Abstract
Phylogenetic relationships were evaluated among 13 species of Neotoma based on DNA sequences from intron 2 of the nuclear alcohol dehydrogenase gene 1 (Adh1-I2). Sequences were analyzed using parsimony, likelihood, and Bayesian methods. Three major clades (I–III) consistently were recovered and relationships among taxa within 2 of the clades remained unchanged between analyses; however, relationships within clade III were largely unresolved. Average genetic divergence values were 2.12% among species, 4% between subgenera (Teonoma and Neotoma), and 5.1% between genera (Hodomys and Neotoma). Adh1-I2 sequences were concatenated with mitochondrial cytochrome-b sequences generated from the same individuals. Examination of the combined data resulted in a phylogeny whose topology was similar to that based only on cytochrome-b sequences.
Keywords: alcohol dehydrogenase, mitochondrial cytochrome b, molecular systematics, Neotoma, nuclear DNA sequences
Woodrats (genus Neotoma) are medium-sized rodents distributed across portions of southern Canada, most of the continental United States, Mexico (including Baja California), and northern Central America (Hall 1981). Since Say and Ord (1825) erected the genus, there has been much debate on how many subgenera and species groups should be included within the genus and which taxa represent distinct species. Morphological data (cranial, skeletal, dental, and pelage) were 1st used to separate various members into a dichotomy, wherein round-tailed taxa were assigned to the genus Neotoma and bushytailed woodrats were placed into Teonoma (Gray 1843). Although Merriam (1894) constructed the 1st revision, Goldman (1910) published the most extensive revision, in which he divided the genus into 3 subgenera (Neotoma, Homodontomys, and Teonoma) and placed N. alleni as a separate genus (Hodomys). Later, Burt and Barkalow (1942) suggested H. alleni be relegated to subgeneric status; however, Hooper (1960) considered N. alleni to be most closely related to Xenomys nelsoni. Hall (1981) divided Neotoma into 4 distinct subgenera (Neotoma, Teonopus, Hodomys, and Teonoma); and consequently relegated Hodomys to subgeneric status. Subsequently, Carleton (1980) and Musser and Carleton (1993) agreed with Goldman’s (1910) assessment and recommended that Hodomys be reelevated to its former generic status and suggested that it is most closely related to X. nelsoni.
Traditionally, the subgenus Neotoma has been arranged into species groups to reflect similarities and relationships among taxa. Goldman (1910) described 6 species groups (albigula, desertorum, floridana, intermedia, mexicana, and pennsylvanica). Burt and Barkalow (1942) identified 6 species groups (albigula, floridana, fuscipes, lepida, mexicana, and micropus), although the species groups differed from Goldman’s (1910) assessment.
In addition to studies pertaining to higher taxonomic questions, various systematic studies have attempted to establish evolutionary relationships among the members of the subgenus Neotoma (Birney 1973; Burt and Barkalow 1942; Carleton 1980; Hayes and Harrison 1992; Hayes and Richmond 1993; Hooper 1960; Koop et al. 1985; Mascarello and Hsu 1976; Patton and Álvarez-Castañeda 2005; Planz et al. 1996; Schwartz and Odum 1957; Shipley et al. 1990; Zimmerman and Nejtek 1977). Several factors (taxon sampling, chromosomal conservatism, and ecological adaptation) have made it difficult to address phylogenetic relationships within Neotoma.
Recently, Edwards et al. (2001) and Edwards and Bradley (2001, 2002a, 2002b) used DNA sequence data from the mitochondrial cytochrome-b gene (Cytb) to evaluate phylogenetic relationships within Neotoma. These studies culminated in the recognition of 24 species of Neotoma, including the elevation of 4 subspecies (N. albigula leucodon, N. floridana magister, N. mexicana isthmica, and N. mexicana picta) to specific status. Edwards and Bradley (2002b) further suggested that these 24 species of Neotoma be placed into 4 species groups: the floridana group including N. albigula, N. floridana, N. goldmani, and N. magister; the lepida group including N. fuscipes, N. lepida complex, and N. stephensi; the mexicana group including N. angustapalata, N. chrysomelas, N. isthmica, N. mexicana, and N. picta; and the micropus group including N. micropus and N. leucodon. Finally, Edwards and Bradley (2002b) agreed with Goldman (1910), Carleton (1980), and Musser and Carleton (1993) in supporting the recognition of Hodomys as a separate genus and tentatively suggested that Neotoma consisted of 3 subgenera (Neotoma, Teonopus, and Teonoma).
Even though mitochondrial genes have provided substantial resolution concerning biological species in nature (Bradley and Baker 2001), and although the study of Edwards and Bradley (2002b) provides an effective hypothesis concerning phylogenetic relationships of the complex, a phylogeny obtained from nuclear data is needed as an independent data set to test those generated by mitochondrial data. Additionally, nuclear genes have been shown to be effective in resolving phylogenetic relationships within other groups of rodents (Adkins et al. 2001; DeBry and Sagel 2001; Flynn and Nedbal 1998; Huchon et al. 2002; Robinson et al. 1997; Walton et al. 2000) and offer an alternative to mitochondrial-based phylogenies.
The primary objective of this study was to evaluate phylogenetic relationships among members of Neotoma based on nucleotide sequence data from intron 2 of the nuclear alcohol dehydrogenase gene 1 (Adh1-I2). The choice of the Adh1-I2 region was based on evidence from classical protein electrophoreses studies that demonstrated that alcohol dehydrogenases were highly polymorphic in rodents and other mammals; by extension, intron regions should contain sufficient levels of polymorphisms for phylogenetic reconstruction. Bradley et al. (1993, 1998) demonstrated that nucleotide sequences from the coding region of Adh1 were species specific and Amman et al. (2006) presented preliminary data demonstrating that Adh1-I2 was informative in an examination of several genera of rodents. A 2nd objective was to concatenate and analyze Adh1-I2 sequences with mitochondrial DNA sequences from Cytb (Edwards and Bradley 2002b). To address these objectives, nuclear DNA sequences from 32 individuals, chosen from the study of Edwards and Bradley (2002b), were used to evaluate the Cytb phylogeny of Neotoma. Specifically, the following questions were addressed: are phylogenetic relationships among Neotoma based on DNA sequences from the nuclear Adh1-I2 congruent with the phylogeny of Edwards and Bradley (2002b) based on DNA sequences from the mitochondrial Cytb, is the genus Neotoma a monophyletic assemblage with the exclusion of Hodomys, and is Hodomys sister to Xenomys?
MATERIALS AND METHODS
Samples
This study used a subset of taxa examined in Edwards and Bradley (2002b). We selected 32 individuals from their study to represent members of primary clades, geographic variation for wide-ranging taxa, and at least 1 individual for each of 13 species of Neotoma including N. albigula (n = 3), N. cinerea (n = 2), N. floridana (n = 4), N. fuscipes (n = 1), N. goldmani (n = 1), N. isthmica (n = 2), N. lepida (n = 2), N. leucodon (n = 2), N. magister (n = 1), N. mexicana (n = 3), N. micropus (n = 4), N. picta (n = 1), and N. stephensi (n = 1). One individual each of H. alleni and X. nelsoni were included as in-group taxa. Outgroup taxa included 1 representative each of Tylomys nudicaudus, Ototylomys phyllotis, and Peromyscus attwateri.
Data collection
Genomic DNA was isolated from frozen liver tissue using a DNeasy tissue kit (Qiagen, Valencia, California). Primers anchored in exons 2 and 3 (EXONII-F and EXONIII-R or EXONII-F and 2340-II—Amman et al. 2006) were used to amplify the entire region of intron 2 of Adh1-I2 (583 base pairs [bp]) using the polymerase chain reaction (PCR—Saiki et al. 1988) with the following thermal profile: initial denaturation of 10 min; 25–30 cycles of 95°C (30 s) denaturing, 53°C → 48°C → 53°C → 73°C ramped at 0.6°C/s annealing, 73°C (1 min, 30 s) extension, followed by 1 cycle of 73°C (4 min). Amplifications were performed in 35-µl reactions using 1.25 U FailSafe PCR Enzyme Mix (Epicentre, Valencia, California) or AmpliTaq Gold (5 U/1 µl; PE Applied Biosystems, Foster City, California). Polymerase chain reaction products were purified using the QIAquick PCR Purification Kit (Qiagen).
Polymerase chain reaction amplicons were sequenced (25–30 cycles) using ABI Prism Big Dye Terminator v3.0 ready reaction mix and a 3100 Avant (both from PE Applied Biosystems) with cycle sequencing conditions of 95°C (30 s) denaturing, 50°C (20 s) annealing, and 60°C (3 min) extension. Six primers (EXONII-F, EXONIII-R, 2340-I, 2340-II, 350F, and 350R—Amman et al. 2006) were used in 1- to 2-µM concentrations in the sequencing protocol.
Sequencher 3.0 software (Gene Codes, Ann Arbor, Michigan) was used to align contiguous fragments and proof nucleotide sequences. Chromatograms were examined to verify all base changes and to check for heterozygous sites. Clustal X software (Thompson et al. 1997) was used for multiple sequence alignments of nucleotides that were adjusted and visually proofed with MacClade 4.0 software (Maddison and Maddison 2000). Sequence alignments produced gaps that represented insertion and deletion events (indels). Gaps were inserted because of primary homology assessment and the fact they often contain historical information suitable for phylogenetic analysis (Giribet and Wheeler 1999). All DNA sequences generated were deposited in GenBank and accession numbers are listed in Appendix I.
Because the Adh1-I2 primers used for amplification were placed in the conserved coding regions of the gene (exon 2 and exon 3), small fragments of exon 2 and exon 3 were amplified, resulting in a 732-bp region including approximately 107 bp of exon 2, 583 bp of intron 2, and 42 bp of exon 3. However, only sequences from the 583 bp of intron 2 were considered in the phylogenetic analyses.
Data analyses
Nucleotide sequence data were analyzed using parsimony, likelihood, and Bayesian models with the software package PAUP*4.0b10 (Swofford 2002) and MrBayes (Huelsenbeck and Ronquist 2001). All nucleotide positions were treated as 5 possible states: A, C, G, T, and gaps (–). Heterozygous sites were designated using the accepted International Union of Biologists polymorphic code.
To avoid the subjectivity inherent with differential weighting, maximum parsimony was performed using unordered, discrete characters with equal weighting (Allard and Carpenter 1996). Optimal trees were estimated using the heuristic search method with tree-bisection-reconnection branch swapping and stepwise addition sequence options. Uninformative characters were excluded from all parsimony analyses. Robustness and nodal support of topologies were assessed using heuristic bootstrapping (Felsenstein 1985) with 1,000 iterations and Bremer support indices (Bremer 1994), calculated with the Autodecay program (Eriksson 1997).
The HKY+G model of evolution was identified as the most appropriate model by Modeltest v3.06 software (Posada and Crandall 1998) and subsequently was used in a maximum-likelihood analysis (heuristic search option in PAUP). Parameters for this model, as determined by the likelihood ratio test, included the gamma shape distribution (γ = 0.9751), transition to transversion ratio of 2.06, and base compositions estimated from the data (A = 0.3207, C = 0.1885, G = 0.1720, and T = 0.3188).
MrBayes software (Huelsenbeck and Ronquist 2001) was used with the following parameters: GTR+I+G model, no data partitions, 4 Markov chains, 2 × 106 generations, and sampled every 100th generation. After a visual inspection of the likelihood scores, the first 100 trees were discarded to allow tree construction based on stabilized scores.
Pairwise genetic distances were generated using the data under the Tamura–Nei model of evolution (Tamura and Nei 1993). This model was chosen so that resulting values could be compared to those of other studies involving woodrats, particularly the Cytb data set (Edwards and Bradley 2001, 2002a, 2002b; Edwards et al. 2001). To obtain comparisons within and among species, mean pairwise values were calculated.
For comparison to the Adh1-I2 data set, Cytb sequences for the 32 samples obtained from Edwards and Bradley (2002b) were analyzed using MrBayes software (Huelsenbeck and Ronquist 2001). Parameters were as above, except that sequences were partitioned into codons, ssgamma value was used, and 300 trees were discarded.
The Adh1-I2 sequences were concatenated with the mitochondrial Cytb sequences from Edwards and Bradley (2002b) and analyzed as a single data set. Sequences for Cytb were generated from the same individuals as the Adh1-I2 sequences with 1 exception; for 1 sample of N. lepida, the Adh1-I2 and Cytb sequences were obtained from different individuals. MrBayes software was used for Bayesian analysis of the combined data set using the GTR+I+G model of evolution with the parameters as described above except that a mixed model was used for nucleotide position (Cytb sequences were partitioned into codons and Adh1-I2 sequences were not partitioned).
RESULTS
Nucleotide sequences were obtained from the nuclear Adh1-I2 locus for 32 individuals representing 6 genera and 18 species. The length of intron 2 ranged from 520 bp (N. floridana) to 572 bp (P. attwateri) among taxa because of indels, resulting in 583 aligned sites. Nucleotide frequencies were: A = 32.1%, C = 18.9%, G = 17.2%, and T = 31.9%. Comparison of nucleotide substitutions revealed that transitions were 2.1 times more common than transversions. Polymorphic sites ranged from 0 (28 taxa) to 2 (4 taxa) polymorphic sites. Indels varied in length and appeared to be conserved for different recognized species of woodrats. For example, a 4-bp deletion was present in all taxa except Peromyscus, members of the genus Neotoma shared 4-bp and 1-bp deletions, Ototylomys a 11-bp deletion, N. cinerea a 7-bp deletion, N. floridana attwateri a 48-bp deletion, N. goldmani a 10-bp deletion, Xenomys a 7-bp deletion, and Ototylomys and Tylomys shared a 9-bp deletion.
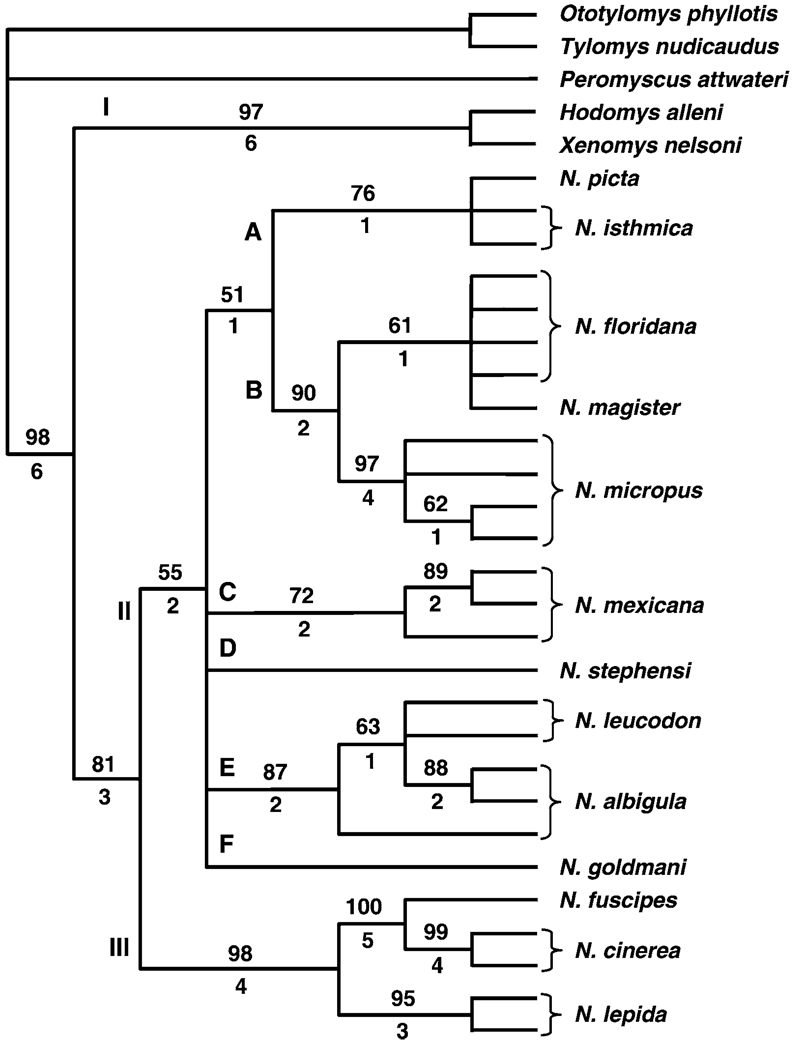
Under a parsimony framework, 499 uninformative characters were excluded resulting in 84 informative characters. Twelve equally most-parsimonious trees were generated (length = 123 steps, consistency index = 0.7561, and retention index = 0.8678). A strict consensus tree (Fig. 1) produced 3 major clades (I–III). Clade I contained X. nelsoni and H. alleni as sister taxa. Clade II included 22 individuals representing 10 species (N. picta, N. isthmica, N. floridana, N. magister, N. micropus, N. mexicana, N. stephensi, N. leucodon, N. albigula, and N. goldmani). Within this clade, 6 minor clades (A–F), primarily reflecting clusters of species, were formed which were unresolved relative to each other. Clade A contained samples of N. picta and N. isthmica; clade B contained 9 samples representing N. floridana, N. magister, and N. micropus; clade C was comprised of 3 individuals of N. mexicana; clade D contained a single species (N. stephensi); clade E contained 5 individuals representing 2 species (N. leucodon and N. albigula); and N. goldmani represented the basal clade (F). Clade III contained 5 individuals representing 3 species (N. fuscipes, N. cinerea, and N. lepida) with N. fuscipes and N. cinerea as sister followed by the addition of N. lepida. Clades II and III were sister, and clade I was basal. Bootstrap and Bremer support values for clades I and III were high, as were values within these clades. The minor clades (A–F) within clade II had low support (bootstrap values less than 50) resulting in these clades being unresolved. However, support within the minor clades was moderate to relatively high.
FIG. 1.
Strict consensus tree based on nucleotide sequences obtained from the nuclear alcohol dehydrogenase gene 1 using parsimony analysis (equal weighted characters and heuristic search option). Bremer support values are below branches and bootstrap values (≥50) are above branches. Roman numerals (I–III) depict major clades and letters (A–F) depict minor clades as defined in text.
Maximum-likelihood analysis based on the HKY+G model of evolution produced a score of −lnL = 2,045.34. The likelihood tree (not shown) was similar to the topology generated by parsimony and Bayesian analysis (discussed below) in that both contained identical major clades and similar minor clades. Clades II and III were sister to each other with clade I joining in a stepwise fashion. Six minor clades (A–F) were present and corresponded to those obtained in the parsimony analysis. Only clades A and B were resolved respective to other minor clades.
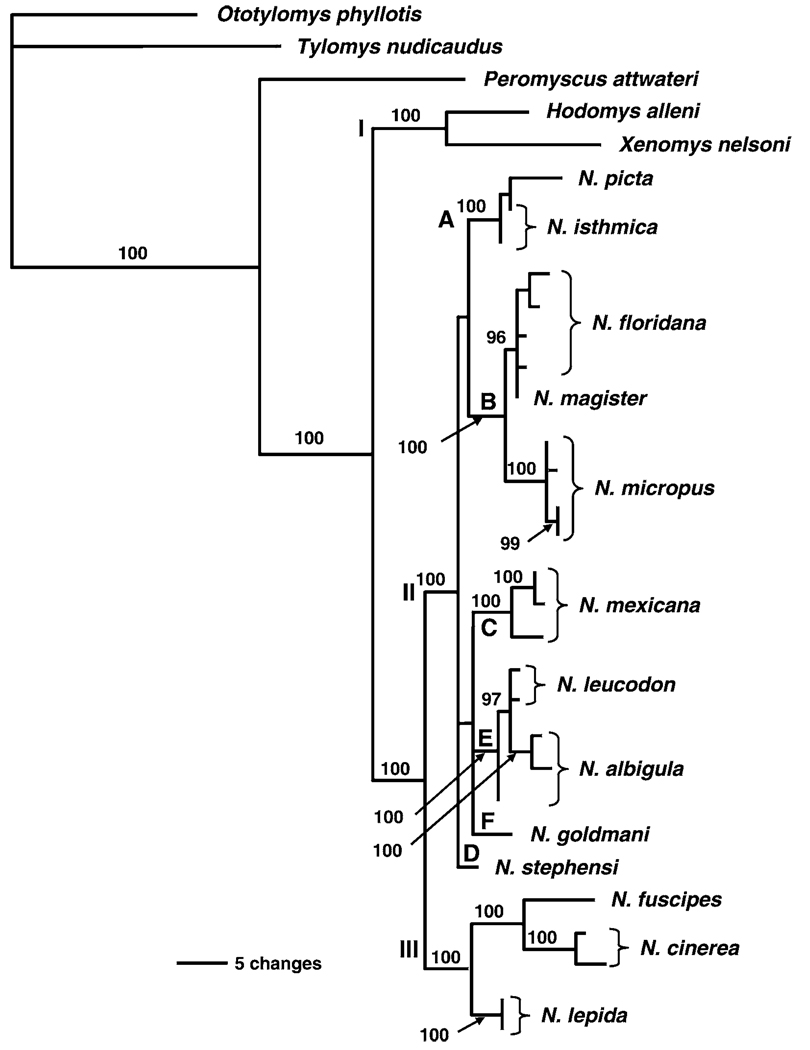
Bayesian analysis resulted in a tree (Fig. 2) similar to that of parsimony and likelihood analyses. Parametric posterior probabilities were generated and are shown on the tree. Three major clades (I–III), representing the same taxa as in the trees generated by parsimony and likelihood, were produced. Clades II and III were sister followed by the addition of clade I in a stepwise fashion. The minor clades (A–F) were unresolved (clade probability values < 95) but contained the same individuals as in the parsimony and likelihood analyses.
FIG. 2.
Phylogenetic tree based on nucleotide sequences obtained from the nuclear alcohol dehydrogenase gene 1 using Bayesian analysis (GTR+I+G model of evolution). Posterior probabilities are shown above branches. Roman numerals (I–III) depict major clades and letters (A–F) depict minor clades as defined in text.
Genetic distances (Table 1) calculated using the Tamura–Nei model of evolution (Tamura and Nei 1993) for members of the subgenus Neotoma ranged from 0.00% for the 2 samples of N. lepida to 2.63% between N. micropus and N. albigula. Between species within the subgenus Neotoma averaged 2.12%. The average genetic distance between Hodomys and the subgenus Neotoma was 5.09%, between Neotoma and Teonoma was 4.01%, and between Hodomys and Teonoma was 6.55%.
TABLE 1.
Average Tamura and Nei (1993) genetic distances for selected comparisons of taxa examined in this study. Genetic distances are based on nucleotide sequences from the nuclear alcohol dehydrogenase gene (Adh1-I2) and mitochondrial cytochrome-b gene (Cytb). Comparisons between taxonomic categories treat Neotoma fuscipes as a subgenus (Homodontomys).
| Taxon | Adh1-I2 | Cytb |
|---|---|---|
| Within N. floridana | 0.0034 | 0.0305 |
| Within N. micropus | 0.0024 | 0.0102 |
| Within N. albigula | 0.0072 | 0.0301 |
| Within N. mexicana | 0.0072 | 0.0175 |
| Within N. lepida | 0.0000 | 0.0106 |
| Within N. leucodon | 0.0036 | 0.0496 |
| Within N. cinerea | 0.0072 | 0.0196 |
| N. floridana versus N. magister | 0.0018 | 0.0856 |
| N. leucodon versus N. albigula | 0.0066 | 0.1189 |
| N. mexicana versus N. picta | 0.0237 | 0.0942 |
| N. mexicana versus N. isthmica | 0.0172 | 0.0882 |
| N. floridana versus N. albigula | 0.0219 | 0.1040 |
| N. micropus versus N. albigula | 0.0263 | 0.1344 |
| N. micropus versus N. mexicana | 0.0238 | 0.1276 |
| N. albigula versus N. mexicana | 0.0200 | 0.1306 |
| N. leucodon versus N. micropus | 0.0245 | 0.0975 |
| Within subgenus Neotoma | 0.0212 | 0.0932 |
| Within subgenus Neotoma | ||
| (without Homodontomys) | 0.0192 | 0.0901 |
| Subgenus Neotoma versus Hodomys | 0.0509 | 0.1871 |
| Subgenus Neotoma versus Homodontomys | 0.0426 | 0.1283 |
| Subgenus Neotoma versus Teonoma | 0.0401 | 0.1733 |
| Subgenus Homodontomys versus Teonoma | 0.0405 | 0.1717 |
| Genus Hodomys versus subgenus Teonoma | 0.0655 | 0.2236 |
| Genus Hodomys versus Homodontomys | 0.0654 | 0.1909 |
The Cytb data set used in the Bayesian analysis (GTR+I+G) produced a topology (not shown) substantially different from the analyses of the Adh1-I2 data (Fig. 1). However, the topology was identical to that reported in Edwards and Bradley (2002b) and similar to that generated in the combined analysis (Fig. 3).
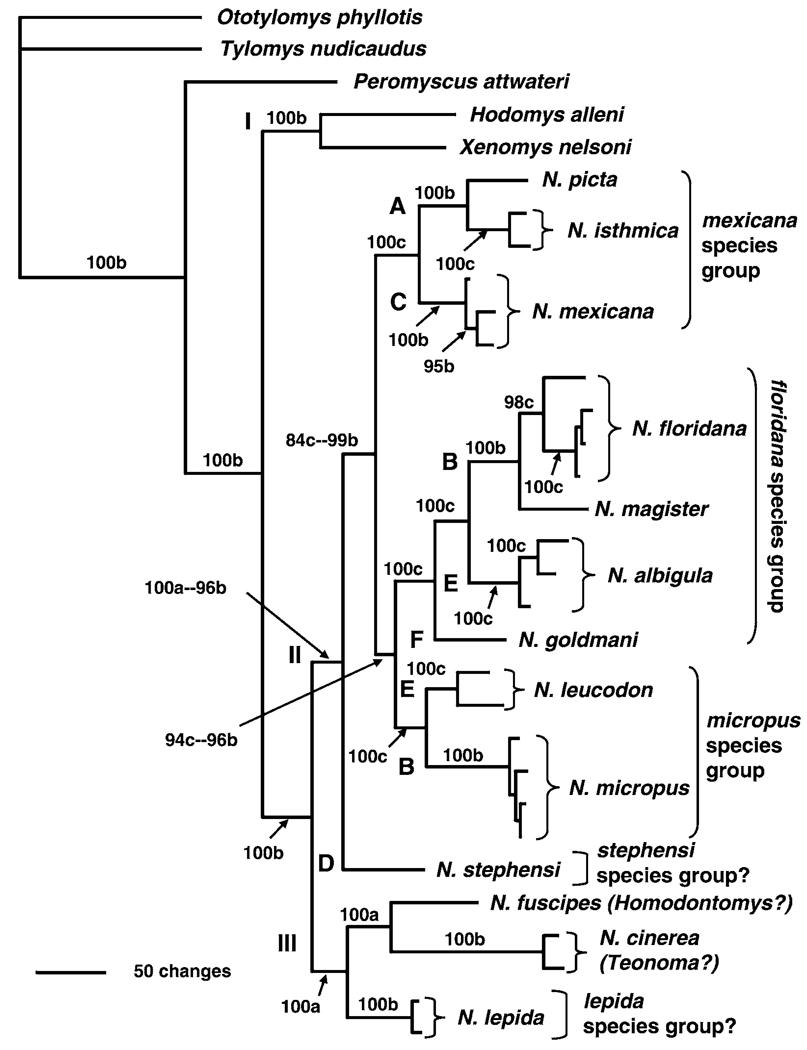
FIG. 3.
Phylogenetic tree based on combined analysis of the nuclear alcohol dehydrogenase gene 1 (Adh1-I2) and cytochrome-b gene (Cytb) using Bayesian analysis (GTR+I+G model of evolution). Posterior probabilities are shown above branches—values contributed by Adh1-I2 data are indicated by “a,” values contributed by Adh1-I2 and Cytb data (both data sets) are indicated by “b,” and values contributed by Cytb data are indicated by “c.” Roman numerals (I–III) depict major clades and letters (A–F) depict minor clades as defined in text.
The combined data set (concatenated sequences for Adh1-I2 and Cytb) analyzed using the Bayesian analysis (GTR+I+G) produced a topology (Fig. 3) similar to that produced in the Cytb only analysis (above); however, minor differences in the combined analysis did reflect contributions from the Adh1-I2 data set. For example, N. cinerea was sister to N. fuscipes with a posterior probability of 100. Several nodes were supported (posterior probabilities ≥ 95) in the combined data analysis.
DISCUSSION
DNA sequences were generated at the nuclear Adh1-I2 locus (583 aligned sites) for 32 individuals, representing 6 genera and 18 species. Although indels were present, they were not problematic for sequence alignment and were often phylogenetically informative. In addition, because heterozygosity was low (11 of 19,239 aligned sites), the effect on phylogenetic analyses was likely minimal. The high A-T content (64.1%) is characteristic of noncoding regions (Li and Graur 1991), but the transition to transversion ratio (approximately 2.1:1) is somewhat higher compared to other vertebrate introns. Prychitko and Moore (1997) reported a ratio of 1.24:1 within intron 7 of the beta fibrinogen gene (Fbg-I7) in woodpeckers, and Carroll and Bradley (2005) reported a value of 1.46:1 in cotton rats. However, Reeder and Bradley (in press) showed a higher ratio of 3:1 in Fbg-I7 within tribal levels of rodents; the higher value may be explained by the higher taxonomic ranks examined.
Similar topologies were obtained in all analyses (parsimony, likelihood, and Bayesian) of the Adh1-I2 data, whereas the topology generated from the Adh1-I2 data set differed substantially from those obtained from the Cytb data. However, the topology obtained from the Cytb data was similar to that reported in Edwards and Bradley (2002b) and similar to that generated in the combined analysis (Fig. 3). A direct comparison of the Adh1-I2 data to the Cytb data (herein and in Edwards and Bradley 2002b) produced the following generalities. First, the Adh1-I2 data failed to support the recognition of N. magister, N. leucodon, N. isthmica, and N. picta as distinct species. In each of these instances, it appears that insufficient genetic variation was present to differentiate the proposed cryptic species. For example, samples of N. leucodon and N. albigula differed by an average genetic distance of 0.0054; and N. isthmica and N. picta differed from N. mexicana (average genetic distances = 0.0194) and from each other by an average genetic distance of 0.0084. Examination of the Adh1-I2 data supports a sister taxon relationship between N. floridana and N. magister followed by the addition of N. micropus, whereas examination of the Cytb data supports the division of N. albigula into 2 species (N. albigula and N. leucodon) with N. albigula as sister to the N. floridana and N. magister clade and N. leucodon as sister to N. micropus. Likewise, the clade containing N. mexicana, N. picta, and N. isthmica obtained in the Cytb study was not supported by examination of the Adh1-I2 data, although N. picta and N. isthmica appeared to be distinct from N. mexicana. The 3 Adh1-I2 analyses did not depict the samples of N. picta, N. isthmica, and N. mexicana as monophyletic.
Second, examination of the Adh1-I2 data indicated that N. fuscipes and N. cinerea were sister taxa and were highly supported with a bootstrap value of 100 and a posterior probability of 100. N. lepida joined the N. fuscipes and N. cinerea clade in a stepwise fashion that also was highly supported with a bootstrap value of 97 and posterior probability of 100. However, examination of the Cytb data placed N. fuscipes as sister to N. lepida.
Third, no support was obtained for the placement of N. goldmani or N. stephensi because of low support values. Examination of the Cytb data placed N. goldmani as sister to a clade containing N. albigula, N. floridana, and N. magister, whereas N. stephensi was placed basally to a clade containing N. picta, N. isthmica, N. mexicana, N. floridana, N. magister, N. albigula, N. goldmani, and N. micropus.
Fourth, the Adh1-I2 data indicated that N. albigula, N. floridana, N. micropus, N. mexicana, N. goldmani, and N. stephensi are members of a single clade (II). Although examination of the Cytb data suggests that the floridana species group includes N. albigula and N. goldmani, examination of the Adh1-I2 data places N. goldmani as a member of the albigula group and sister to the N. mexicana clade. Adh1-I2 also supports the close alignment of these species by placing them in the same major clade (II).
Previously, Edwards and Bradley (2002b) suggested that the subgenus Teonoma (N. cinerea) might be a separate genus because of its placement as the basal clade of Neotoma and its high genetic distance value of 17.3% to the subgenus Neotoma. Similarly, the Adh1-I2 genetic distance values (4.01%) for a comparison of Teonoma to subgenus Neotoma indicated a relatively high level of divergence. In addition, examination of the Adh1-I2 data showed that the subgenus Neotoma and N. fuscipes were highly divergent, with an average genetic distance value of 4.3%, which was higher than for the previously mentioned subgenus Teonoma. Given the level of genetic divergence between Teonoma and Neotoma, an argument can be made to recognize N. fuscipes in the subgenus Homodontomys (Goldman 1910).
Combined Adh1-I2 and Cytb data
The combined data set (Adh1-I2 and Cytb sequences) supported most of the topology and phylogenetic relationships obtained from the Cytb data (Edwards and Bradley 2002b). In most cases, nodes had Bayesian posterior support values of ≥95, although this may be a consequence of the combined data set being more heavily weighted by the larger number of characters supplied by the Cytb data set. Minor differences existed in the inclusion of subgenus Teonoma (N. cinerea) in the lepida group (as defined by Edwards and Bradley [2002b]) and the placement of N. stephensi; however, the 3 major clades were retained. In addition, clade probability values were increased at all nodes except for the node that joined N. albigula to N. floridana and at the node joining the subspecies of N. micropus. Clade I placed Hodomys as a sister taxon to Xenomys. Clade II involved a reshuffling of the 6 minor clades (A–F) obtained in the analyses based only on the Adh1-I2 data. Clades A (N. picta and N. isthmica) and C (N. mexicana) formed sister clades and represented the mexicana species group as described by Edwards and Bradley (2002b). Clade B(N. floridana and N. magister) was sister to clade E (N. albigula), with N. goldmani joining in a stepwise fashion, and represented the floridana species group (Edwards and Bradley 2002b). Portions of Adh1-I2 clades B (N. micropus) and E (N. leucodon) formed sister taxa and supported the composition of the micropus species group of Edwards and Bradley (2002b). Clade D contained N. stephensi as the basalmost member of clade II. Clade III contained 3 species, with N. fuscipes and N. cinerea forming a sister taxon relationship followed by the addition of N. lepida.
Average genetic distances (Table 1) obtained using the Tamura–Nei model of evolution (Tamura and Nei 1993) indicated low levels of genetic divergence for the Adh1-I2 locus compared to those obtained from the Cytb sequences; in most cases, the Cytb genetic distances were about 4 times higher on average. For example, the average genetic distance between Hodomys and Xenomys was 4.3% and 16.4%, between Hodomys and the subgenus Neotoma was 5% and 19.1%, between Hodomys and the subgenus Teonoma was 6.6% and 22.4%, and between Teonoma and Neotoma was 4% and 17.2%, respectively. Values for comparisons between species within the subgenus Neotoma averaged 2.12% for the Adh1-I2 locus, whereas genetic distances obtained for the Cytb locus averaged 9.3%.
Taxonomic conclusions
The combined data set of Adh1-I2 and Cytb, for the most part, supports the recognized species groups (floridana, micropus, and mexicana) of Edwards and Bradley (2002b). However, the combined data set and the Adh1-I2 topologies and genetic distances warrant the placement of N. cinerea (subgenus Teonoma) within the N. lepida–N. fuscipes clade. If this arrangement is correct, these data argue for a reevaluation of N. lepida and N. fuscipes within the subgenus Neotoma. If Teonoma is a valid subgenus, then perhaps N. fuscipes should be recognized as a member of Homodontomys, following Goldman (1910). Although the genetic distance (4%) approached values that were seen between recognized genera (5%, Hodomys versus Neotoma) in the Adh1-I2 data, it is probably best to retain Teonoma as a subgenus within Neotoma, until a member of Teonopus (the 3rd subgenus in Neotoma) can be analyzed. Hodomys was placed as a sister taxon to Xenomys and appears to be a valid genus following Carleton (1980) and Edwards and Bradley (2002b).
The Adh1-I2 alone was not phylogenetically informative at the species-group level. The rate of evolution for the nuclear Adh1-I2 is slower (approximately 0.25%) than that reported for Cytb in Neotoma (Edwards and Bradley 2002b). The paucity of informative characters (86) may indicate that this region is evolving too slowly for phylogenetic relationships to be reconstructed at lower levels. This especially was true at the middle and terminal nodes where there were few characters (see Bremer support values; Fig. 1). Better support was evident near the base of the tree (Fig. 1) and between taxa representing older radiations (species groups, subgeneric, and generic levels). For example, in the combined data analyses, examination of the Adh1-I2 data helped increase nodal support (near the base) where the Cytb data displayed lower support values. Results from the combined data set as well as Adh1-I2 data are encouraging toward the goal of generating reliable phylogenetic relationships from nuclear genes, and the potential of combining data sets to produce comprehensive trees and relationships that are verified by high support values.
ACKNOWLEDGMENTS
We thank B. R. Amman, B. D. Baxter, R. R. Chambers, N. D. Durish, J. D. Hanson, M. L. Haynie, and F. Mendez-Harclerode for reviewing earlier versions of this manuscript. Tissue samples were provided by R. J. Baker (Natural Science Research Laboratory, Museum of Texas Tech University), T. L. Yates (Museum of Southwestern Biology, University of New Mexico), and C. F. Fulhorst (University of Texas Medical Branch at Galveston). Support for this research was provided by a National Institutes of Health grant (DHHS A141435-01 to RDB) and the Texas Tech University Center for Zoonoses and Epidemiology.
APPENDIX I
Specimens examined.—Collection localities, museum acronyms, and GenBank accession numbers are provided for each specimen examined in this study. Specimens are from the United States unless otherwise noted. Abbreviations for museum acronyms (in parentheses and to the left of the semicolon) follow Hafner et al. (1997): Angelo State University Museum Natural History Collections (ASNHC; San Angelo, Texas), Museum of Texas Tech University (TTU; Lubbock, Texas), The Museum of Southwestern Biology (MSB; Albuquerque, New Mexico), and Universidad Nacional Autónoma de México (UNAM; Mexico City, Distrito Federal, Mexico). GenBank accession numbers (AF or AY) for the nuclear alcohol dehydrogenase gene 1 (Adh1-I2) and cytochrome-b gene (Cytb) are provided in parentheses to the right of the semicolon and separated by a comma, respectively. TK is a special number of the Museum of Texas Tech University. UTM is universal transverse mercator coordinates.
Hodomys alleni.—MEXICO: Michoacan; 2 km N, 2 km W Caleta de Campos (TTU catalogue number unavailable, TK45042; AY817627, AF186801).
Neotoma albigula.—MEXICO: Chihuahua; 14 km E Cuidad de Chihuahua (MSB60812; AY817651, AF186804). Arizona; Apache County (MSB77371; AY817650, AF186808); Yuma County, 3.7 km S, 5.6 km W Somerton, UTM 11 708569E 3608362N (TTU78451; AY817648, AF376477). New Mexico; Otero County, UTM 13 403600E 3599552N (TTU76474; AY817649, AF186803).
Neotoma cinerea.—Colorado; Moffat County, 40°38′N, 108°19′W (MSB74610; AY817636, AF186800). Utah; San Juan County, Owaehamo Bridge (MSB121427; AY817635, AF186799).
Neotoma floridana.—Kansas; Lyon County, Ross Natural History Preserve, 6.4 km W, 1.6 km S Americus (TTU catalogue number unavailable, TK28244; AY817640, AF186818). Oklahoma; Creek County, Heyburn State Recreational Area (TTU catalogue number unavailable, TK27751; AY817639, AF294341). South Carolina; Richland County, Congaree Swamp National Monument, 33°49′N, 80°50′W (MSB74955; AY817637, AF294335). Texas; Anderson County, Gus Engeling Wildlife Management Area, UTM 15 226785E 3537370N (TTU75413; AY817638, AF186819).
Neotoma fuscipes.—California; Riverside County, Rancho Capistrano (Ortega Mountains) (TTU81391; AY817632, AF376479).
Neotoma goldmani.—MEXICO: Nuevo Leon; 1 km S Providencia (TTU45227; AY817656, AF186829).
Neotoma isthmica.—MEXICO: Chiapas; 14.4 km N Ocozocoautla, UTM 15 524171E 1852486N (TTU82666; AY817631, AF305567). Oaxaca; Las Minas, UTM 15 191165E 1824954N (TTU82665; AY817630, AF329079).
Neotoma lepida.—California; Orange County, Irvine Lake, 1.3 km E Fremont Canyon on Lake View access road (TTU79131; AY817633, AF307835); Mission Viejo, 1.6 km N Crown Valley Parkway on Camino Capistrano (TTU79134; AY817634, no Cytb data); Riverside County, 24.5 km NE Mecca on Box Canyon Road D (TTU41898; no Adh1-I2 data, AF376467).
Neotoma leucodon.—MEXICO: Durango; 2.4 km SE Las Herreras (TTU75440; AY817644, AF186809). Texas; Kerr County, Kerr Wildlife Management Area, UTM 14 452336E 3330772N (TTU71198; AY817643, AF186806).
Neotoma magister.—Virginia; Madison County, Shenendoah National Park, White Oak Canyon, 38°34′36″N, 78°22′30″W (MSB74952; AY817641, AF294336).
Neotoma mexicana.—Colorado; Las Animas County, Lake Dorothey State Wildlife Area, 37°00′11.3″N, 104°22′14″W (TTU catalogue number unavailable, TK51346; AY817647, AF186821). New Mexico; Los Alamos County, City of Los Alamos (TTU79129; AY817646, AF294345). Texas; Jeff Davis County, Mount Livermore Preserve, UTM 13 579953E 3389871N (TTU101643; AY817645, AF294346).
Neotoma micropus.—New Mexico; Otero County, Fort Bliss Military Base (TTU79096; AY817653, AF376474); Roosevelt County, 26.4 km S, 4.8 km E Taiban (TTU catalogue number unavailable, TK31643; AY817652, AF186822). Texas; Dimmitt County, Chaparral Wildlife Management Area, UTM 14 459023E 3133552N (TTU80855; AY817654, AF186826); (TTU80856; AY817655, AF186827).
Neotoma picta.—MEXICO: Guerrero; 6.4 km SSW Filo de Caballo (UNAM catalogue number unavailable, TK93390; AY817629, AF305569).
Neotoma stephensi.—Arizona; Navaho County, 4.8 km S Woodruff, UTM 12 588361E 3844338N (TTU78505; AY817642, AF308867).
Ototylomys phyllotis.—HONDURAS: Atlantida; Lancetilla Botanical Gardens, UTM 16 451012E 1740282N (TTU84371; AY817624, no Cytb data); MEXCIO: Quintana Roo, 1 km N Noh-bec (ASNHC7254; no Adh1-I2 data, AY009788).
Peromyscus attwateri.—Oklahoma; McIntosh County, 5 km E Dustin (TTU55688; AY817626, AF155384).
Tylomys nudicaudatus.—GUATEMALA: Izabal, Cerro San Gil (TTU62082; AY817625, AF307839).
Xenomys nelsoni.—MEXICO: Jalisco; 6 km SE Chamela (TTU37790; AY817628, AF307838).
LITERATURE CITED
- Adkins RM, Gelke EL, Rowe D, Honeycutt RL. Molecular phylogeny and divergence time estimates for major rodent groups: evidence from multiple genes. Molecular Biology and Evolution. 2001;18:777–791. doi: 10.1093/oxfordjournals.molbev.a003860. [DOI] [PubMed] [Google Scholar]
- Allard MW, Carpenter JM. On weighting and congruence. Cladistics. 1996;12:183–198. doi: 10.1111/j.1096-0031.1996.tb00008.x. [DOI] [PubMed] [Google Scholar]
- Amman BR, Hanson JD, Longhofer LK, Hoofer SR, Bradley RD. Intron 2 (Adh1-I2) of the alcohol dehydrogenase gene: a potential nuclear DNA phylogenetic marker for mammals. Occasional Papers, Museum of Texas Tech University. 2006;256:1–16. [Google Scholar]
- Birney EC. Systematics of three species of woodrats. Miscellaneous Publications, Museum of Natural History, University of Kansas. 1973;58:1–173. [Google Scholar]
- Bradley RD, Adkins RM, Honeycutt RL, Mcdonald JH. Nucleotide polymorphism at the alcohol dehydrogenase locus of pocket gophers, genus Geomys. Molecular Biology and Evolution. 1998;15:709–717. doi: 10.1093/oxfordjournals.molbev.a025974. [DOI] [PubMed] [Google Scholar]
- Bradley RD, Baker RJ. A test of the genetic species concept: cytochrome-b sequences and mammals. Journal of Mammalogy. 2001;82:960–973. doi: 10.1644/06-MAMM-F-038R2.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley RD, Bull JJ, Johnson AD, Hillis DM. Origin of a novel allele in a mammalian hybrid zone. Proceedings of the National Academy of Sciences. 1993;90:8939–8941. doi: 10.1073/pnas.90.19.8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer K. Branch support and tree stability. Cladistics. 1994;10:295–304. [Google Scholar]
- Burt WH, Barkalow FS., Jr A comparative study of the baculum of woodrats (subfamily Neotominae) Journal of Mammalogy. 1942;23:287–297. [Google Scholar]
- Carleton MD. Phylogenetic relationships in neotomine–peromyscine rodents (Muroidea) and a reappraisal of the dichotomy within New World Cricetinae. Miscellaneous Publications, Museum of Zoology, University of Michigan. 1980;157:1–140. [Google Scholar]
- Carroll DS, Bradley RD. Systematics of the genus Sigmodon: DNA sequences from beta-fibrinogen and cytochrome b. Southwestern Naturalist. 2005;50:342–349. [Google Scholar]
- Debry RW, Sagel RM. Phylogeny of Rodentia (Mammalia) inferred from the nuclear-encoded gene IRBP. Molecular Phylogenetics and Evolution. 2001;19:290–301. doi: 10.1006/mpev.2001.0945. [DOI] [PubMed] [Google Scholar]
- Edwards CW, Bradley RD. Molecular phylogenetics of the Neotoma floridana species group. Journal of Mammalogy. 2001;82:791–798. [Google Scholar]
- Edwards CW, Bradley RD. Molecular systematics and historical phylobiogeography of the Neotoma mexicana species group. Journal of Mammalogy. 2002a;83:20–30. [Google Scholar]
- Edwards CW, Bradley RD. Molecular systematics of the genus Neotoma. Molecular Phylogenetics and Evolution. 2002b;25:489–500. doi: 10.1016/s1055-7903(02)00294-4. [DOI] [PubMed] [Google Scholar]
- Edwards CW, Fulhorst CF, Bradley RD. Molecular phylogenetics of the Neotoma albigula species group: further evidence of a paraphyletic assemblage. Journal of Mammalogy. 2001;82:267–279. [Google Scholar]
- Eriksson T. Autodecay, version 3.03. Stockholm, Sweden: Botaniska Institution, Stockholm University; 1997. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Flynn JJ, Nedbal MA. Phylogeny of the Carnivora (Mammalia): congruence vs incompatibility among multiple data sets. Molecular Phylogenetics and Evolution. 1998;9:414–426. doi: 10.1006/mpev.1998.0504. [DOI] [PubMed] [Google Scholar]
- Giribet G, Wheeler WC. On gaps. Molecular Phylogenetics and Evolution. 1999;13:132–143. doi: 10.1006/mpev.1999.0643. [DOI] [PubMed] [Google Scholar]
- Goldman EA. Revision of the wood rats of the genus Neotoma. North American Fauna. 1910;31:1–124. [Google Scholar]
- Gray JE. List of the specimens of Mammalia in the collection of the British Museum. London, United Kingdom: British Museum (Natural History) Publications; 1843. [Google Scholar]
- Hafner MS, Gannon WL, Salazar-Bravo J, Alvarez-Castañeda ST. Mammal collections in the Western Hemisphere: a survey and directory of existing collections. Lawrence, Kansas: Allen Press; 1997. [Google Scholar]
- Hall ER. The mammals of North America. 2nd ed. New York: John Wiley & Sons, Inc.; 1981. [Google Scholar]
- Hayes JP, Harrison RG. Variation in the mitochondrial DNA and the biogeographic history of woodrats (Neotoma) in the eastern United States. Systematic Biology. 1992;41:331–344. [Google Scholar]
- Hayes JP, Richmond ME. Clinal variation and morphology of woodrats (Neotoma) of the eastern United States. Journal of Mammalogy. 1993;74:204–-216. [Google Scholar]
- Hooper ET. The glans penis in Neotoma (Rodentia) and allied genera. Occasional Papers of the Museum of Zoology, University of Michigan. 1960;618:1–20. [Google Scholar]
- Huchon D, et al. Rodent phylogeny and a timescale for the evolution of Glires: evidence from an extensive taxon sampling using 3 nuclear genes. Molecular Biology and Evolution. 2002;19:1053–1065. doi: 10.1093/oxfordjournals.molbev.a004164. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Koop BF, Baker RJ, Mascarello JT. Cladistic analysis of chromosomal evolution within the genus Neotoma. Occasional Papers, The Museum, Texas Tech University. 1985;96:1–9. [Google Scholar]
- Li W, Graur D. Fundamentals of molecular evolution. 2nd ed. Sunderland, Massachusetts: Sinauer Associates, Inc., Publishers; 1991. [Google Scholar]
- Maddison WP, Maddison DR. MacClade, version 4: analysis of phylogeny and character evolution. Sunderland, Massacusetts: Sinauer Associates, Inc., Publishers; 2000. [Google Scholar]
- Mascarello JT, Hsu TC. Chromosome evolution in woodrats, genus Neotoma (Rodentia: Cricetiae) Evolution. 1976;30:152–169. doi: 10.1111/j.1558-5646.1976.tb00892.x. [DOI] [PubMed] [Google Scholar]
- Merriam CH. Abstract of a study of the American wood rats, with descriptions of fourteen new species and a subspecies of the genus Neotoma. Proceedings of the Biological Society of Washington. 1894;9:117–128. [Google Scholar]
- Musser GG, Carleton MD. Family Muridae. In: Wilson DE, Reeder DM, editors. Mammal species of the world: a taxonomic and geographic reference. 2nd ed. Washington, D.C: Smithsonian Institution Press; 1993. pp. 501–755. [Google Scholar]
- Patton JL, Álvarez-Castañeda ST. Phylogeography of the desert woodrat, Neotoma lepida, with comments on systematics and biogeographic history. In: Sánchez-Cordero V, Medellín RA, editors. Contribuciones mastozoológicas en homenaje a Bernardo Villa. Ciudad de México, Distrito Federal, México: Instituto de Biología e Instituto de Ecología, Universidad Nacional Autónoma de México; 2005. pp. 375–388. [Google Scholar]
- Planz JV, Zimmerman EG, Spradling TA, Akins DR. Molecular phylogeny of the Neotoma floridana species group. Journal of Mammalogy. 1996;77:519–535. [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–118. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Prychitko TM, Moore WS. The utility of DNA sequences of an intron from the beta-fibrinogen gene in phylogenetic analysis of woodpeckers (Aves: Picidae) Molecular Phylogenetics and Evolution. 1997;8:193–204. doi: 10.1006/mpev.1997.0420. [DOI] [PubMed] [Google Scholar]
- Reeder SA, Bradley RD. Phylogenetic relationships of neotomine–peromyscine rodents using DNA sequences from intron 7 of the beta fibrinogen gene. In: Kelt DA, Lessa E, Patton JL, Salazar-Bravo JA, editors. The quintessential naturalist: honoring the life and legacy of Oliver P. Pearson. University of California Publications in Zoology; In press. [Google Scholar]
- Robinson M, Catzeflis F, Briolay J, Mouchiroud D. Molecular phylogeny of rodents, with special emphasis on murids: evidence from nuclear gene LCAT. Molecular Phylogenetics and Evolution. 1997;8:423–434. doi: 10.1006/mpev.1997.0424. [DOI] [PubMed] [Google Scholar]
- Saiki RK, et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Say T, Ord G. A new genus of Mammalia proposed, and a description of the species upon which it is founded. Journal of the Academy of Natural Sciences, Philadelphia. 1825;4:345–349. [Google Scholar]
- Schwartz A, Odum EP. The woodrats of the eastern United States. Journal of Mammalogy. 1957;38:197–206. [Google Scholar]
- Shipley MM, Stangl FB, Jr, Cate RL, Hood CS. Immunoelectrophoretic relationships among 4 species of woodrats (Cricetidae: Neotoma) Southwestern Naturalist. 1990;35:173–176. [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods) version 4.0b10. Sunderland, Massachusetts: Sinauer Associates, Inc., Publishers; 2002. [Google Scholar]
- Tamura K, Nei M. Model selection in the estimation of the number of nucleotide substitutions. Molecular Biology and Evolution. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;24:4876–4482. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton AH, Nedbal MA, Honeycutt RL. Evidence from intron 1 of the nuclear transthyretin (prealbumin) gene for the phylogeny of African mole-rats (Bathyergidae) Molecular Phylogenetics and Evolution. 2000;16:467–474. doi: 10.1006/mpev.2000.0808. [DOI] [PubMed] [Google Scholar]
- Zimmerman EG, Nejtek ME. Genetics and speciation of 3 semispecies of Neotoma. Journal of Mammalogy. 1977;58:391–402. [Google Scholar]