Abstract
Molecular modeling and dynamics simulations have been performed to study how cocaine inhibits dopamine transporter (DAT) for the transport of dopamine. The computationally determined DAT-ligand binding mode is totally different from previously proposed overlap binding mode in which cocaine- and dopamine-binding sites are the same (Beuming, T. et al. Nature Neurosci. 2008, 11, 780–789). The new cocaine-binding site does not overlap with, but close to, the dopamine-binding site. Analysis of all results reveals that when cocaine binds to DAT, the initial binding site is likely the one modeled in this study, as this binding site can naturally accommodate cocaine. Then, cocaine may move to the dopamine-binding site after DAT makes some necessary conformational change and expands the binding site cavity. It has been demonstrated that cocaine may inhibit the transport of dopamine through both blocking the initial DAT-dopamine binding and reducing the kinetic turnover of the transporter following the DAT-dopamine binding. The relative contributions to the phenomenological inhibition of the transport of dopamine from blocking the initial binding and reducing the kinetic turnover can be different in different types of assays. The obtained general structural and mechanistic insights are consistent with available experimental data and could be valuable for guiding future studies towards understanding cocaine inhibiting other transporters.
Introduction
Cocaine has been recognized as one of the most psychostimulant drugs abused by millions of people worldwide. The disastrous consequences of cocaine addiction has brought tremendous burden to our society through increasing crimes, medical expenses, and loss of lives.1,2,3,4 Biological studies revealed that the primary target for cocaine in human body is dopamine transporter (DAT).5,6 It has been proposed that the rewarding and reinforcing effects of cocaine are mediated predominantly by its inhibition of DAT.4, 7 Cocaine analog, (−)-2β-carbomethoxy-3β-(4-iodophenyl) tropane (β-CIT, see Figure S1 of Supporting Information for its structure), was found8 to be competitive with substrate dopamine in the binding with DAT. Considering that cocaine and analog β-CIT bind with DAT at the same site,9 cocaine can be regarded as a competitive inhibitor of DAT according to the binding assay. However, results obtained from various kinetic inhibition assays on the inhibition of the transport of dopamine have been confusing, as different kinetic assays10, 11, 12 demonstrated different patterns of inhibition by cocaine, including competitive, noncompetitive, and uncompetitive. Apparent patterns of inhibition of transport vary with the type of transport experiment being conducted. The inhibition of the transport of dopamine by cocaine appears to result from the external binding of cocaine to the transporter.9 Hence, it is important to understand how the transport of dopamine through DAT is inhibited by cocaine binding with DAT.
Like other members of neurotransmitter sodium symporters (NSS) family, DAT binds with dopamine released from synaptic cleft and transports dopamine into pre-synaptic neurons. The process of the transport of dopamine must be assisted by the binding of two Na+ ions and one Cl− ion, in which DAT frequently switches between different conformational states. 6,7, 13, 14, 15 The determination of X-ray structure of LeuTAa in complex with its substrate Leucine and two Na+ ions (PDB entry of 2A65 at 1.65 Å resolution) has been viewed as a milestone in understanding structural and functional relationships of NSS members.16 Since then, computational and experimental studies17,18,19,20,21 have addressed several critical questions of NSS members including DAT, e.g. the binding site of Cl− ion, the selectivity of cationic ions, and the mechanism for inward-releasing of Na+ and substrate. These studies16,17,18,19,20,21 have made the Na+/Cl−-dependent transporting more visible. Meanwhile, co-crystal structures of LeuTAa with antidepressant drugs revealed that noncompetitive ligands bind in a vestibule open toward extracellular side (called “extracellularly-open” state/conformation below for convenience). Such a binding site for noncompetitive ligands is about 11 Å above the binding sites of substrate and two Na+ ions, thus stabilizing the extracellularly-open conformation of the transporter.22,23 These progresses16,22,23 provide fundamental information about ligands interacting with NSS members such as DAT, making possible exploring the inhibitory mechanism of cocaine inhibiting DAT at atomic resolution.
So far, considerable experimental and computational studies, including Zn2+-site engineering, site-directed mutagenesis, and molecular dynamics (MD) simulations, have been performed to explore the possible mechanism of cocaine inhibiting DAT.15,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39 Most of these studies were focused on exploring possible binding site of cocaine in DAT and the extent of overlap between cocaine- and dopamine-binding sites.15,25,26,27,31,32,33,35,36,38,39 Many of the studies aimed to identify key residues of DAT that can differentiate the binding of cocaine and other non-addictive ligands. Identification of such residues would be valuable for rational design of novel agents that can selectively inhibit cocaine binding as antagonists, without (or with little) effect on the dopamine uptake.28,29,30,35,36,38,39 However, no satisfactory antagonist has been identified yet. As indicated by the X-ray crystal structures of LeuTAa complexed with its substrate and typical antidepressant drugs like clomipramine (CMI), the transporting of substrate was inhibited by large-size inhibitors (e.g. clomipramine, its structure is shown in Figure S1 of Supporting Information) due to the binding of these inhibitors at a site located in the substrate-entry tunnel.16,22,23 A recent study21 suggested that LeuTAa has a second binding site for its substrate Leucine, which is located at the similar position as the reported binding sites for antidepressant drugs.22,23 All these studies21,22,23 demonstrated that the substrate cannot be transported once the second binding site was occupied by an inhibitor. In our previous study,40 we found that DAT shared the same mechanism of substrate binding as that of LeuTAa. Our previously modeled structure of DAT also has a substrate-entry tunnel which is formed by transmembrane (TM) helices 1, 3, 6, 8, and 10 and the tunnel opens toward extracellular side. However, Beuming et al.38 proposed that the binding site of cocaine and its analog (−)-2β-carbomethoxy-3β-(4-fluorophenyl)tropane (β-CFT, Figure S1 in Supporting Information) overlaps with the binding site of substrate dopamine based on the fact that the cocaine and the analog were observed to be competitive with dopamine in the binding assays.
Based on the assumption that cocaine binds with DAT in the dopamine-binding site, Beuming et al.38 built a homology model of DAT similar to our aforementioned homology model40 (by using the same template LeuTAa with substrate bound, PDB entry of 2A65) and constructed a 3D model of the DAT-cocaine binding complex. In order to obtain a reasonable DAT-cocaine binding structure, Beuming et al.38 had to expand the binding site cavity as the directly modeled dopamine-binding site cavity is not large enough to accommodate a larger ligand like cocaine. Nevertheless, the proposed DAT-cocaine binding mode, denoted by “overlap binding mode” here for convenience, was supported by their new site-directed mutagenesis and cross-linking experiments.38 In particular, according to the overlap binding mode, cocaine should be close to Val152, Asn157, Val328, and Ser422 and mutations on these residues indeed significantly decreased the inhibitory activity of cocaine and binding affinity of β-CFT.38 However, the overlap binding mode cannot explain some of the previously reported experimental observations very well. For example, the Trp84Leu mutation significantly increased the binding affinity of cocaine with DAT (~8-fold decrease of the apparent equilibrium dissociation constant).28,39 The Leu104Val/Phe105Cys/Ala109Val mutant of DAT was ~70-fold more insensitive to cocaine inhibition than the wild-type, but the mutant retained >50% transporting activity in cultured cells.4 According to the overlap binding mode,38 these four amino acid residues (Trp84, Leu104, Phe105, and Ala109) should be all far away from cocaine, making these experimental observations4,28,39 hard to be explained as allosteric effects on the binding of cocaine.
Based on our computational studies on DAT-ligand binding in combination with the findings of recent reports,9,16,21,22,23,39,40 we demonstrate in the present study that DAT has a new, alternative cocaine-binding site which is near the extracellular end of substrate-entry tunnel. Our computational results and analysis of available data reveal that, at least for its initial binding with DAT, cocaine can bind to this new binding site without the need of expanding the binding site cavity. We first constructed a 3D model of the complex of DAT with cocaine in the presence of dopamine started from our previously modeled DAT-dopamine complex structure.40 MD simulations have been performed on the complex structure in an environment consistent with the physiological condition. Our molecular modeling and MD simulations reveal that cocaine is bound in the alternative/initial binding site located on the substrate-entry tunnel, typically composed of residues from TM helices 1, 3, 6, 8, and 10. The benzoyl ester group of cocaine is buried by conserved residues Leu80, Ala81, Ile159, Phe155, Tyr156, and Phe265. The cationic head group of cocaine is partially covered by extracellular loop 4 (EL-4) and in contact with residues from TM helices 3 and 6. Based on the modeling results, a detailed molecular mechanism for cocaine inhibiting the transport of dopamine is proposed to reasonably interpret aforementioned different patterns of the inhibition.8,10,11,12
Computational Methods
Model refinement and molecular docking
According to the experimental studies,7,24,25,32 EL-2 of DAT does not directly affect dopamine entry into DAT or dopamine binding with the transporter. It has been demonstrated that there is a disulfide bond inside EL-2.33 EL-2 is also involved in the formation of endogenous Zn2+-binding site around extracellular loops including part of EL-4.7,25 In our previous modeling of DAT structure,40 the initial structure of the DAT model was constructed according to the structural information available at that time. The initial structure of DAT was relaxed by carrying out MD simulation in a physiological environment. Since then, further experimental studies have resulted in new structural insights reported recently.17,18,25,33 These newly reported structural insights were used to refine our DAT model further in the present study. First, a disulfide bond was assigned between residues Cys180 and Cys189 from EL-2, in light of a recent experimental finding reported by one of us.33 According to the latest experimental studies on the binding site of Cl− ion in homologous LeuTAa,17,18 one Cl− ion was incorporated into the binding site formed by side chains of Tyr102, Ser321, Asn353, Ser354, and Ser357 of DAT. Further modeling allowed the side chains of His193 from EL-2 and His375 and Glu396 from EL-4 to be close to each other and mimic the Zn2+-binding site, which is consistent with the recent experimental observation.25 Finally, the DAT model was energy-minimized by using the Sander module of Amber8 program suite with a nonbonded cutoff of 12 Å and a conjugate gradient energy-minimization method.41
Recent studies have shown that cocaine inhibits dopamine reuptake through its binding with the extracellularly-open state of DAT.9,10,11,12,27,28,29,32,35,36,38,39 Starting from the newly refined DAT structure bound with dopamine, herein denoted by DAT-DA, molecular docking was carried out by using AutoDock3.0.5 program42 to explore the mode of cocaine binding with DAT in the presence of dopamine. Briefly, the atomic charges of protonated cocaine, i.e. the biologically active (−)-cocaine, used in molecular docking were the RESP charges determined and used in our previous studies on cocaine hydrolysis by butyrylcholinesterase (BChE).43,44,45,46,47,48,49 During the process of docking with the AutoDock3.0.5 program, the Lamarckian genetic algorithm42 was used to account for the translation and orientation of the ligand with respect to the protein, and the search for local conformation of cocaine molecule itself in the binding site was performed by using the Solis and Wets search method.50 During the docking process, the grid size was set to be 60 × 60 × 60 and the grid space was the default choice of 0.375 Å. The possible binding site of cocaine was first roughly assigned as that of the ligand in the co-crystal structures of LeuTAa with antidepressant drugs,22,23 i.e. cocaine binds at the vestibule around extracellular end of helices 1, 3, 6, 8, and 10. The binding site of cocaine was further explored by changing the center and the size of docking grid. All the obtained possible complex structures were evaluated and ranked in terms of geometric matching quality and binding affinity by using the standard scoring function implemented in the AutoDock 3.0.5 program.42 By visualizing the docking structures, the microscopic binding structure which has the best score and best geometric matching with the vertical vestibule of DAT-DA was selected as the initial complex structure for energy minimization using the Sander module of Amber8 program.41
Molecular dynamics simulation
In order to further relax the structure of DAT-DA-cocaine complex selected from molecular docking, MD simulations were carried out on the complex by using the Sander module of Amber8 program.41 The complex structure was inserted into a pre-equilibrated palmitoyloleoyl phosphatidylcholine (POPC) phospholipid bilayer structure and then solved by two layers of water molecules at each side of the phospholipid bilayer. Atomic charges and force field parameters for POPC molecules were directly adopted from our previous study,40 which were developed based on the ab initio electronic structure calculations at the HF/6-31G* level. The final system size was ~133 Å × 131 Å × 112 Å and consisted of 141,039 atoms, including 351 phospholipid molecules and 28,540 water molecules. The DAT-DA-cocaine complex was energy-minimized in a similar way as described above for energy minimization of the DAT model. After the energy minimization, MD simulations were performed on water molecules in NTP ensemble for 200 ps, and then on phospholipid molecules for 82 ps. Then, the whole solvated DAT-DA-cocaine complex system was slowly heated to 300 K by weak-coupling method51 and further equilibrated for additional 40 ps. For MD simulations, a 12 Å non-bonded interaction cutoff was used and the non-bonded list was updated every 25 steps and the translational motion for the center of mass of the system was removed every 1,000 steps. The particle-mesh Ewald (PME) method52 was applied to treat long-range electrostatic interactions. The lengths of covalent bonds involving hydrogen atoms were fixed with the SHAKE algorithm,53 enabling the use of a 2-fs time step to numerically integrate the equations of motion. Finally, the production MD was kept running for 4.5 ns with a periodic boundary condition in the NTP ensemble at T = 300 K with Berendsen temperature coupling, and at P = 1 atm with anisotropic molecule-based scaling (ntp=2 option in the Amber program41). The surface tension of membrane molecules was kept constant (by using the ntp=2 option available in the Amber8 program) throughout the MD simulation process.54
Most of the MD simulations were performed on a supercomputer (e.g. IBM X-series Cluster with 340 nodes or 1,360 processors) at University of Kentucky Center for Computational Sciences. Some other modeling and computations were carried out on SGI Fuel workstations and a 34-processor IBM x335 Linux cluster in our own lab.
Results and Discussion
Binding mode
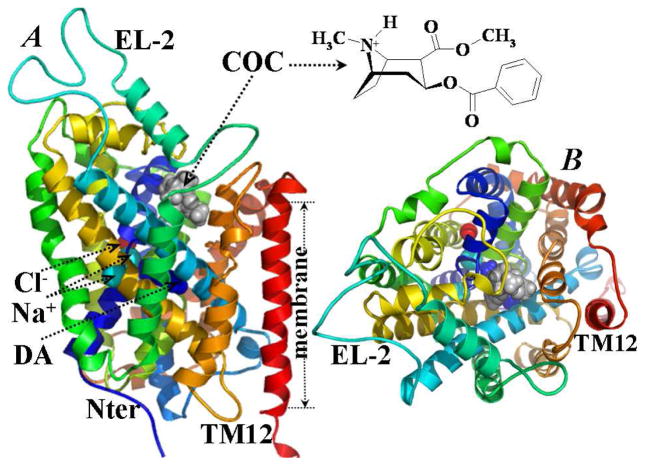
In the selected best candidate structure of the initial DAT-DA-cocaine complex, cocaine molecule was orientated perpendicularly inside the vertical vestibule formed by helices 1, 3, 6, 8, and 10 of DAT-DA, with its cationic head pointed toward the extracellular end of the vestibule. Such orientation of cocaine in the selected DAT-DA-cocaine complex represented the most common binding pose among the conformational clusters of the docked results (see Figure S2 of Supporting Information). As depicted in Figure 1, the overall DAT structure in the energy-minimized DAT-DA-cocaine complex is essentially the same as that of our previous modeled DAT structure40 in terms of the 12 helices, particularly the pseudo-two-fold axis between helices 1 to 5 and 6 to 10. The first 10 helices act as an essential core of Na+/Cl−-dependent dopamine transporting.6,7 An obvious difference from our previous DAT-DA model exists in the relative positions of EL-2 and EL-4, as the side chain of His193 is close to the side chains of His375 and Glu396. Another difference is the binding site for Cl− ion, which consists of the side chains of residues Tyr102, Ser321, Asn353, Ser354, and Ser357 of DAT.
Figure 1.
The energy-minimized model of DAT-DA-cocaine complex structure. (A) Side view along the normal of the membrane. (B) Top view of the complex structure. The DAT-DA protein is represented as colored ribbons. Na+ ions are shown in cyan spheres, and Cl− ion in red sphere. Dopamine (DA) is shown in space-filled spheres and colored in blue. Cocaine molecule (COC) in the complex is shown in space-filled spheres and colored gray. Also labeled are the EL-2, TM12, and the membrane position. This figure is generated by using PyMol (DeLano Scientific LLC, San Carlos, California, USA).
We also compared our modeled DAT structure in the DAT-DA-cocaine complex with the X-ray crystal structure of LeuTAa bound with tricyclic antidepressant CMI (PDB entry code 2Q6H at 1.85 Å resolution)22 by superimposing the Cα atoms of 12 transmembrane helices in the DAT-DA-cocaine complex with the corresponding atoms in the X-ray crystal structure of the LeuTAa-CMI complex (see Figure S3 of Supporting Information). The root-mean-square deviation (RMSD) of the positions of all Cα atoms within the 12 transmembrane helices in the energy-minimized DAT-DA-cocaine from those in the X-ray crystal structure of the LeuTAa-CMI complex22 was calculated to be 1.99 Å. The two Na+ ions in the energy-minimized DAT-DA-cocaine structure are located at similar binding sites as those in the X-ray structure of LeuTAa bound with substrate and antidepressant CMI (PDB entry 2Q6H).22 One Na+ ion in our DAT-DA-cocaine structure is ~0.81 Å away from the position of the corresponding Na+ ion in the X-ray structure, while the other Na+ ion in DAT-DA-cocaine structure is ~1.41 Å away from the position of the corresponding Na+ ion in the X-ray structure.22 In this energy-minimized DAT-DA-cocaine structure, the binding site of cocaine can be superimposed well with the binding site for antidepressant CMI in the X-ray structure of LeuTAa (PDB entry 2Q6H)22 (Figure S3 of Supporting Information). As we proposed previously, the dopamine entry starts from the extracellular side and the molecule slides down along the vertical vestibule between helices 1, 3, 6, 8, and 10 of DAT.40 The docked cocaine plugs in this vestibule with its benzoyl ester group steering down to the bottom, while its cationic head group stays straight up along helices 1, 3, and 6, and is partially covered by Gly386 and Pro387 from EL-4 of DAT.
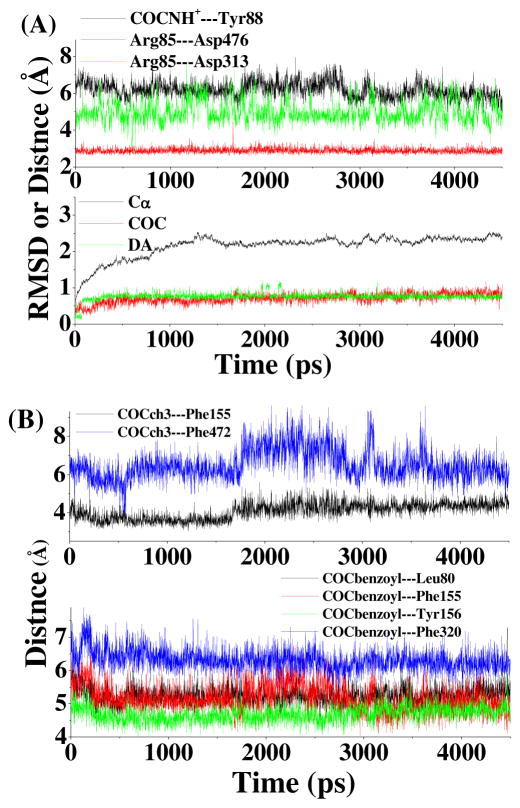
The energy-minimized DAT-DA-cocaine structure was used to carry out MD simulation in order to examine the stability of the binding mode. The time-dependent RMSD of the positions of all Cα atoms of DAT or all atoms of ligand (cocaine or DA) in the MD-simulated DAT-DA-cocaine structure from those of the respective atoms in the energy-minimized DAT-DA-cocaine structure (i.e. the starting structure of MD simulation) is depicted in Figure 2A. The tracked RMSD curve (lower panel in Figure 2A) for the Cα atoms during the MD simulation became flat after ~1.3 ns, indicating that the DAT-DA-cocaine binding structure was stabilized after ~1.3 ns. When tracking the changes of key internuclear distances between cocaine and the surrounding residues of DAT, it was interesting to note that the cationic head group of cocaine was always located nearby the aromatic side chain of Tyr88 from helix 1 (upper panel of Figure 2A), but there was no direct cation-π interactions as the distance between the cationic head of cocaine and the aromatic ring of Tyr88 was around 6 Å during the MD simulation. The positively charged Arg85 from helix 1 formed a stable salt bridge with the negatively charged side chain of Asp476 from helix 10 (Figure 2A), but not with Asp313 from helix 6. Our previous MD simulations on DAT-dopamine complex and free DAT structures40 revealed that the Arg85-Asp476 salt bridge existed only in the DAT-dopamine complex. The MD simulation on DAT-DA-cocaine complex in the present study further indicates that the Arg85-Asp476 salt bridge can also exist when DAT binds with both cocaine and dopamine at the same time.
Figure 2.
Plots of RMSDs and key distances versus the simulation time during the MD simulations on the DAT-DA-cocaine complex. (A) Lower panel: the RMSDs for DAT (Cα atoms only), cocaine, and dopamine; upper panel: COCNH+---Tyr88 represents the distance from the cationic nitrogen atom of cocaine to the center of the aromatic ring of Tyr88 side chain, Arg85---Asp476 refers to the shortest distance from atom NH1 or NH2 of Arg85 side chain to atom OD1 or OD2 of Asp476 side chain, and Arg85---Asp313 has the similar meaning of the internuclear distance. (B) Lower panel: the distances from the center of the aromatic ring of cocaine to the centers of side chains of Leu80, Phe155, Tyr156, and Phe320; upper panel: the distances from the methyl group on the methyl ester moiety of cocaine to the centers of the side chains of Phe155 and Phe472.
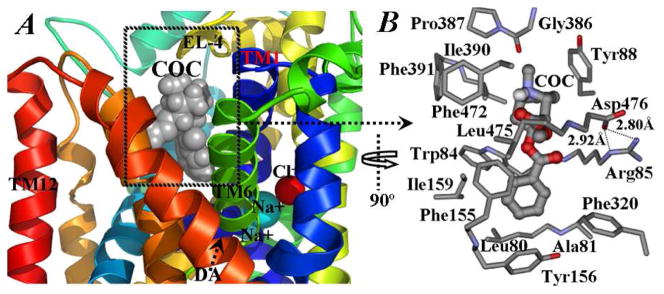
Tracked changes of the distances along the MD trajectories reveal that the benzoyl ester group of cocaine is constantly packed with the hydrophobic side chains of Leu80, Ala81, and Ile159 and aromatic side chains of Phe155, Tyr156, and Phe320 of DAT (Figure 2B). The benzoyl ester group of cocaine is perpendicular to the aromatic side chain of Tyr156 (Figure 3). Such positioning of the cocaine benzoyl ester group makes it impossible for the escape of substrate dopamine from its binding pocket, as Tyr156 is located right above the aromatic tail of dopamine. Phe320 side chain is expected to play a similar, but much less, role as compared to the side chain of Tyr156. The oxygen atoms of the benzoyl ester group of cocaine have close contacts with side chains of Trp84 and Arg85 residues, while the 2β-methyl ester group interacts with the side chains of Phe155, Phe472, and Leu475 (Figures 2B and 3). The cationic head group of cocaine is packed with Tyr88, Ile390, Phe391, and Phe472, and partly sheltered by Gly386 and Pro387 from EL-4 (Figure 3).
Figure 3.
Local view of representative structure for DAT-DA-cocaine complex, taken from the last snapshot of the MD simulations. (A) The coloring scheme for the complex structure is the same as that used in Figure 1A, except 180° rotation along the normal of the membrane. (B) Molecular interactions between cocaine (COC) and DAT-DA. Cocaine is shown in ball-and-stick, and residues within 5 Å of cocaine are labeled and shown in stick. The hydrogen bonding interactions between the side chain atoms of Arg85 and Asp476 are shown in dashed lines along with the distances.
The binding of dopamine with DAT in the MD-simulated DAT-DA-cocaine structure was essentially the same as that in our previously reported MD-simulated DAT-dopamine structure.40 The very similar binding of dopamine indicates that the binding mode of dopamine with DAT does not change significantly after a cocaine molecule also binds to DAT. Other interesting structural features of the MD-simulated DAT-DA-cocaine binding include information about the coordination of the Na+ and Cl− ions. The transporting of dopamine by DAT is Na+/Cl−-dependent, 8,15 and typical antidepressant drugs were found to bind with LeuTAa that has already bound with Na+ ions and substrate Leucine.22,23 Therefore, it is interesting to compare the changes of coordinating atoms for the two Na+ ions after the binding of cocaine. The atoms coordinating the first Na+ ion were the carbonyl oxygen of Ala77 backbone, OD2 of Asp79 side chain, OD1 of Asn82 side chain, OD1 of Asn353 side chain, and carbonyl oxygen of Ser321 backbone. The OG atom of Ser321 side chain became the sixth atom coordinating the first Na+ ion with a coordination fraction of 0.773 (i.e. the OG atom coordinated this Na+ ion for 77.3% of the simulation time). Ser321 and Asn353 interacting with the first Na+ also used their side chain atoms (i.e. the HG of Ser321 and HD21 of Asn353) to coordinate Cl− ion, providing two of the five atoms coordinating Cl− ion. In the MD-simulated DAT-DA-cocaine structure, the hexacoordination fraction of the first Na+ ion was as large as 0.698 and its pentacoordination fraction was 0.295. The other Na+ ion was coordinated mainly by the same five atoms as reported in our previous paper,40 i.e. the backbone carbonyl oxygen atoms of Gly75, Val78, and Leu418 and OD1 and OD2 atoms of Asp421 side chain. In addition, in the MD-simulated DAT-DA-cocaine structure, the OG atom of Ser422 side chain served as a part-time (sixth) atom coordinating this Na+ ion, with a coordination fraction of only 0.137.
Comparison with available experimental data
It is important for validation of the modeling to appropriately compare our MD-simulated DAT-DA-cocaine structure with the available experimental results. The modeled DAT-DA-cocaine binding complex structure shows that cocaine inhibits the transport of dopamine from the extracellular face of the transporter. The DAT-DA-cocaine complex structure is consistent with accumulative indications about the location of the cocaine-binding site obtained from experimental studies (Table 1).9,10,11,12,16,17,18,26,27,28,29,32,34,35,36,39 For example, it was shown that the lipid-insoluble Hg2+ ion was able to inhibit cocaine binding10 and the transport of dopamine,9 indicating that cocaine may bind at a site near the extracellular side of the transmembrane domain of DAT.
Table 1.
Comparison between the experimental observations/insights and insights from our simulated 3D model of the DAT-DA-cocaine complex.
| Experiments | Observations or insights from the experiment | Ref. | Insights from our DAT-DA-COC model |
|---|---|---|---|
| Hg2+ binding test | Binding of cocaine was inhibited. | 9, 10 | Cocaine-binding site is near the extracellular side of TM domain of DAT. |
| Epitope-specific immunoprecipitation studies | Cocaine binding site should be near helix 6 of DAT | 35 | Cocaine-binding site is formed by α-helices 1, 3, 6, and 8 of DAT |
| Trp84Leu mutation | Cocaine-binding affinity was increased (~8-fold decrease for the apparent dissociation constant). | 28, 39 | The mutation provides more space to accommodate the cationic head of cocaine at the binding site. |
| Phe154Ala and Phe155Ala mutations | Cocaine-binding affinity was decreased. | 26 | The mutation weakens the packing between the two residues and the benzoyl ester group of cocaine. |
| Phe391Ala mutation | Cocaine-binding affinity was decreased by ~6-fold. | 26 | The mutation weakens packing between the residue and the cationic head of cocaine. |
| Asp313Asn and Asp313Gln mutations | Cocaine-binding affinity was increased (~3-fold decrease for the apparent dissociation constant). | 28, 39 | Arg85-Asp476 salt bridge matches better around cationic head of cocaine after the mutation. |
| Leu104Val/Phe105C ys/Ala109Val mutation | The mutant was ~70-fold insensitive to cocaine inhibition compared to the wild-type DAT. | 4, 29 | Hydrophobic interactions with Phe105 is destroyed and α-helix 6 stays away from the cocaine-binding site in the mutant. |
| Asp79Glu mutation | The mutation considerably decreased DAT binding with dopamine, but with little change in DAT-cocaine binding. | 30 | The negatively charged side chain of the residue has direct contact with the cationic head of dopamine, but has not direct contact with that of cocaine. |
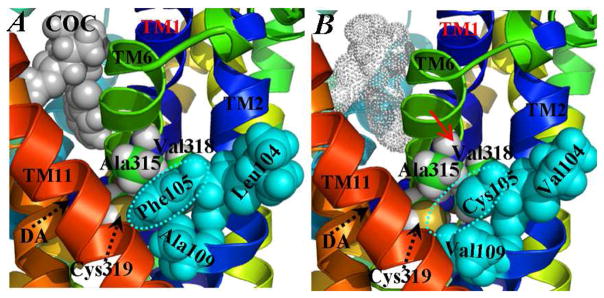
In a recent study using epitope-specific immunoprecipitation method,35 the cocaine-binding site was found near helix 6 but not helices 4, 5, and 7 of human DAT. It was reported that the Trp84Leu mutation on DAT significantly increased the binding affinity of cocaine with the transporter (~8-fold decrease in the apparent equilibrium dissociation constant).28,39 This can be explained in terms of steric effect of Trp84 residue. As the bulky side chain of Trp84 is replaced by a smaller hydrophobic side chain such as that of Leu or Ala, the mutation brings more space for the cationic head of cocaine to be buried more deeply by residues near the extracellular end of helix 1. On the contrary, the Phe154Ala and Phe155Ala mutations26 on helix 3 have negative steric effects on the binding pocket of cocaine (Figure 3), because these two residues are packed closely with Tyr156 residue and cocaine benzoyl ester group through hydrophobic interactions. The aromatic side chain of Tyr156 is packed with the beneath dopamine40 and also the above benzoyl ester group of cocaine. As shown in Figure 3, Phe391 from EL-4 interacts directly with the cationic head group of cocaine. The Phe391Ala mutation makes the DAT-cocaine binding weaker, which is consistent with the experimental observation that the Phe391Ala mutation caused a 6-fold decrease of binding affinity of cocaine with DAT.26 As shown in Figure 2A, the shortest distance between charged atoms of Asp313 side chain and charged atoms of Arg85 side chain was fluctuated around 5 Å during the MD simulation. Asp313 interacts with the Arg85-Asp476 salt bridge through water molecules between them. Based on the modeled binding structure, if Asp313 is changed into a hydrophilic residue such as Asn or Gln, the long-range electrostatic interactions between Asp313 and Arg85 should be decreased, leading to a somewhat better match between the Arg85-Asp476 salt bridge and the cationic head of cocaine. Therefore, the Asp313Asn or Asp313Gln mutation is expected to increase the binding affinity of cocaine with DAT, which is consistent with the experimental observation that the Asp313Asn mutation did increase the binding affinity of cocaine (~3-fold decrease of the apparent dissociation constant).28,39 Recently, it was found that both Trp84Leu and Asp313Asn mutants of DAT have higher binding affinities with not only cocaine but also cocaine analogs including β-CFT, 3β-benzoyltropane, and 2β,3α-allococaine (see structures of these ligands in Figure S1 of Supporting Information).39 These experimental measurements support that Trp84 and Asp313 of DAT may directly interact with cocaine or be around the binding site of cocaine. It was reported that the Phe105Met27 and Leu104Val/Phe105Cys/Ala109Val mutants4,29 became much more cocaine-insensitive than the wild-type DAT although these mutants retained >50% transport activity. As observed in our MD-simulated DAT-DA-cocaine structure, these residues from helix 2 form a local hydrophobic core along with hydrophobic residues Ala315, Val318, and Cys319 from helix 6 (Figure 4A), making TM6 packed tightly with TM2. When aromatic residue Phe105 is mutated into a smaller residue like Cys or Met, a local hydrophobic cavity around residue #105 will be created (Figure 4B). Such structural perturbation is expected to loose the hydrophobic packing between TM6 and TM2. As a result, TM6 will probably be away from the cocaine-binding site, thus the binding affinity of cocaine will be decreased. The influence of Phe98Ala, Pro101Ala, Phe114Cys, and Gln122Cys mutations26,31 on the binding of cocaine with DAT may be considered similar to that of the Phe105Cys mutation.
Figure 4.
Representative structural effect of Leu104Val/Phe105Cys/Ala109Val mutation on the binding of cocaine with DAT-DA. (A) Wild-type DAT-DA. The aromatic side chain of Phe105 (circled and colored in cyan) is packed with the surrounding residues as Leu104, Ala109, Ala315, Val318, and Cys319. (B) The Leu104Val/Phe105Cys/Ala109Val mutant. The same cyan colored circle shows the local hydrophobic cavity around position #105 created by the mutation. The red arrow represents the probable leaning away motion of TM6 caused by the triple mutation. The cocaine molecule is represented as dotted spheres as its binding affinity with DAT-DA is significantly reduced. In both (A) and (B), the coloring scheme and the orientation of the complex structure are the same as that in Figure 3A. Key residues are labeled and shown in space-filled spheres.
As demonstrated in an experimental study,30 the Asp79Glu mutation had a dramatic influence on the binding of dopamine, and decreased the inhibitory potency of cocaine on dopamine uptake by only 2-fold. As demonstrated in our previous computational study,40 the Asp79Glu mutation did significantly weaken the interaction of DAT with the cationic head of dopamine. According to the modeled DAT-DA-cocaine complex structure in the present study, the Asp79Glu mutation will have no obvious effect on the binding of cocaine because the binding site of cocaine is far away from Asp79, and the mutation does not change the net charge of the transporter and, therefore, should not dramatically change the long-range electrostatic attraction between DAT and cocaine. The Glu491Ala mutation32 (i.e. the mutation on a residue at the intracellular end of helix 10) does affect the cocaine binding through weakening of the long-range electrostatic attractions as cocaine bears +1 net charge and Glu491 side chain is negatively charged. Mutations on residues from helix 6, such as Thr316Ala, Gln317Ala, and Phe332Ala, are expected to have negative, allosteric effects on the binding of cocaine as these residues are located just nearby the cocaine-binding site.
In summary, our modeled new, alternative DAT-cocaine binding mode can reasonably explain all of the previously reported experimental data.4,9,10,11,12,18,24,26,27,28,29,30,31,32,35,36,39 However, this new binding mode cannot explain very well the most recently reported data38 for the mutations on Val152, Asn157, Val328, and Ser422 residues. According to our new binding mode, cocaine is far away from Val152, Asn157, Val328, and Ser422 residues, whereas the Val152Ala, Val152Ile, Val152Met, Asn157Cys, Val328Ile, Val328Phe, and Ser422Ala mutations significantly decreased the inhibitory activity of cocaine and binding affinity of β-CFT.38 It would be hardly convincing to simply consider the effects of these mutations as the result of the mutation-caused conformational changes of the transporter.
Further, compared to our modeled new binding mode, the recently proposed overlap binding mode38 can more reasonably explain the experimental data for the mutations on Val152, Asn157, Val328, and Ser422 residues, but the overlap binding mode cannot reasonably explain the experimental data for the Trp84Leu and Leu104Val/Phe105Cys/Ala109Val mutants. The Trp84Leu mutation significantly increased the DAT-cocaine binding affinity (~8-fold decrease in the apparent equilibrium dissociation constant)28 and the Leu104Val/Phe105Cys/Ala109Val mutation resulted in a ~70-fold more insensitive mutant for cocaine compared to the wild-type DAT, but this mutant is still capable of transporting dopamine.4 Cocaine interacts directly with Trp84 and indirectly with these three amino acid residues (Leu104, Phe105, and Ala109; see Figure 4) according to our modeled alternative binding mode, whereas these four residues are all far away from cocaine according to the overlap binding mode.38 So, none of the two binding modes alone can perfectly explain all of the mutational data. In fact, residues Trp84, Leu104, Phe105, and Ala109 are far away from Val152, Asn157, Val328, and Ser422 based on the modeled DAT structure. It would be impossible to find a site in DAT which allows cocaine to be close to all of these crucial residues at the same time. In consideration of the all available mutational data, the most likely scenario is that both of the two binding modes exist so that cocaine can bind with all of these residues at different time. When cocaine binds to DAT, the initial binding site is likely the one modeled in the present study, as this initial binding mode does not require DAT to make a large conformational change. Then, cocaine may further advance to the dopamine-binding site after DAT makes some necessary conformational change and expands the binding site cavity. The existence of these two different binding modes for DAT-cocaine binding is similar to that (non-prereactive and prereactive protein-cocaine complexes) proposed for cocaine binding with butyrylcholinesterase (BChE).55,56 The concept of the two BChE-cocaine binding modes has eventually led to the successful design and discovery of promising high-activity mutants of human BChE for anti-cocaine medication development.43,47,49,57
It is also interesting to compare the DAT-cocaine binding modeled in the present study with norepinephrine transporter (NET)-inhibitor binding proposed in a recent study,34 as NET is a functionally close homolog of DAT. Paczkowski et al.34 tested various mutants of NET for its interaction with an inhibitor, e.g. desipramine (DMI), and concluded that DMI binds with NET just above the norepinephrine-binding site.34 Their proposed NET-DMI binding mode is consistent with the DAT-cocaine binding mode modeled in the present study.
Mechanisms for cocaine blocking the transport of dopamine
In our previous study,40 we proposed a substrate-entry tunnel for dopamine entry into the binding site, i.e. through a funnel-like tunnel between helices 1, 3, 6, 8, and 10. Along the tunnel, Phe155, Phe320, and Phe326 act as a gate and form the bottom of this extracellular vestibule. An inhibitor like cocaine or its analog with limited structural diversity can use this vestibule as the alternative binding site. After dopamine is bound in DAT, the gate will gradually be closed and further conformational changes will occur in order to transport the bound dopamine into the intracellular side. However, right before DAT changes its conformation from extracellularly-open state, cocaine molecule may readily be captured near the mouth at the extracellular side of DAT. Once the cocaine molecule is captured, it will prevent transporting-related conformational change of DAT. Further attraction by hydrophobic interactions from Leu80, Phe155, Tyr156, Ile159, and Phe320 may direct the benzoyl ester group of cocaine moving toward the bottom of the vestibule depicted in Figures 1 and 3. Modulated by the local electrostatic and hydrophobic interactions from the Arg85-Asp476 salt bridge, Asp313, and Tyr88 (Figures 2A and 3), the cationic head group of cocaine will be located around the upper half of helices 1, 3, 6, and 8 as depicted in Figures 1 and 4A. Due to the electrostatic and hydrophobic characters of the cationic head group of cocaine, side chains of some residues nearby EL-4 may be attracted and dragged to cover the cationic head group of cocaine. Thus, hydrophobic side chains of Ile390 and Phe391 form close contacts with cocaine (Figure 3). As the extracellularly-open conformation of DAT is stabilized by the binding of cocaine, the transporter protein is not expected to undergo the conformational change necessary for the transport of dopamine in the presence of cocaine inside the protein.
According to our new (initial) DAT-DA-cocaine binding mode described above, cocaine and dopamine can bind with DAT in different sites and the initial/alternative binding site of cocaine does not overlap with the binding site of substrate dopamine. Although cocaine and dopamine both interact with a common residue Tyr156, the modes of interaction with this same residue are different. Accounting for the initial binding site alone, one might naturally consider cocaine to be a noncompetitive inhibitor of DAT for the transport of dopamine. In order to better address this critical issue, we also modeled the binding of cocaine with DAT in the absence of dopamine and found that the binding mode of cocaine with DAT in the absence of dopamine is essentially the same as that of cocaine with DAT-DA. The predicted existence of the DAT-cocaine binding complex suggests that cocaine in the initial DAT-cocaine binding mode can inhibit the transport of dopamine by blocking the formation of the DAT-dopamine complex, as cocaine blocks the dopamine-entry tunnel. So, dopamine cannot bind with DAT once cocaine binds with DAT, whether it is still in the initial binding mode or the overlap binding mode. The predicted existence of the DAT-DA-cocaine binding complex suggests that cocaine can also inhibit the transport of dopamine by blocking the conformational change of the formed DAT-dopamine complex required for the transport of dopamine after dopamine binds with DAT, thus reducing the kinetic turnover of the transporter.
Further, due to the electrostatic repulsion between the positively charged dopamine and positively charged cocaine, the affinity of cocaine binding with DAT-DA, i.e. DAT in the presence of dopamine, is expected to be significantly lower than that of cocaine binding with DAT in the absence of dopamine. We estimated the binding free energy difference, ΔΔGbind, between the DAT-DA-cocaine binding and the DAT-cocaine binding (i.e. in the absence of dopamine) by using the same molecular mechanics-Poison-Boltzmann surface area (MM-PBSA) approach as used in our previous study.40 Based on the binding free energy calculations, we obtained ΔΔGbind = ~4.0 kcal/mol, indicating that the dissociation constant (Kd3) of cocaine binding with DAT-DA is ~1000-fold larger than the Kd2 value of cocaine binding with DAT (i.e. Kd3 ≈ ~1000Kd2). This suggests that cocaine binding with DAT-DA is significantly weaker than cocaine binding with DAT itself.
When cocaine and dopamine coexist with DAT, there are three essential equilibrium binding processes. i.e. Eqs. (1) to (3), in solution:
| (1) |
| (2) |
| (3) |
The corresponding dissociation constants can be expressed as
| (4) |
| (5) |
| (6) |
Based on Eqs. (4) to (6), the overall concentration fraction of dopamine-bound DAT (fDA) in the presence of cocaine can be defined as
| (7) |
Substitution of Eqs.(4) to (6) into Eq. (7) gives
| (8) |
According to Eq.(8), when the cocaine concentration used in an experimental assay is low such that [cocaine] ≪ Kd3, Eq.(8) can be simplified as
| (9) |
Equation (9) indicates that the concentration fraction of dopamine-bound DAT (fDA) is dependent on the concentrations of cocaine and dopamine. In this situation, the contribution to fDA from DAT-DA-cocaine binding is negligible in comparison with that from the corresponding DAT-cocaine binding. As discussed above, the relative binding free energies obtained from the MM-PBSA calculations suggest that the concentration of the DAT-cocaine complex should be at least ~1000-fold higher than that of the DAT-DA-cocaine complex. When the DAT-DA-cocaine binding is negligible, the main experimental observation would be that DAT binds with either cocaine or dopamine. Thus, the practical experimental observation of cocaine blocking DAT-dopamine binding should be close to that of a competitive inhibitor.
However, when the cocaine concentration used in an experimental assay is significantly higher than the Kd3 value, the DAT-DA-cocaine binding becomes significant. Under this extreme experimental condition, [cocaine] ≫ Kd3, Eq.(8) can be simplified as
| (10) |
Since [cocaine] ≫ Kd3 and Kd3 ≈ ~1000Kd2, Kd1Kd2Kd3 in Eq.(10) is negligible compared to Kd1Kd3[cocaine]. Thus, under the extreme condition of [cocaine] ≫ Kd3, Eq.(10) can be simplified further as
| (11) |
Equation (11) shows that fDA, i.e. the concentration fraction of dopamine-bound DAT, is independent of the concentration of cocaine. So, when [cocaine] ≫ Kd3, the experimental observation of cocaine blocking DAT-dopamine binding should be close to that of a noncompetitive inhibitor, as cocaine can always bind with DAT no matter whether dopamine is bound in DAT or not. The computational insight is consistent with the reported experimental observations that different assays led to different patterns of cocaine inhibiting DAT for the transport of dopamine. For example, in the binding assay,8 dopamine and β-CIT (a cocaine analog in a low concentration) were found to competitively bind with DAT assuming that cocaine and the analog bind with DAT in the same site, whereas a variety of kinetic inhibition assays have demonstrated both the competitive and noncompetitive kinetic inhibition mechanisms for the transport of dopamine, depending on how the assays were carried out.9
In addition, an uncompetitive inhibition mechanism was observed in the zero trans entry experiment which determines the inhibition by cocaine of the apparent initial rate of the transport of striatal dopamine.9 Based on their observed uncompetitive inhibition of the initial rate, Meiergerd and Schenk9 concluded that cocaine appears to inhibit the transport of dopamine by reducing the kinetic turnover of the transporter without affecting the kinetics of association of dopamine with the extracellularly-open conformation of DAT. Their proposed molecular mechanism, i.e. cocaine inhibits the transport of dopamine by reducing the kinetic turnover of the transporter after dopamine binding with DAT, is consistent with our MD-simulated DAT-DA-cocaine binding structure. The MD-simulated DAT-DA-cocaine binding suggests that cocaine still can bind with DAT-DA immediately after dopamine binds with DAT in the initial process of the transport of dopamine. Once cocaine also binds with DAT in the presence of dopamine, i.e. DAT-DA, the necessary conformational change of DAT-DA required for the kinetic turnover of the transporter cannot occur.
Conclusion
The three-dimensional structural model of dopamine transporter (DAT) has been refined to model and simulate DAT binding with cocaine and dopamine. Based on molecular docking and molecular dynamics (MD) simulations, at least for its initial binding with DAT, cocaine is bound inside the vestibule pocket formed by helices 1, 3, 6, and 8 near the extracellular side of DAT. The benzoyl ester group of cocaine is buried by hydrophobic residues Leu80, Phe155, Tyr156, Ile159, and Phe330, while the methyl ester group of cocaine contacts closely with side chains of Phe155, Phe472, Leu475, and Trp84. The cationic head group of cocaine is packed with side chains of several residues including Tyr88, Ile390, Phe391, and Phe472.
The computationally determined new DAT-ligand binding mode is totally different from the overlap binding mode proposed recently in literature. It has been demonstrated that both of the two binding modes can exist. When cocaine binds to DAT, the initial binding site is likely the one modeled in the present study, as this binding site can naturally accommodate cocaine without the need of a large conformational change in DAT. Then, cocaine may move further to the dopamine-binding site (the overlap binding mode) after DAT makes some necessary conformational change and expands the binding site cavity. The original dopamine-binding site cavity is not large enough to accommodate cocaine. The existence of these two binding modes is completely consistent with available experimental data concerning the binding site of cocaine.
According to the new, initial DAT-ligand binding mode determined in the present study, the initial cocaine-binding site does not overlap with, but close to, the dopamine-binding site. Dopamine cannot reach its binding site if cocaine binds with DAT prior to dopamine binding as cocaine blocks the dopamine-entry tunnel. However, cocaine can always bind with DAT whether dopamine is bound in the transporter protein or not. On the other hand, the affinity of cocaine binding with DAT in the presence of dopamine is much lower than that of cocaine binding with DAT in the absence of dopamine. When both dopamine and cocaine bind with DAT, the transporter protein is not expected to undergo the conformational change necessary for the transport of dopamine. So, cocaine may inhibit the transport of dopamine through both blocking the initial binding of dopamine with DAT and reducing the kinetic turnover of the transporter after dopamine binds with DAT. The relative contributions to the phenomenological inhibition of the transport of dopamine from blocking the initial binding and reducing the kinetic turnover can be different in different types of kinetic inhibition assays.
Supplementary Material
Acknowledgments
This work was supported in part by NIH (grants R01DA013930 and R01DA025100 to C.-G. Zhan, and R01DA014610 to H. H. Gu). The authors also acknowledge the Center for Computational Sciences (CCS) at University of Kentucky for supercomputing time on IBM X-series Cluster consisting of 340 nodes or 1,360 processors.
Footnotes
Supporting Information Available. Four figures for molecular structures of dopamine, cocaine, β-CFT, β-CIT, 3β-benzoyltropine, (2β,3α)-allococaine, and clomipramine; the conformational clusters of the docked results for the docking of cocaine into the binding site of DAT-DA; the energy-minimized DAT-DA-cocaine complex superimposed with the X-ray structure of LeuTAa bound with antidepressant clomipramine (PDB entry 2Q6H); and plots of the time-dependence of internuclear distances related to some important hydrogen bonds in the MD-simulated DAT-DA complex, and DAT-DA-cocaine complex. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Mendelson JH, Mello NK. Management of cocaine abuse and dependence. New Eng J Med. 1996;334:965–972. doi: 10.1056/NEJM199604113341507. [DOI] [PubMed] [Google Scholar]
- 2.Sparenborg SE, Vocci F, Zukin S. Peripheral cocaine-blocking agents: new medications for cocaine dependence. Drug Alcolhol Depend. 1997;48:149–151. doi: 10.1016/s0376-8716(97)00072-0. [DOI] [PubMed] [Google Scholar]
- 3.Singh S. Chemistry, design, and structure-activity relationship of cocaine antagonists. Chem Rev. 2000;100:925–1024. doi: 10.1021/cr9700538. [DOI] [PubMed] [Google Scholar]
- 4.Chen R, Tilley MR, Wei H, Zhou F, Zhou FM, Ching S, Quan N, Stephens RL, Hill ER, Nottoli T, Han DD, Gu HH. Abolished cocaine reward in mice with a cocaine-insensitive dopamine transporter. Proc Natl Acad Sci US A. 2006;103:9333–9338. doi: 10.1073/pnas.0600905103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaintdinov RR, Sotmikova TD, Caron MG. Monoamine transporter pharmacology and mutant mice. Trends Pharmacol Sci. 2002;231:367–373. doi: 10.1016/s0165-6147(02)02044-8. [DOI] [PubMed] [Google Scholar]
- 6.Torres GE, Gainetdinov RR, Caron MG. Plasma membrane monoamine transporters: structure, regulation and function. Nat Rev Neurosci. 2003;4:13–25. doi: 10.1038/nrn1008. [DOI] [PubMed] [Google Scholar]
- 7.Loland CJ, Norgaard-Nielsen K, Gether U. Probing dopamine transporter structure and function by Zn2+-site engineering. Eur J Pharmacol. 2003;479:187–197. doi: 10.1016/j.ejphar.2003.08.068. [DOI] [PubMed] [Google Scholar]
- 8.Gu H, Wall S, Rudnick G. Stable expression of biogenic amine transporters reveals differences in inhibitor sensitivity, kinetics, and ion dependence. J Biol Chem. 1994;269:7124–7130. [PubMed] [Google Scholar]
- 9.Meiergerd SM, Schenk JO. Kinetic evaluation of the commonality between the site(s) of action of cocaine and some other structurally similar and dissimilar inhibitors of the striatal transporter for dopamine. J Neurochem. 1994;63:1683–1692. doi: 10.1046/j.1471-4159.1994.63051683.x. [DOI] [PubMed] [Google Scholar]
- 10.Cao CJ, Yong MM, Wong JB, Mahran LG, Eldefrawi ME. Putative cocaine receptor in striatum is a glycoprotein with active thiol function. Membr, Biochem. 1989;8:207–220. doi: 10.3109/09687688909026815. [DOI] [PubMed] [Google Scholar]
- 11.McElvain JS, Schenk JO. A multisubstrate mechanism of striatal dopamine uptake and its inhibition by cocaine. Biochem Pharmacol. 1992;43:2189–2199. doi: 10.1016/0006-2952(92)90178-l. [DOI] [PubMed] [Google Scholar]
- 12.Wayment H, Meiergerd SM, Schenk JO. Relationships between the catechol substrate binding site and amphetamine, cocaine, and mazindol binding sites in a kinetic model of the striatal transporter of dopamine in vitro. J Neurochem. 1998;70:1941–1949. doi: 10.1046/j.1471-4159.1998.70051941.x. [DOI] [PubMed] [Google Scholar]
- 13.Giros B, Meistikawy SE, Godinot N, Zheng K, Han H, Yang-Feng T, Caron MG. Cloning, pharmacological characterization, and chromosome assignment of the human dopamine transporter. Mol Pharmacol. 1992;42:383–390. [PubMed] [Google Scholar]
- 14.Hastrup H, Sen N, Javitch JA. The human dopamine transporter forms a tetramer in the plasma membrane. J Biol Chem. 2003;278:45045–45048. doi: 10.1074/jbc.C300349200. [DOI] [PubMed] [Google Scholar]
- 15.Chen N, Reith MEA. Na+ and the substrate permeation pathway in dopamine transporters. Eur J Pharmacol. 2003;479:213–221. doi: 10.1016/j.ejphar.2003.08.070. [DOI] [PubMed] [Google Scholar]
- 16.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 17.Forrest LR, Tavoulari S, Zhang YW, Rudnick G, Honig B. Identification of a chloride ion binding site in Na+/Cl−-dependent transporters. Proc Natl Acad Sci USA. 2007;104:12761–12766. doi: 10.1073/pnas.0705600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zomot E, Bendahan A, Quick M, Zhao Y, Javitch J, Kamner B. Mechanism of chloride interaction with neurotransmitter sodium symporters. Nature. 2007;449:726–730. doi: 10.1038/nature06133. [DOI] [PubMed] [Google Scholar]
- 19.Beuming T, Shi L, Javitch JA, Weinstein H. A comprehensive structure-based alignment of prokaryotic and eukaryotic neurotransmitter/Na+ symporters (NSS) aids in the use of the LeuT structure to probe NSS structure and function. Mol Pharmacol. 2006;70:1630–1642. doi: 10.1124/mol.106.026120. [DOI] [PubMed] [Google Scholar]
- 20.Noskov S, Roux B. Control of ion selectivity in LeuT: two Na+ binding sites with two different mechanisms. J Mol Biol. 2008;377:804–818. doi: 10.1016/j.jmb.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi L, Quick M, Zhao Y, Weinstein H, Javitch JA. The mechanism of a neurotransmitter: sodium symporter-inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol Cell. 2008;30:667–677. doi: 10.1016/j.molcel.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh SK, Yamashita A, Gouaux E. Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature. 2007;448:952–956. doi: 10.1038/nature06038. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Z, Zhen J, Karpowich NK, Goetz RM, Law CJ, Reith MEA, Wang DN. LeuT-desipramine structure reveals how antidepressants block neurotransmitter reuptake. Science. 2007;317:1390–1393. doi: 10.1126/science.1147614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norregaard L, Frederiksen D, Nielsen EØ, Gether U. Delineation of an endogenous zinc-binding site in the human dopamine transporter. The EMBO J. 1998;17:4266–4273. doi: 10.1093/emboj/17.15.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bjorklund NL, Volz TJ, Schenk JO. Differential effects of Zn2+ on the kinetics and cocaine inhibition of dopamine transport by the human and rat dopamine transporters. Eur J Pharmacol. 2007;565:17–25. doi: 10.1016/j.ejphar.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 26.Uhl GR, Lin Z. The top 20 dopamine transporter mutants: structure-function relationships and cocaine actions. Eur J Pharmacol. 2003;479:71–82. doi: 10.1016/j.ejphar.2003.08.058. [DOI] [PubMed] [Google Scholar]
- 27.Wu X, Gu HH. Cocaine affinity decreased by mutations of aromatic residue phenylalanine 105 in the transmembrane domain 2 of dopamine transporter. Mol Pharmacol. 2003;63:653–658. doi: 10.1124/mol.63.3.653. [DOI] [PubMed] [Google Scholar]
- 28.Chen N, Zhen J, Reith MEA. Mutation of Trp84 and Asp313 of the dopamine transporter reveals similar mode of binding interaction for GBR12909 and benztropine as opposed to cocaine. J Neurochem. 2004;89:853–864. doi: 10.1111/j.1471-4159.2004.02386.x. [DOI] [PubMed] [Google Scholar]
- 29.Chen R, Han DD, Gu HH. A triple mutation in the second transmembrane domain of mouse dopamine transporter markedly decreases sensitivity to cocaine and methylphenidate. J Neurochem. 2005;94:352–359. doi: 10.1111/j.1471-4159.2005.03199.x. [DOI] [PubMed] [Google Scholar]
- 30.Ukairo OT, Bondi CD, Newman AH, Kulkarni SS, Kozikowski AP, Pan S, Surratt CK. Recognition of benztropine by the dopamine transporter (DAT) differs fromm that of the classical dopamine uptake inhibitors cocaine, methylphenidate, and mazindol as a function of a DAT transmembrane 1 aspartic residue. J Pharmacol Exp Ther. 2005;314:575–583. doi: 10.1124/jpet.105.085829. [DOI] [PubMed] [Google Scholar]
- 31.Sen N, Shi L, Beuming T, Weinstein H, Javitch JA. A pincer-like configuration of TM2 in the human dopamine transporter is responsible for indirect effects on cocaine binding. Neuropharmacol. 2005;49:780–790. doi: 10.1016/j.neuropharm.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 32.Dar DE, Metzger TG, Vandenbergh DJ, Uhl GR. Dopamine uptake and cocaine binding mechanisms: the involvement of charged amino acids from transmembrane domains of the human dopamine transporter. Eur J Pharmacol. 2006;538:43–47. doi: 10.1016/j.ejphar.2006.03.048. [DOI] [PubMed] [Google Scholar]
- 33.Chen R, Wei H, Hill ER, Chen L, Jiang L, Han DD, Gu HH. Direct evidence that two cysteines in the dopamine transporter form a disulfide bond. Mol Cell Biochem. 2007;298:41–48. doi: 10.1007/s11010-006-9348-7. [DOI] [PubMed] [Google Scholar]
- 34.Paczkowski FA, Sharpe IA, Dutertre S, Lewis RJ. X-conotoxin and tricyclic antidepreseeant interactions at the norepinephrine transporter define a new transporter model. J Biol Chem. 2007;282:17837–17844. doi: 10.1074/jbc.M610813200. [DOI] [PubMed] [Google Scholar]
- 35.Vaughan RA, Sakrikar DS, Parnas ML, Adkins S, Foster JD, Duval RA, Lever JR, Kulkarni SS, Hauck-Newman A. Localization of cocaine analog [125I]RTI82 irreversible binding to transmembrane domain 6 of the dopamine transporter. J Biol Chem. 2007;282:8915–8925. doi: 10.1074/jbc.M610633200. [DOI] [PubMed] [Google Scholar]
- 36.Loland CJ, Desai RI, Zou M-f, Cao J, Grundt P, Gerstbrein K, Sitte HH, Newman AH, Katz JL, Gether U. Relationship between conformational changes in the dopamine transporter and cocaine-like subjective effects of uptake inhibitors. Mol Pharmacol. 2008;73:813–823. doi: 10.1124/mol.107.039800. [DOI] [PubMed] [Google Scholar]
- 37.Kniazeff J, Shi L, Loland CJ, Javitch JA, Weinstein H, Gether U. An intracellular interaction network regulates conformational transitions in the dopamine transporter. J Biol Chem. 2008;283:17691–17701. doi: 10.1074/jbc.M800475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beuming T, Kniazeff J, Bergmann ML, Shi L, Gracia L, Raniszewska K, Newman AH, Javitch JA, Weinstein H, Gether U, Loland CJ. The binding site for cocaine and dopamine in the dopamine transporter overlap. Nature Neurosci. 2008;11:780–789. doi: 10.1038/nn.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitt KC, Zhen J, Kharkar P, Mishra M, Chen N, Dutta AK, Reith MEA. Interaction of cocaine, benztropine, and GBR12909-like compounds with wild-type and mutant human dopamine transporters: molecular features that differentially determine antagonist binding properties. J Neurochem. 2008;107:928–940. doi: 10.1111/j.1471-4159.2008.05667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang X, Zhan C-G. How dopamine transporter interacts with dopamine: insights from molecular modeling and simulation. Biophys J. 2007;93:3627–3639. doi: 10.1529/biophysj.107.110924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, Crowley M, Brozell S, Tsui V, Gohlke H, Mongan J, Hornak V, Cui G, Beroza P, Schafmeister C, Caldwell JW, Ross WS, Kollman PA. AMBER. Vol. 8. University of California; San Francisco: 2004. [Google Scholar]
- 42.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson JA. Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- 43.Pan Y, Gao D, Yang W, Cho H, Yang GF, Tai HH, Zhan CG. Computational redesign of human butyrylcholinesterase for anti-cocaine medication. Proc Natl Acad Sci USA. 2005;102:16656–16661. doi: 10.1073/pnas.0507332102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhan CG, Gao D. Catalytic mechanism and energy barriers for butyrylcholinesterase-catalyzed hydrolysis of cocaine. Biophys J. 2005;89:3863–3872. doi: 10.1529/biophysj.105.070276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao D, Zhan CG. Modeling effects of oxyanion hole on the ester hydrolyses catalyzed by human cholinesterases. J Phys Chem B. 2005;109:23070–23076. doi: 10.1021/jp053736x. [DOI] [PubMed] [Google Scholar]
- 46.Gao D, Zhan CG. Modeling evolution of hydrogen bonding and stabilization of transition states in the process of cocaine hydrolysis catalyzed by human butyrylcholinesterase. Proteins. 2006;62:99–110. doi: 10.1002/prot.20713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao D, Cho H, Yang W, Pan Y, Yang GF, Tai HH, Zhan CG. computational design of a human butyrylcholinesterase mutant for accelerating cocaine hydrolysis based on the transition-state simulations. Angew Chem Int Ed Engl. 2006;45:653–657. doi: 10.1002/anie.200503025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan Y, Gao D, Zhan CG. Modeling the catalysis of anti-cocaine catalytic antibody: Competing reaction pathways and free energy barriers. J Am Chem Soc. 2008;130:5140–5149. doi: 10.1021/ja077972s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng F, Yang W, Ko M-C, Liu J, Cho H, Gao D, Tong M, Tai H-H, Woods JH, Zhan C-G. Most efficient cocaine hydrolase designed by virtual screening of transition states. J Am Chem Soc. 2008;130:12148–12155. doi: 10.1021/ja803646t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Solis FJ, Wets RJB. Minimization by random search techniques. Math Oper Res. 1981;6:19–30. [Google Scholar]
- 51.Morishita T. Fluctuation formulas in molecular dynamics simulations with the weak coupling heat bath. J Chem Phys. 2000;113:2976–2982. [Google Scholar]
- 52.Toukmaji A, Sagui C, Board J, Darden T. Efficient particle-mesh Ewald based approach to fixed and induced dipolar interactions. J Chem Phys. 2000;113:10913–10927. [Google Scholar]
- 53.Ryckaert J, Ciccotti PG, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 54.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 55.Zhan CG, Zheng F, Landry DW. Fundamental reaction mechanism for cocaine hydrolysis in human butyrylcholinesterase. J Am Chem Soc. 2003;125:2462–2474. doi: 10.1021/ja020850+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hamza A, Cho H, Tai HH, Zhan CG. Molecular dynamics simulation of cocaine binding with human butyrylcholinesterase and its mutants. J Phys Chem B. 2005;109:4776–4782. doi: 10.1021/jp0447136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pan Y, Gao D, Yang W, Cho H, Zhan CG. Free energy perturbation (FEP) simulation on the transition-states of cocaine hydrolysis catalyzed by human butyrylcholinesterase and its mutants. J Am Chem Soc. 2007;129:13537–13543. doi: 10.1021/ja073724k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.