using the BigDye™version 3.1 terminator cycle sequencing kit according to the manufacturer's protocol (Applied Biosystems, CA) on an ABI 3100 automated sequencer.
Table 2.
Primers for human DISP1 mutational analysis
| Exon | Primer | T(°C) PCR | Size (bp) | Tm(°C) dHPLC |
|---|---|---|---|---|
| 1 | 5′ TCTTACTTAGAGTCAAGAAATTGG 3 | 53 | 509 | 60.5 |
| 5′ TGAATGCTAAAAGCAAAACTTTCG 3′ | ||||
| 2 | 5′ ATGTTATGATGTTTATGATGCTCTG 3′ | 50 | 250 | 54 |
| 5′ GAATTCCTCAAGCAGCCAACTCATG 3′ | ||||
| 3 | 5′ AGTTATGCAGCTCTGATAGCCGAC 3′ | 54 | 216 | 59 |
| 5′ CAATATTTGGAGATGATTTTAGGC 3′ | ||||
| 4–5 | 5′ ACTAATGAGCACCTGTAATTTTGC 3′ | 51 | 648 | 55 |
| 5′ TGGTTTGTTCATCTACAATGTCAC 3′ | ||||
| 6 | 5′ TGAATTATTTCCAAATCCTGAGTC 3′ | 45 | 233 | 53 |
| 5′ TAATACAACCTTATTTGTGCTAAG 3′ | ||||
| 7A | 5′ CCTTCTGCTTGTCTCTATCTCTGC 3′ | 53 | 525 | 58 |
| 5′ AGTATCCATTAGAAGATAATCCTG 3′ | ||||
| 7B | 5′ GATTGAGTTTGGTATCAAACACAG 3′ | 53 | 541 | 57.5 |
| 5′ TATATTAAGAAGATACCGCTCATG 3′ | ||||
| 7C | 5′ CATGGCTTCCAGCAGTTGTTGTGC 3′ | 53 | 565 | 58 |
| 5′ GTTTGGTTTCTCAGTTTTTGACAG 3′ | ||||
| 7D | 5′ ACATCGCCAGCCCAGCTTCCCAGG 3′ | 58 | 563 | 57.5 |
| 5′ CCAAGTTGTCAGCAGCATCACGCT 3′ | ||||
| 7E | 5′ CATGGGGCTGTCAGTTGCTGTTGC 3′ | 58 | 556 | 59 |
| 5′ GTATGTGTTTTGCTTTGTCCCTTG 3′ | ||||
| 7F | 5′ TACAGTGCAGTGCCTTTTCCCATG 3′ | 58 | 568 | 59 |
| 5′ AGGCAGGGACAGTGGTGGATGTGC 3′ | ||||
| 7G | 5′ TGAAGGCCACACACCAAGCTGTCG 3′ | 58 | 632 | 59 |
| 5′ AGGCACTGGTTCTGAATTGAATGC 3′ |
Functional analysis of human DISP1 alterations
The export of a Renilla luciferase-tagged Shh molecular probe in a transfected Drosophila S2R+ cell line is dependent on the transporter activity of co-transfected murine DispA (or synthetic versions designed to mimic mutations in the human DISP1 gene). Note that murine or human A genes are equivalent to the 1 genes, and are the only forms that are co-expressed with hedgehog genes in tissues producing the ligand. The murine or human B genes are indicative of the Disp2 orthologs; there is currently no known function for these B = 2 genes in vertebrate development. An export ratio index was measured essential as previously described (Ma et al. 2002), such that a ratio of 1 indicates that independent of the presence (or absence) of transfected Disp protein there is no effect on the measured export of the renilla-tagged Shh ligand; however, a positive ratio indicates that the export is enhanced by Disp bioactivity. Briefly, the S2R+ cells were maintained in Schneiders Drosophila medium supplemented with 10% fetal bovine serum and co-transfected with a modified Dispatched gene to be tested and a Shh-tagged reporter by the calcium phosphate technique. Three days after transfection the cells were collected by centrifugation and lysed directly in Passive Lysis Buffer (Promega Dual Luciferase, Promega). Conditioned media was further cleared by centrifugation at 21,000×g prior to analysis. Each Disp construct (murine or human) contained an N-terminal firefly luciferase tag to measure successful co-transfection (quantified on the X-axis as luciferase activity). Note that the ratio rapidly reaches a plateau value of approximately fourfold. A relative export efficiency index ratio was calculated using the following formula:
Results
Some DISP1 genetic variants are found only among HPE patients
The entire coding region of the human DISP1 gene was examined for genetic variants that might contribute directly towards defective signaling or potentially modify the effects of other factors influencing signaling strength. As illustrated in Table 1, we identified 12 alterations that were provisionally unique (i.e., not identified in healthy controls of Northern European ancestry or previously identified as a common variant in public databases) to the HPE patient group and served as candidate variants for more detailed analysis. Note that several of these variants were identified among self-identified Hispanic probands for whom ethnically matched samples were not studied. Since five of these changes occur in regions of the protein that are not conserved between the murine and human genes and the Drosophila disp (Burke et al. 1999) orthologs we focused our study on the remaining seven changes as suitable for functional analysis independent of the ethnicity of the subjects. Variants present equally in affected individuals and controls were not considered for detailed functional analysis (Fig. 1).
Table 1.
Genetic variation at the human DISP1 locus
| Provisionally unique | Coding position | Suitable for testing | Comment |
|---|---|---|---|
| Yes (Hispanic Caucasian) | A19V | No | Not conserved or present in Disp |
| Yes (Hispanic Caucasian) | H159Y | Yes | N-terminal intracellular domain |
| Yes (Caucasian, USA) | L469S | Yes | N-terminal intracellular domain |
| Yes (Caucasian, Italy) | W475X | Yes | Termination between loops 1–2 |
| Yes (Hispanic Caucasian, Mexico) | Q674H | Yes | Intracellular between loops 6–7 |
| Yes (Hispanic Caucasian) | Y734X | Yes | Termination within transmembrane 7 |
| Yes (Caucasian, USA) | F816V | Yes | Loop 7–8 |
| Yes (Caucasian, USA) | Q928R | Yes | Loop 7–8 |
| Yes (Caucasian, USA) | V1079M | No | Methionine normally present in murine Disp |
| Yes (Hispanic Caucasian) | Del(ELEH)>D | No | Not conserved |
| Yes (Caucasian, USA) | P1255L | No | Not conserved or present in Disp |
| Yes (Hispanic Caucasian) | A1471P | No | Not conserved or present in Disp |
| No | P25P | No | Common SNP including controls |
| No | E103N | No | Rs2789975 |
| No | V514V | No | Common SNP including controls |
| No | K945K | No | Rs2609355 |
| No | A1247T | No | Rs9441940 |
| No | P1274P | No | Rs9441941 |
| No | V1261M | No | Common SNP including controls |
| No | A637A | No | Synonymous change |
| Yes | D847D | No | Synonymous change |
| No | S1163G | No | Glycine the normal position in murine Disp |
| Yes | A182A | No | Synonymous change |
| No | IVS2-3T>G | No | Common SNP |
| No | IVS4+84T>C | No | Common SNP |
| No | IVS5-14T> C | No | rs2609359 |
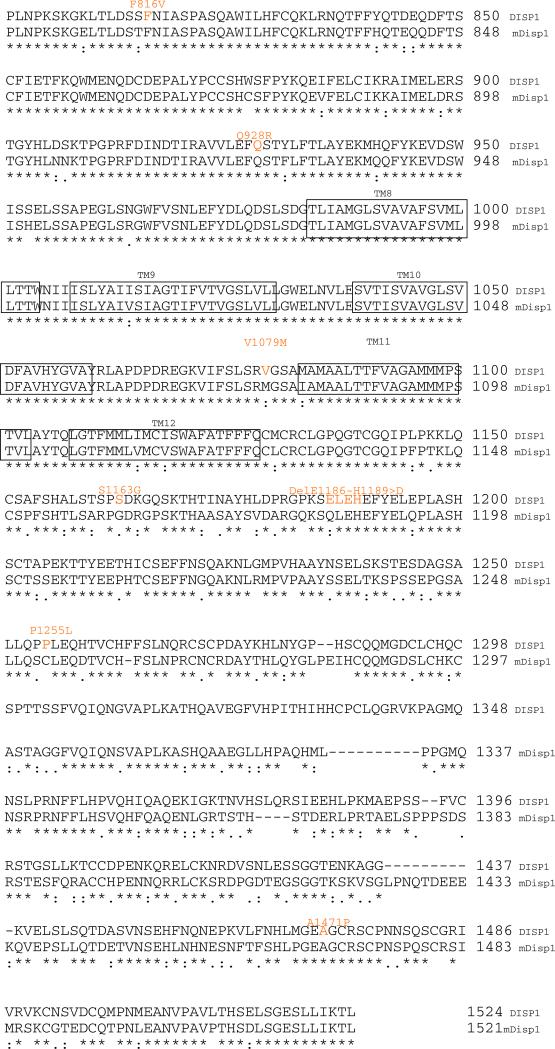
Fig. 1.
Protein alignment of human DISP1 and murine Disp1 proteins based on the Clustal W algorithm (http://www.ebi.ac.uk/Tools/clustalw2/index.html). Both of these vertebrate Disp are orthologs of Drosophila disp (not shown) are predicted to share a common topology with a core 12 membrane-spanning structure (each transmembrane element is boxed to indicate the membrane embedded amino acids) including the segment between boxed transmembrane regions (TM) 2–5 comprising the sterol-sensing domain. Key differences between genetic variants of the human and murine proteins are highlighted in orange (mis-sense or in-frame deletion) or red (truncating mutations)
Despite being apparently patient-specific, most novel variants are benign
As shown in Fig. 2a, the export of a Renilla-tagged Shh biomarker into the culture medium is an effective method to estimate the function of Disp alleles (Ma et al. 2002). Unfortunately, when we tested the ability of the human DISP1 gene to function in these cells we could not detect any enhancement of marker export. Presumably, the human gene product lacks some unknown feature that allows it to function efficiently in Drosophila cells. However, five of the mis-sense changes could readily be incorporated into the murine Disp1 gene for testing. None of these alterations clearly diminished the ability of the murine gene to act normally and are likely to represent benign variants. These cases may well prove to be variants typical of ethnic groups not commonly studied in our control group or publicly funded databases. Alternatively, these may affect the function of the gene in ways not successfully measured in our system.
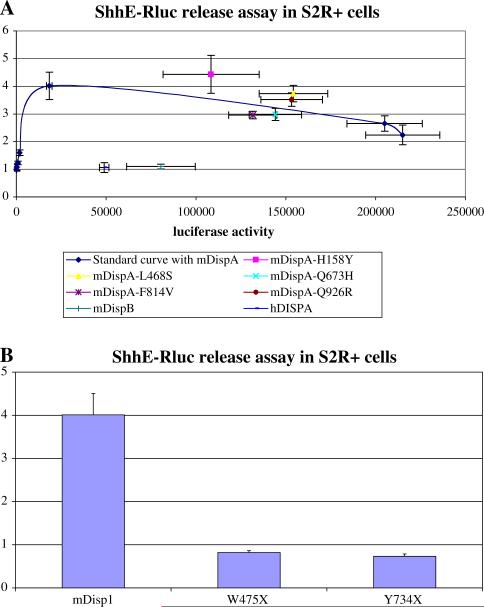
Fig. 2.
Determination of relative functional activity of genetic variants by assessment of the transporter function of Disp on a renilla-tagged Shh reporter in Drosophila S2R+ cells. a The standard dose–response curve with transfected murine DispA documents that increasing amounts of murine DispA (determined by measurement of the firefly luciferase activity on the X-axis) results in a maximum stimulation of ShhE-Rluc export of fourfold. In contrast, the murine DispB or the human DISPA fail to enhance ShhE-Rluc reporter export under these conditions (in spite of adequate expression of the N-firefly luciferase-tagged proteins). Five of the mis-sense changes incorporated within the murine DispA backbone cDNA could be studied; however, none of these variants appreciably affected the transporter-like function of the Disp test protein. b Normally, a bioactive murine Disp1 construct will enhance the export of the tagged Shh ligand by 4- to 10-fold (Ma et al. 2002). Both truncation mutations (murine construct versions mimicking W475X and Y734X) abrogate the ability of the Disp1 test molecule to influence ShhE-Rluc export. Values reported are the maximal release values obtained at a fixed input of Disp1 construct used in all three examples
Loss-of-function is associated with the truncation alleles
As shown in Fig. 2b, both truncation alleles eliminate the effective transporter activity when introduced into the murine Disp1 test construct, i.e., have an export ratio of less than unity. We conclude that individuals with similar truncations in the human gene would consequently have only a single functional copy of the DISP1 gene.
In the first family, a clinically un-affected mother has transmitted the W475X allele to her daughter who has a history of seizures, developmental delay including speech, a midline cleft-lip/palate and mild decortication. She was evaluated in the genetics clinic and diagnosed as HPE sequence with a normal head CT scan. Neither the incomplete penetrance nor the apparent absence of brain findings is inconsistent with microforms of the HPE spectrum (Muenke and Beachy 2000; Cohen 2006; Ming and Muenke 2002). Molecular evaluations included normal chromosome analysis, no apparent uniparental disomy for D2S44 (HPE2/SIX3) or deletion of D7S22 (HPE3/SHH), and no known coding region mutations in SHH, SIX3, TGIF, ZIC2, PATCHED, SMOOTHENED, DKK1, SIL or GLI2 genes.
The second family is quite similar with the presence of a clinically normal mother transmitting the Y734X mutation to her daughter who is of normal intelligence, has a normal karyotype, normal MRI, and normal molecular studies (vide supra). The proband has clear signs of microform HPE with a repaired bilateral cleft-lip/palate, hypotelorism, upslanting palpebral fissures and a solitary maxillary central incisor. All of these findings are typical of the mild end of the HPE spectrum and consistent with diminished signaling by Sonic Hedgehog as the likely mechanism.
Discussion
Our families with loss-of-function truncating mutations in human DISP1 are highly reminiscent of the newly described deletion syndrome that encompasses this gene (Shaffer et al. 2007). In this study seven different de novo cases of deletion of chromosome 1q41q42 that were identified and confirmed by comparative genomic hybridization were compared (range 2.72–9.07 Mb with the smallest region of deletion overlap estimated to be ~1.17 Mb and encode 4 additional known genes, other than DISP1). Common features included significant developmental delay, seizures, craniofacial dysmorphisms, microcephaly, cleft palate, clubfeet, and short stature. Interestingly, the authors noted that none of their cases were diagnosed with HPE. At the extreme end of the clinical spectrum of this series of cases were two who carried the diagnosis of Fryns syndrome.
Our two independent families underscore that the craniofacial and neuro-developmental features resulting from a single functional copy of DISP1 are clearly independent of the brain anomalies associated with more typical HPE. Interestingly, although at the present time the number of described cases are probably too small to generalize, we find it intriguing that only one of the five families with human GLI2 haploinsufficiency have overt brain findings of HPE either. Thus, there are actually numerous instances known of a single functional gene in an essential component in the Sonic Hedgehog pathway yet only a mild phenotype detected.
We note a trend in studies of model organisms that often the best characterized phenotype(s) reflect the extremes with complete absence of Disp1 activity and are also unfortunately embryonic lethals. However, it is intriguing that when extrapolated from an initial and admittedly limited set of experiments in the mouse (Tian et al. 2008), it can be inferred that hypomorphic alleles of Disp can be generated and that craniofacial abnormalities manifest themselves before frank central nervous system (i.e., cyclopia) abnormalities as the levels of Disp1 function are progressively decreased. Therefore, we think that our cases of DISP1 truncations reflect the fact that the activity of the Sonic Hedgehog pathway is significantly impaired, but by no means eliminated. A model of HPE that depends on the concerted action of multiple genetic and/or environmental insults (Ming and Muenke 2002) (i.e., the “multiple hit hypothesis”) could provide the best framework for a more comprehensive understanding of the complex interplay among HPE predisposing factors.
Acknowledgments
We thank the families who participated in these studies and for their ongoing support. This work was supported, in part, by the Division of Intramural Research, the National Human Genome Research Institute, NIH, and by research grants to P.A.B.
Footnotes
Present Address: P. A. Beachy Stanford University, Stanford, USA
Contributor Information
Erich Roessler, Medical Genetics Branch, National Human Genome Research Institute, National Institutes of Health, 35 Convent Drive-MSC 3717, Building 35, Room 1B-203, Bethesda, MD 20892-3717, USA.
Yong Ma, Johns Hopkins University, Baltimore, USA.
Maia V. Ouspenskaia, Medical Genetics Branch, National Human Genome Research Institute, National Institutes of Health, 35 Convent Drive-MSC 3717, Building 35, Room 1B-203, Bethesda, MD 20892-3717, USA
Felicitas Lacbawan, Medical Genetics Branch, National Human Genome Research Institute, National Institutes of Health, 35 Convent Drive-MSC 3717, Building 35, Room 1B-203, Bethesda, MD 20892-3717, USA.
Claude Bendavid, Laboratoire de Génétique Moléculaire, CHU Pontchaillou, IFR 140, Rennes, France; Génétique et Développement, Université de Rennes 1, IFR 140, Rennes, France.
Christèle Dubourg, Laboratoire de Génétique Moléculaire, CHU Pontchaillou, IFR 140, Rennes, France; Génétique et Développement, Université de Rennes 1, IFR 140, Rennes, France.
Philip A. Beachy, Johns Hopkins University, Baltimore, USA
Maximilian Muenke, Medical Genetics Branch, National Human Genome Research Institute, National Institutes of Health, 35 Convent Drive-MSC 3717, Building 35, Room 1B-203, Bethesda, MD 20892-3717, USA.
References
- Bendavid C, Haddad BR, Griffin A, Huizing M, Dubourg C, Gicquel I, Cavalli LR, Pasquier L, Long R, Ouspenskaia MV, Odent S, Lacbawan F, David V, Muenke M. Multicolor FISH and quantitative PCR can detect submicroscopic deletions in holoprosencephaly patients with a normal karyotype. J Med Genet. 2005a;43:496–500. doi: 10.1136/jmg.2005.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendavid C, Dubourg C, Gicquel I, Pasquier L, Saugler-Veber P, Durou M-R, Jaillard S, Frebourg T, Haddad BR, Henry C, Odent S, David V. Molecular evaluation of foetuses with holoprosencephaly shows high incidence of microdeletions in the HPE genes. Hum Genet. 2005b;119:1–8. doi: 10.1007/s00439-005-0097-6. [DOI] [PubMed] [Google Scholar]
- Burke R, Nellen D, Bellotto M, Hafen E, Senti K-A, Dickson BJ, Basler K. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell. 1999;99:803–815. doi: 10.1016/s0092-8674(00)81677-3. [DOI] [PubMed] [Google Scholar]
- Caspary T, Garcia-Garcia MJ, Huangfu D, Eggenschwiler JT, Wyler MR, Rakeman AS, Alcorn HL, Anderson KV. Mouse Dispatched homolog1 is required for long-range, but not juxtacrine, Hh signaling. Cur Biol. 2002;12:1628–1632. doi: 10.1016/s0960-9822(02)01147-8. [DOI] [PubMed] [Google Scholar]
- Cohen MM., Jr Holoprosencephaly: clinical, anatomic, and molecular dimension. Birth Defects Res Part A Clin Mol Teratol. 2006;76:658–673. doi: 10.1002/bdra.20295. [DOI] [PubMed] [Google Scholar]
- Cordero D, Marcucio R, Hu D, Gaffield W, Tapadia M, Helms JA. Temporal perturbations in sonic hedgehog signaling elicit the spectrum of holoprosencephaly phenotypes. J Clin Invest. 2004;114:485–494. doi: 10.1172/JCI19596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Kawcak TN, Zhang W, Hu Y, Chuang P-T. Mouse dispatched mutants fail to distribute hedgehog proteins and are defective in hedgehog signaling. Development. 2002;129:5753–5765. doi: 10.1242/dev.00178. [DOI] [PubMed] [Google Scholar]
- Krauss RS. Holoprosencephaly: new models, new insights. Expert Rev Mol Med. 2007;9(26):1–17. doi: 10.1017/S1462399407000440. doi:1.1017/S1462399407000440. [DOI] [PubMed] [Google Scholar]
- Ma Y, Erkner A, Gong R, Yao S, Taipale J, Basler K, Beachy PA. Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of Dispatched. Cell. 2002;111:63–75. doi: 10.1016/s0092-8674(02)00977-7. [DOI] [PubMed] [Google Scholar]
- Maity T, Fuse N, Beachy PA. Molecular mechanisms of Sonic hedgehog mutant effects in holoprosencephaly. Proc Natl Acad Sci USA. 2005;102:17026–17031. doi: 10.1073/pnas.0507848102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga E, Shiota K. Holoprosencephaly in human embryos: epidemiological studies on 150 cases. Teratology. 1997;16:261–272. doi: 10.1002/tera.1420160304. [DOI] [PubMed] [Google Scholar]
- Ming JE, Muenke M. Multiple hits during early embryonic development: digenic disease and holoprosencephaly. Am J Hum Genet. 2002;71:1017–1032. doi: 10.1086/344412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming JE, Kaupas ME, Roessler E, Brunner HG, Golabi M, Tekin M, Stratton E, Bale SJ, Muenke M. Mutations in PATCHED-1, the receptor for SONIC HEDGEHOG, are associated with holoprosencephaly. Hum Genet. 2002;110:297–301. doi: 10.1007/s00439-002-0695-5. [DOI] [PubMed] [Google Scholar]
- Muenke M, Beachy PA. Holoprosencephaly. In: Scriver CR, et al., editors. The metabolic and molecular bases of inherited disease. McGraw-Hill; New York: 2000. pp. 6203–6230. [Google Scholar]
- Nakano Y, Kim HR, Roy S, Schier AF, Ingham PW. Inactivation of dispatched 1 by the chameleon mutation disrupts hedgehog signaling in the zebrafish embryo. Dev Biol. 2004;269:381–392. doi: 10.1016/j.ydbio.2004.01.022. [DOI] [PubMed] [Google Scholar]
- Nanni L, Ming JE, Bocian M, Steinhaus K, Bianchi DW, de Die-Smulders C, Giannotti A, Imaizumi K, Jones KL, Del Campo M, Martin RA, Meinecke P, Pierpont MEM, Robin NH, Young ID, Roessler E, Muenke M. The mutational spectrum of the Sonic Hedgehog gene in holoprosencephaly: SHH mutations cause a significant proportion of autosomal dominant holoprosencephaly. Hum Mol Genet. 1999;8:2479–2488. doi: 10.1093/hmg/8.13.2479. [DOI] [PubMed] [Google Scholar]
- Roessler E, Belloni E, Gaudenz K, Jay P, Berta P, Scherer SW, Tsui L-C, Muenke M. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat Genet. 1996;14:357–360. doi: 10.1038/ng1196-357. 14. [DOI] [PubMed] [Google Scholar]
- Roessler E, Du YZ, Mullor JL, Casas E, Allen WP, Gillessen-Kaesbach Roeder ER, Ming JE, Ruiz i Altaba A, Muenke M. Loss-of-function mutations in the human GLI2 gene are associated with pituitary anomalies and holoprosencephaly-like features. Proc Natl Acad Sci USA. 2003;100:13424–13429. doi: 10.1073/pnas.2235734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Ermilov AN, Grange DK, Wang A, Grachtchouk M, Dlugosz AA, Muenke M. A previously unidentified amino-terminal domain regulates transcriptional activity of wild-type and disease-associate human GLI2. Hum Mol Genet. 2005;14:2181–2188. doi: 10.1093/hmg/ddi222. [DOI] [PubMed] [Google Scholar]
- Shaffer L, Theisen A, Bejjani BA, Ballif BC, Aylesworth AS, Lim C, McDonald M, Ellison JW, Kostiner D, Saitta S, Shaikh T. The discovery of microdeletion syndromes in the post-genomic era: review of the methodology and characterization of a new 1q41q42 microdeletion syndrome. Genet Med. 2007;9:607–616. doi: 10.1097/gim.0b013e3181484b49. [DOI] [PubMed] [Google Scholar]
- Tian H, Jeong J, Tabin CJ, McMahon AP. Mouse Disp1 is required in sonic hedgehog-expressing cells for paracrine activity of the cholesterol-modified ligand. Development. 2004;132:133–142. doi: 10.1242/dev.01563. [DOI] [PubMed] [Google Scholar]
- Tian H, Jeong J, Tabin CJ, McMahon AP. Mouse Disp1 is required in sonic hedgehog-expressing cells for paracrine activity of the cholesterol-modified ligand. Development. 2008;135:3471–3472. doi: 10.1242/dev.01563. Corrigendum. [DOI] [PubMed] [Google Scholar]
- Traiffort E, Dubourg C, Faure H, Rognan D, Odent S, Durou M-R, David V, Ruat M. Functional characterization of Sonic Hedgehog mutations associated with holoprosencephaly. J Biol Chem. 2004;279:42889–42897. doi: 10.1074/jbc.M405161200. [DOI] [PubMed] [Google Scholar]