SUMMARY
Protein-DNA interactions (PDIs) mediate a broad range of functions essential for cellular differentiation, function, and survival. However, it is still a daunting task to comprehensively identify and profile sequence-specific PDIs in complex genomes. Here, we have used a combined bioinformatics and protein microarray-based strategy to systematically characterize the human protein-DNA interactome. We identified 17,718 PDIs between 460 DNA motifs predicted to regulate transcription and 4,191 human proteins of various functional classes. Among them, we recovered many known PDIs for transcription factors (TFs). We also identified a large number of new PDIs for known TFs, as well as for previously uncharacterized TFs. Remarkably, we found that over three hundred proteins not previously annotated as TFs also showed sequence-specific PDIs, including RNA binding proteins, mitochondrial proteins, and protein kinases. One of such unconventional DNA-binding proteins, MAPK1, acts as a transcriptional repressor for interferon gamma-induced genes.
INTRODUCTION
A major challenge in the post-genome era is decoding the functional elements in the human genome. Aided by the sequencing of multiple genomes, computational approaches have identified a large number of evolutionarily conserved DNA elements that include many previously characterized cis-regulatory elements (Xie et al., 2005; Xie et al., 2007). Additional studies have identified DNA motifs that are highly enriched in promoters of co-expressed genes (Elemento et al., 2007; Elemento and Tavazoie, 2005; Yu et al., 2006). However, the proteins that recognize these elements cannot be reliably predicted computationally, and the target preferences of only a small minority of DNA binding proteins have been characterized. Therefore, the identification of interaction networks among the functional elements is the next major step following the identification of the parts list in the human genome.
Protein-DNA interactions (PDIs) are perhaps the most important regulatory interactions involving these functional elements. The most intensively studied subset of PDIs is those between transcription factors (TFs) and their specific DNA target sequences. There are over 1,400 known and predicted human TFs, which fall into multiple subfamilies (Kummerfeld and Teichmann, 2006; Messina et al., 2004). Aside from the interactions between conventional TFs and DNA, the larger set of potential DNA-binding proteins has not been extensively explored. Some proteins that lack any known DNA-binding domains have been found to bind specific DNA sequences (Boggon et al., 1999; Kipreos and Wang, 1992). For instance, Arg5,6, a yeast protein which has traditionally been regarded as a metabolic enzyme with no additional biological functions, recognizes specific DNA sequences and regulates the transcription of genes in the mitochondria (Hall et al., 2004). In general, most proteins that display sequence-specific DNA binding are thought to act as TFs (Teichmann and Babu, 2004); however, some sequence-specific DNA-binding proteins play central roles in such processes as DNA replication, DNA repair, and chromosome dynamics, and are not thought to act as TFs (Petukhova et al., 2005; Tokai-Nishizumi et al., 2005; Zhu et al., 2003).
In the past biochemical approaches have been used to characterize PDIs, but such approaches are generally laborious and slow. Recent years have witnessed the development of large-scale, unbiased technologies to characterize PDIs. These approaches can be either DNA-centered, in which an individual protein is used to identify target sequences, or protein-centered, in which a DNA sequence is used to screen for uncharacterized DNA-binding proteins. Two recent large-scale, DNA-centered approaches have employed the double-stranded DNA microarrays and the bacterial one-hybrid system to characterize PDIs for homeodomain TFs in mice and Drosophila, respectively (Berger et al., 2008; Noyes et al., 2008). Conversely, protein microarrays have been used both to characterize PDI networks (Ho et al., 2006) and to identify unconventional DNA-binding proteins in yeast (Hall et al., 2004).
In the present study, by using a microarray of 4,191 non-redundant human proteins comprising of known and predicted TFs, as well as representative proteins from other functional classes, we have systematically identified proteins that selectively bind DNA sequences that are either highly evolutionarily conserved or found in the promoters of co-expressed genes. We were able to extensively identify PDIs for known as well as previously uncharacterized human TFs, and we unexpectedly also found that many proteins of other functional classes showed sequence-specific PDIs. We further characterized the DNA-binding activity of MAPK1, one of these unconventional DNA binding proteins, using in vitro and in vivo assays and demonstrated that MAPK1 acts as a transcriptional repressor regulating interferon gamma signaling in mammalian cells.
RESULTS
Experimental Design
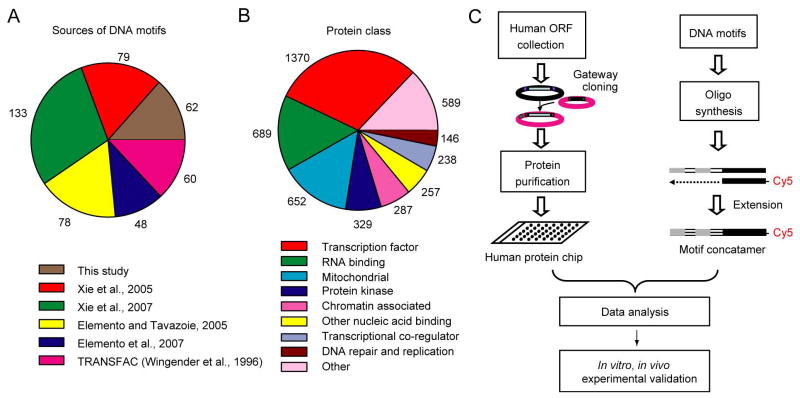
To systematically identify proteins that can specifically recognize predicted functional human DNA elements, a combined approach was employed (Figure 1). First, we obtained 752 predicted DNA motifs from previously published studies (Elemento et al., 2007; Elemento and Tavazoie, 2005; Xie et al., 2005; Xie et al., 2007). Second, we used algorithms generated in our laboratories to identify different sets of DNA elements enriched in promoter sequences of tissue-specific genes (Supplemental Method). Third, we retrieved 60 sequences from the TRANSFAC database corresponding to experimentally-verified binding sites for known TFs (Wingender et al., 1996). After combining these three sources, we removed highly similar motif sequences using a clustering algorithm to produce 460 sequence-diverse DNA motifs with lengths ranging from 6 to 34 base pairs (Figure 1A, Supplemental Method, Figures S1 and S2, and Table S1). Double-stranded DNA (dsDNA) probes based on these sequences were then synthesized as previously described (Ho et al., 2006).
Figure 1. Overall Experimental Design for Analyzing Human PDIs.
(A) Sources of the DNA motifs used for probe construction.
(B) Distribution of human proteins selected for protein microarray construction. Some proteins belong to more than one functional class and thus may be counted more than once.
(C) Overall scheme used to identify PDIs in humans using DNA probe binding to protein microarrays.
We next assembled a list of proteins that are likely to recognize these predicted DNA motifs (Table S2 and Supplemental Method). The proteins can be categorized into multiple functional classes (Figure 1B): 1) 1,370 known and predicted TFs, representing around 80% of annotated human TFs (Ashburner et al., 2000); 2) proteins known to bind to nucleic acids but without known sequence-specific PDIs, such as RNA binding proteins, chromatin-associated proteins, and DNA repair enzymes; 3) proteins that regulate transcription but are not known to directly bind DNA, such as transcriptional co-regulators; 4) mitochondria-encoded and -targeted proteins and protein kinases, for which previous experimental evidences had suggested that these classes of protein may regulate gene expression (Hall et al., 2004; Pokholok et al., 2006); and 5) an assortment of proteins from a broad range of other functional classes (Table S3).
Human ORFs on this list were selected from the Invitrogen Ultimate ORF collection (Liang et al., 2004) or subcloned in our own laboratories. Using Gateway site-specific recombination (Hartley et al., 2000), ORFs were shuttled to a yeast expression vector that produces N-terminal GST fusions of each protein, and purified from yeast using a previously described strategy (Zhu et al., 2001). To ensure that recombinant proteins were of good quality, we performed immunoblot analysis using anti-GST antibodies, along with silver staining on a randomly selected subset of 200 proteins. Detectable levels of full-length forms of over 90% of the proteins were observed using both methods. Silver staining confirmed the absence of detectable contaminating yeast proteins after purification (Figure S3). Following printing onto nitrocellulose-coated slides (FAST), the complete protein array was probed multiple times with anti-GST antibodies, and more than 98% of the spots produced a signal above background (Figure S4). Pair-wise correlation coefficients of signal intensities ranged from 0.90 – 0.95 between these slides, illustrating consistency in the array quality.
Data Quality Assessment
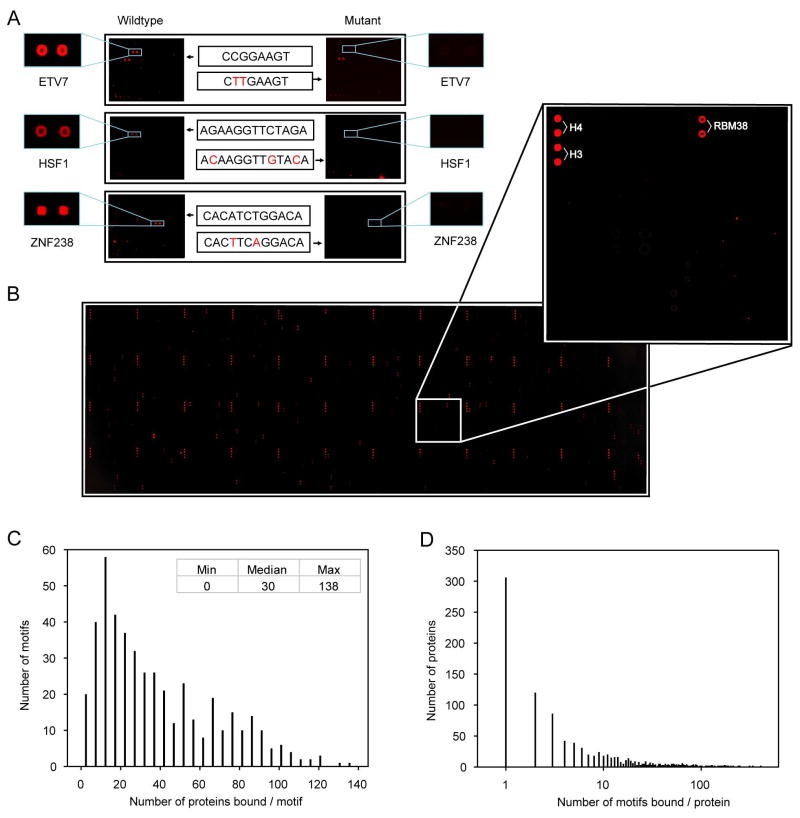
To assess the specificity and sensitivity of our approach, we first probed the protein microarrays with three DNA motifs corresponding to consensus-binding sequences for three TFs. These motifs produced highly specific signals, binding selectively to their target proteins with minimal background (Figure 2A). We further tested the specificity of these interactions by probing the array with mutant motifs and observed that they no longer showed specific PDIs (Figure 2A). To eliminate non-specific PDIs, we also probed the array with Cy5-labeled oligos corresponding to the T7 primer that was used to generate the dsDNA probes. We identified 134 proteins that bound this probe and excluded them from further analysis. On the basis of our earlier observation that bovine histones H3 and H4 bound intensely and nonspecifically to every DNA probe tested, we printed these proteins multiple times on each array as landmarks for orientation and as positive controls for hybridization (Figure 2B). Experimental variability for microarray hybridization was determined by conducting replicate hybridizations of the same probe to four slides. Pair-wise correlation coefficients of signal intensities ranged from 0.68–0.84 for the four slides, with greater consistency for strong signal intensities (Figure S5). On the basis of these control experiments, we concluded that our approach could detect known PDIs sensitively, specifically, and reproducibly.
Figure 2. Human Protein-DNA Interactions Detected via Protein Microarrays.
(A) Binding specificity of three previously characterized PDIs. Three Cy5-labeled, known dsDNA motifs are separately probed to the protein microarrays and can be specifically recognized by their known TFs, whereas the mutant motifs can no longer bind to their known TFs. Mutated positions are indicated in red.
(B) A typical example of a DNA-binding assay. The DNA motif selectively recognizes RBM38, a predicted RNA-binding protein (inset). Histones H3 and H4, which serve as landmarks and positive controls, are printed in duplicate at a corner of each of the 48 printed blocks
(C) Histogram showing the number of proteins on the array that were bound by each DNA probe tested.
(D) Histogram showing the number of DNA probes bound by each protein on the array.
Global Properties of Observed PDIs
We next used the protein array to analyze PDIs for all of the designed dsDNA motif probes. DNA binding signals were acquired, analyzed, and normalized using the procedures described in Supplemental Method. From histogram analysis of each hybridization reaction, we observed that a small number of proteins showed strong positive signals with signal intensities many standard deviations (SD) above background, while the vast majority of proteins produced only small background levels of intensity (Figures 2A and B, Figure S6). To increase our confidence in our PDI identification, we applied a stringent cut-off value of 6 SD above background (Table S4).
A total of 17,718 PDIs were detected, with a median number of 30 proteins interacting with each DNA motif probe. Only a single motif did not bind specifically to any of the proteins on the array (Figure 2C). Motif length did not correlate with either the binding intensity or the number of binding proteins observed with a given motif probe (Figure S7). Many proteins on the array bound to only a few probes, while only relatively few proteins bound to a large fraction of probes, a behavior that followed a power-law distribution (Figure 2D). In fact, more than 85.7% of the proteins bound to fewer than 30 of the motifs, confirming that most of the observed PDIs are sequence-specific. For the remaining analysis performed in this study, we focus on only those proteins that fall into this class. It is notable that proteins from different functional classes showed different levels of sequence binding specificity, where RNA-binding proteins have the least sequence specific binding (Figure S8).
TF Binding Specificity
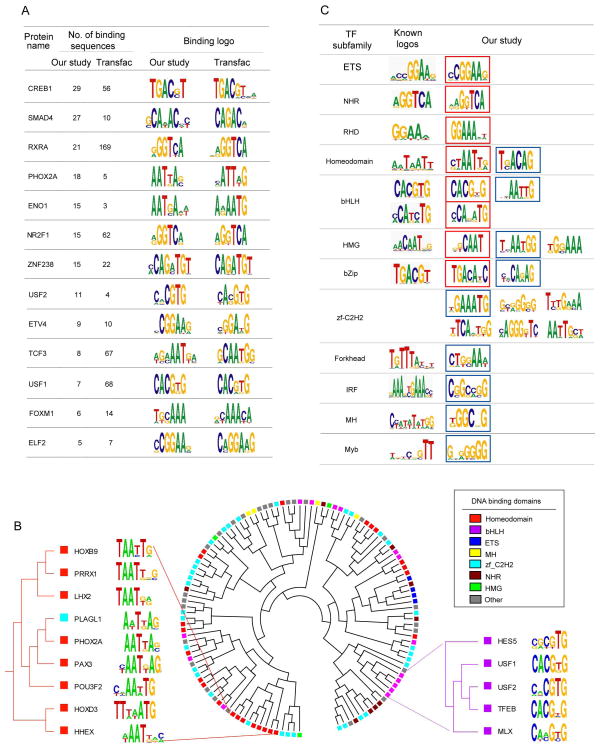
To comprehensively characterize sequence-specificity of the human TFs, we first attempted to identify consensus sequences (logos) that were preferentially bound by individual TFs. We were able to extract significant consensus sequences for 201 TFs (Table S5). These often show considerable overlap with those extracted from TRANSFAC, indicating that our approach can recover reliable consensus sequences using the test motifs (Figure 3A and Table S6). Among all consensus sequences, there are 166 novel ones for TFs which have no known binding sites listed in TRANSFAC. Our analysis considerably expands our knowledge of binding specificity of human TFs, almost doubling the number of human TFs for which consensus binding sites have been identified.
Figure 3. PDIs for Known and Predicted TFs.
(A) Comparison between TF binding logos identified in this study and those listed in TRANSFAC SITE.
(B) Clustering of TFs based on similarity of their DNA logos identified in this study. Only TFs containing known DNA binding domains were used to construct the cluster. Seven DNA binding domains are explicitly indicated in the cluster and the other domains are indicated as “Other”.
(C) Familial logos identified for the 12 TF subfamilies. Known logos were obtained from JASPAR database (Sandelin et al., 2004). Familial logos recovered in this study that are similar to the known familial logos are outlined in red. Logos validated with EMSA assays are outlined in blue.
We next clustered the TFs based on the similarity of their consensus sequences (Figure 3B). For some TFs with certain DNA-binding domains (e.g., ETS, homeodomain and bHLH), they showed more conserved DNA-binding specificity. For example, in a clade all but one TF contain the homeodomain and recognize a TAAT consensus sequence (Figure 3B). Interestingly, we found that while some TFs in the same subfamilies showed DNA binding profiles that were distinct from other members of that gene family (e.g., zf-C2H2), many TFs with highly divergent protein sequences bound to highly similar or even identical target DNA sequences (Figure 3B and Table S7). This observation suggests that global primary protein sequence identity does not necessarily correlate with DNA binding specificity.
Finally, we examined the PDIs on the TF subfamily level. We extracted familial logos for the 12 major TF subfamilies (Figure 3C). When compared to the known familial logos from the TRANSFAC and JASPAR databases (Sandelin et al., 2004; Wingender et al., 1996), our analysis identified 8 of the 12 previously reported familial logos. Furthermore, multiple logos were identified for five subfamilies, suggesting that a considerable diversity of DNA binding specificity can be found in members of a given TF subfamily, as has recently been shown for mouse and Drosophila homeodomain proteins (Berger et al., 2008; Noyes et al., 2008).
The zf-C2H2 subfamily serves as an illustration of the ability of our approach. This subfamily contains over 400 members, but no familial logos have been previously reported because of the limited number of confirmed PDIs. With the large number of PDIs characterized in this study, we identified six significant logos. For the homeodomain subfamily, we identified not only the canonical consensus site, but also the atypical site recently reported for the TGIF (Drosophila) and Meis1 (mouse) groups (Berger et al., 2008; Noyes et al., 2008). On the other hand, only a single familial logo was identified for the NHR, ETS, and RHD subfamilies. These logos closely matched the reported familial logo for each subfamily. Finally, in the case of the Forkhead, IRF, MH1, and Myb subfamilies, we identified novel familial logos that did not closely resemble the reported ones.
To confirm the specificity of novel PDIs identified for TFs, we carried out electrophoretic mobility shift assays (EMSA) to test the PDIs for 22 annotated and 9 predicted TFs. Notably, 27 of the 31 TFs tested (87.1%) demonstrated specific PDIs, indicating a low false-positive rate for the PDIs identified by protein microarray analysis (Table S8). Figure S9 shows representative examples of 9 of the subfamilies for which novel familial logos were identified, along with an example of a predicted TF that does not belong to any of these subfamilies. The proteins used in EMSA were tested with silver staining to eliminate the possibility of yeast protein contamination (Figure S10). For the four subfamilies (Forkhead, IRF, MH1, and Myb) that did not match the known logos, we were able to validate the new logos using EMSA.
Identification of Unconventional DNA-binding Proteins (uDBPs)
Surprisingly, we were able to detect many PDIs between DNA motifs and proteins of other functional classes not previously known to show sequence-specific PDIs. We also extracted consensus sequences for individual uDBPs (Table S9) as well as significant familial logos for each functional class (Figure S11).
For each class of proteins queried, we observed different percentages of proteins showing DNA-binding activity (Table 1). The percentages of proteins in different classes that showed DNA-binding activity varied greatly – from 4.3% of the protein kinases to 29.7% of the RNA-binding proteins. As a comparison, 41.2% of the annotated TFs showed PDIs, the highest among all protein classes tested. In total, we identified 634 unique uDBPs (Table 1, complete set; note that some proteins belong to multiple functional classes, so that the number of proteins in each functional class listed on Table 1 adds up to more than this total number). This represents 22.4% of all the 2820 non-TF proteins tested, implying that an unexpectedly large fraction of human proteins possess sequence-specific DNA binding activity.
Table 1.
Statistics of human PDIs detecting in this study.
| Protein class | Total No. of proteins | DNA binding proteins | |||
|---|---|---|---|---|---|
| Complete set* | High-confidence set* | ||||
| No. | Ratio(%) | No. | Ratio(%) | ||
| Known TFs | 1106 | 456 | 41.2 | 382 | 34.5 |
| Predicted TFs | 264 | 37 | 14.0 | 20 | 7.6 |
| Protein kinases | 329 | 14 | 4.3 | 7 | 2.1 |
| Chromatin-associated proteins | 287 | 73 | 25.4 | 63 | 22.0 |
| RNA-binding proteins | 698 | 207 | 29.7 | 124 | 17.8 |
| Transcriptional co-regulators | 238 | 43 | 18.1 | 25 | 10.5 |
| Other nucleic acid-binding proteins | 257 | 50 | 19.5 | 38 | 14.8 |
| DNA repair & replication | 146 | 50 | 34.2 | 42 | 28.8 |
| Mitochondrial proteins | 652 | 97 | 14.9 | 64 | 9.9 |
| All other categories | 589 | 132 | 22.4 | 42 | 7.1 |
Complete set of DNA binding proteins denotes proteins showing DNA binding activity on the protein microarrays. High-confidence set denotes proteins in the complete set which are also annotated as nuclear-localized proteins in GO database, expect for mitochondrial proteins, whose cellular localization is annotated as either nuclear and/or mitochondrial in GO.
We noticed that some of these proteins are not known to be located in the nucleus, implying that some observed unconventional PDIs might not occur in vivo. To increase the confidence, we further refined this data set to consider only proteins annotated as having nuclear localization in the GO database (Table 1, high-confidence set). Since mitochondrial transcription is actively regulated, all PDIs annotated in GO as showing either nuclear or mitochondrial localization were considered high-confidence. Filtering our initial results in this manner, we obtained 367 unique uDBPs (the high-confidence set, Table 1 and Figure 4B).
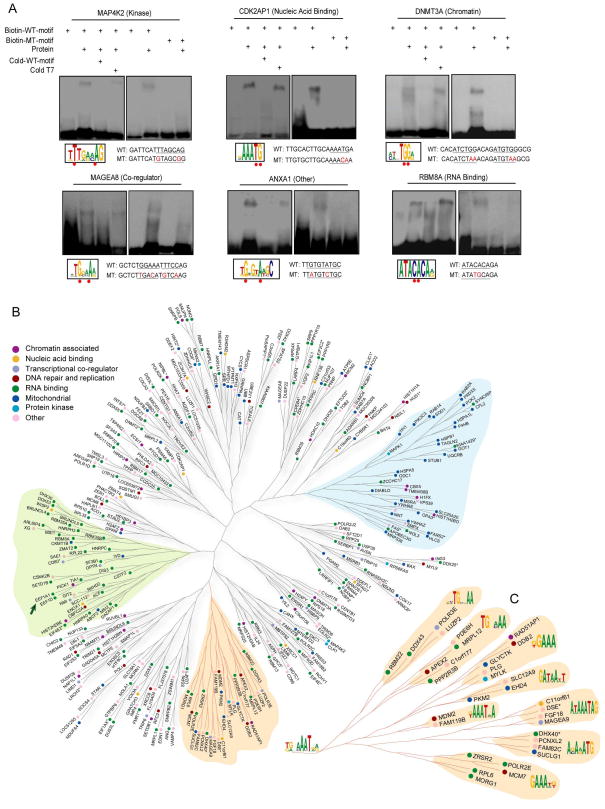
Figure 4. DNA-Binding Specificity of uDBPs.
(A) Validation of unconventional PDIs with EMSA analysis. Representative examples are shown. Newly identified consensus sites for different proteins are boxed and underlined in the DNA motif sequences used for the EMSA analysis. Mutated positions are indicated in red in motif sequences used for EMSA and underscored with red dots in the predicted consensus sequences.
(B) Clustering of uDBPs based on target sequence similarity. Proteins of different function classes are color-coded. Branches highlighted in green and blue are enriched for RNA-binding and mitochondrial-targeted proteins, respectively. Asterisks indicate that multiple proteins bind to identical target sequences; in this case, a single representative protein is shown (see Table S12 for detail). The arrow indicates an example of two proteins that interact as part of a protein complex but do not share protein sequence homology.
(C) Magnified view of the orange branch in (A), where the consensus sequences for each sub-branch are shown.
Validation of uDBPs
We first used EMSA assays to confirm direct binding of representative uDBPs to the corresponding DNA motifs in vitro. Over 91% (41/45) of the tested uDBPs showed direct PDIs with the corresponding DNA motifs identified from the protein microarray data (Figure 4A, Table S10). To experimentally validate the calculated familial logos, we designed mutant DNA sequences with differing sequences at two conserved nucleotide positions. Of the 13 tested proteins, 12 (92.3%) showed significant decreases in PDIs with the mutant motifs. Proteins demonstrating sequence-specific PDIs in this assay came from diverse functional categories, including mitochondrial-targeted proteins, RNA-binding proteins, and protein kinases (Figure 4A and Figure S12). Furthermore, no contaminating yeast proteins were observed following silver-staining analysis of the purified recombinant proteins that were used for EMSA, implying that any observed PDIs are highly unlikely to result from the presence of any contaminating yeast TFs (Figure S10).
It is notable that the EMSA assays confirmed highly sequence-specific PDIs for several RNA-binding proteins, many of which were believed to bind RNA and/or DNA molecules indiscriminately. To further validate their binding specificity, we performed additional EMSA assays with single-stranded DNA (ssDNA) as competitors for two representative RNA-binding proteins. The sequence-specific PDIs showed no apparent difference with or without competition from ssDNA (Figure S13), confirming that observed specific PDIs for these RNA binding proteins indeed result from binding to dsDNA. Taken together, these results indicate that the majority of the uDBPs identified in this study can indeed interact with DNA motifs directly and specifically.
Many uDBPs Associate with DNA in vivo
The most surprising result to us is the observation of sequence-specific PDIs for sugar and protein kinases. To determine whether these uDBPs associate with DNA in vivo, we selected antibodies against phosphoenolpyruvate carboxykinase 2 (PCK2) and mitogen-activated protein kinase 1 (MAPK1/Erk2) to perform chromatin-immunoprecipitation (ChIP). Using primers designed to flank genomic binding sites for these proteins predicted from our protein microarray PDI data, we obtained positive PCR products for both proteins (Figures 5D and Figure S14), indicating that they do indeed associate with these predicted target sequences in vivo. We next conducted a thorough literature search and found that an additional 12 of the 367 uDBPs identified in this study have been shown to associate with DNA in vivo using ChIP (Table S11), although these previous studies had interpreted these data to indicate that these proteins did not directly bind DNA. More importantly, we found that ChIPed DNA products in every case included sequences that match the predicted consensus DNA binding sites for these uDBPs. Taken together, a total of 14 uDBPs are associated in vivo with DNA fragments that contain our predicted DNA logos.
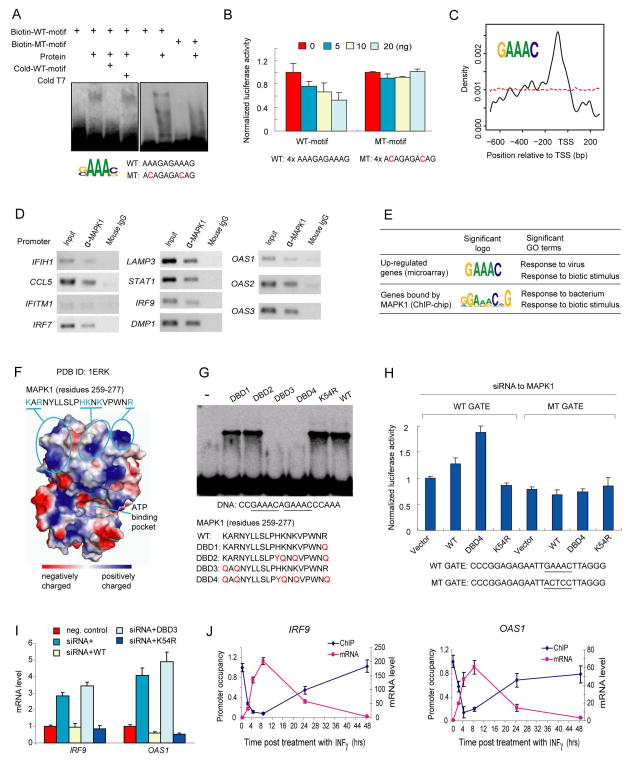
Figure 5. MAPK1 as a Transcriptional Repressor.
(A) Validation of MAPK1-DNA interaction with EMSA analysis. Mutated positions are indicated in red in motif sequences used for EMSA.
(B) Dose-dependent transcriptional repression by MAPK1 using cell-based luciferase assays. Four tandem repeats of the wild-type (WT) motif shown to complex with MAPK1 were cloned into pTK-Luc vector and co-transfected into GT1-7 cells with varying amount of plasmids that expressed MAPK1. The mutant motif that abolished gel-shifting also abolished dose-dependent transcriptional repression by MAPK1.
(C) Positioning distribution of MAPK1-binding sites in promoters. Application of an in silico motif discovery algorithm to the promoter regions of 82 up-regulated genes in a MAPK1 knockdown experiment revealed a similar consensus sequence (inset) to that determined by the protein microarray analysis (panel A). The promoter region extends from -700 to 300 bp relative to the transcription start site (TSS). The red dashed line shows the relative position of 1000 random 5-mer DNA sequences to the TSS.
(D) In vivo validation of MAPK1 and DNA interactions using ChIP coupled with PCR analysis. An anti-MAPK1 monoclonal antibody was used to ChIP the endogenous MAPK1 proteins in HeLa cells. Specific primer pairs were designed to PCR-amplify the promoter regions of the predicted targets of MAPK1. Mouse IgG was used as a negative control for immunoprecipitation. Of the 21 up-regulated genes assessed, 11 (52.3%) showed higher levels of immunoprecipitation with the anti-MAPK1 antibody than with the IgG control.
(E) Comparison of consensus sites and enriched GO terms in MAPK1 knockdown and ChIP-chip experiments.
(F) Structural analysis for DNA binding domain in MAPK1. Calculated using PyMol, the electrostatics surface potential of MAPK1 is color-coded. A surface patch (residues 259–277) comprised of three positively charged clusters are indicated with the amino acid sequence showing above. The ATP-binding pocket is also shown.
(G) Mapping the DNA binding domain in MAPK1. Five mutant forms of MAPK1 were constructed and the corresponding proteins were purified. As determined with EMSA analysis, mutations in DNA-binding-deficient (DBD) mutants 3 and 4 completely abolished the DNA binding activity, indicating that K259 and R261 are required. In contrast, K54R mutation (kinase-dead) did not affect the DNA binding activity, indicating that the two activities are independent. The DNA sequence used in the EMSA assay is also shown.
(H) Specific interactions between GATE element and the DNA-binding domain in MAPK1. Using a previously reported luciferase reporter system (Weihua et al., 1997), the effects of overexpressing MAPK1 in various mutant forms are monitored in cells that the endogenous MPAK1 is knocked down.
(I) Regulation of IFNγ-induced gene expression by the DNA-binding activity of MAPK1. Changes in IRF9 and OAS1 expression are normalized to those in negative control cells.
(J) Dynamics of promoter occupancy by MAPK1 in reverse correlation to mRNA expression levels of IRF9 and OAS1 after IFNγ treatment.
Global Classification of uDBPs
Given the existence of this new group of uDBPs, we set out to classify and organize these new proteins. We assessed protein relatedness on the basis of the DNA motif sequences to which the proteins bound. DNA-binding profiles were constructed for each protein to include the binding intensity of the protein to each of the 460 distinct DNA binding motifs (Supplemental Method). A hierarchical tree was then built based only on the similarity of the binding profiles of these unconventional DNA-binding proteins (Figure 4B). Two disparate trends were observed: On the one hand, in some clades there was a clear enrichment of proteins traditionally known to be part of a specific functional class. For example, two clades (Figure 4B, blue and green shading) were significantly over-represented for mitochondria proteins (p<4.78e-11) and RNA-binding proteins (p<4.15e-9), respectively. Another interesting example is that eukaryotic translation elongation factor 1 alpha 1 (EEF1A1) and delta (EEF1D), which belong to the translational elongation complex but share no sequence homology, were found to recognize similar DNA motif sequences. Such clustering indicates that some proteins that are similar either in terms of sequence homology or functional annotation may have similar DNA-binding characteristics. On the other hand, a mixture of functionally divergent proteins without sequence homology were also observed to share similar DNA binding motifs in some clades (Figures 4B and C), indicating that these proteins of highly divergent structure and function may cooperate to control the same DNA-binding targets.
MAPK1 Acts as a Transcriptional Repressor
As demonstrated above, many uDBPs directly and specifically bind DNA in vitro and 14 of them are found to associate with DNA in vivo. Therefore, we predicted that these uDBPs might play a physiological role in transcriptional regulation in vivo. We decided to focus on in-depth characterization of this property in MAPK1, an extensively studied protein that is known to be involved in a variety of biological processes, including proliferation, differentiation, and development.
Our protein microarray-based PDI analysis revealed that MAPK1 can bind to a G/CAAAG/C consensus sequence. We investigated this directly using EMSA analysis using both wild-type oligonucleotides matching the consensus site and mutant probes that departed from this consensus. We found that this binding is sequence-specific, since mutant oligonucleotides no longer showed binding activity (Figure 5A). Silver-staining analysis of MAPK1 showed that no contaminating yeast proteins were observed (Figure S10). In addition, we performed EMSA assays with MAPK1 protein purified from E. coli and still observed the sequence-specific PDI, further ruling out any possible contamination from yeast TFs (Figure S15).
To determine whether MAPK1 could act as a transcriptional regulator in vivo through sequence-specific DNA binding, we next employed cell-based luciferase analysis. The corresponding wild-type and mutant motif sequences were cloned upstream of a minimal promoter in a luciferase reporter construct. We found that MAPK1 tested with the wild-type motif sequence showed repression of luciferase expression in a dose-dependent manner, but showed little or no change in luciferase expression when assayed with the mutant motif, which did not bind to MAPK1 protein in the EMSA assay (Figure 5B).
To identify targets of MAPK1 and thereby gain clues to its function, we compared the gene-expression profiles of HeLa cells to those of the cells in which MAPK1 is knocked down using siRNA (Huang et al., 2008). Because MAPK1 showed a dose-dependent repression of luciferase activity in the assays described above, we collected the promoter sequences of 82 genes that showed at least a two-fold up-regulation of expression following siRNA-mediated knockdown of MAPK1 when compared to the control. Application of an in silico motif discovery algorithm to these sequences revealed a similar consensus sequence (GAAAC) to that determined by the protein microarray analysis (Figure 5C and Supplemental Method). In fact, the promoter regions of 78 of the 82 genes contained a total of 270 GAAAC sites, a clear indication of significant enrichment for these up-regulated genes (p = 1.5e-9). The distribution of the MAPK1 binding sites relative to the transcription start site showed a sharp peak around –90 bp, a typical distribution for many TFs (Figure 5C). MAPK1 consensus sequences were not enriched in the promoter sequences of down-regulated genes in MAPK1 siRNA-treated cells, consistent with our observation that MAPK1 represses gene expression in luciferase assays (Figure 5B).
To determine whether MAPK1 binds in vivo to the promoters of any of these genes whose expression is up-regulated in HeLa cells lacking MAPK1 and that contain GAAAC logos upstream, 21 of these genes were tested for MAPK1 binding by using ChIP. Eleven of 21 genes (52.3%) showed higher levels of immunoprecipitation with the anti-MAPK1 antibody relative to controls (Figure 5D). Such enrichment was not observed for any of the six down-regulated or the six unaffected genes tested (Figure S16). Thus, MAPK1 associates with GAAAC sequences in vivo to regulate expression of a large number of genes.
DNA-Binding Activity of MAPK1 Is Independent of Kinase Activity
Because the protein kinase activity of MAPK1 has been well studied, it is possible that its DNA-binding activity serves a distinct cellular function. To explore the possibility, we examined the 82 up-regulated genes for potential functional enrichment. These genes are enriched for proteins involved in response to biotic stimuli (p=1.0e-16) and to viral infection (p=1.0e-24) (Figure 5E). Furthermore, by analyzing the results of our ChIP-chip analysis for MAPK1, we discovered a similar consensus sequence and a functional enrichment for response to biotic stimuli (p=0.03) and response to bacterial infection (p=0.02) (Figure 5E). These functions are not known for MAPK1 in previous studies. In contrast, we found that the 53 confirmed substrates of MAPK1 (Diella et al., 2008) are not enriched for the same functions (Figure S17). Thus, it is very likely that sequence-specific DNA binding activity of MAPK1 is independent of its kinase activity.
To examine the structural basis of this hypothesis, we analyzed the crystal structure of MAPK1 and identified one surface patch as a potential DNA-binding domain, which is comprised of three clusters of positively charged residues close to the C-terminus at considerable distance from the ATP-binding pocket and the substrate groove (Figure 5F). Using site-directed mutagenesis, we investigated whether these residues might be required for sequence-specific DNA binding by MAPK1. We found that mutations in DBD3 and DBD4 completely abolished sequence-specific DNA binding by MAPK1 using EMSA analysis, indicating that K259 and R261 are the two key residues required for its DNA-binding activity (Figure 5G). In contrast, the kinase-dead mutant (K54R) did not show any effect on DNA binding (Robinson et al., 1996). We further confirmed that the kinase activity of MAPK1 was not essential for DNA binding by performing EMSA analysis with purified MAPK1 proteins co-expressed with MEK1 in E. coli. We observed that DNA binding was unaffected by the presence of staurosporine, a kinase inhibitor (Figure S15).
MAPK1 Directly Represses Expression of Interferon Gamma-Induced Genes via DNA-Binding Activity
Finally, we set out to determine the physiological function of the DNA-binding activity of MAPK1. Interestingly, 9 out of the 11 genes whose promoters could be ChIPed with the anti-MAPK1 antibody in HeLa cells are known to be induced by interferon. Furthermore, previous studies have shown that a transcription factor, CCAAT/enhancer binding protein-β (C/EBP-β, binds to a so-called GATE element in the proximal promoters of one of these genes, IRF9, and activates its transcription upon interferon gamma (IFNγ) stimulation (Roy et al., 2000). We found that the consensus site for MAPK1 is embedded in GATE element. These evidences suggest that MAPK1 might be involved in IFNγ signaling via its DNA-binding activity.
To test specific interactions between GATE element and the newly identified DNA-binding domain in MAPK1, we conducted luciferase analysis in transfected HeLa cells, using a wild-type GATE element reporter and a mutant element that lacks the consensus MAPK1 binding site (Weihua et al., 1997). We find that co-transfection of the siRNA-resistant wild-type MAPK1, along with siRNAs directed against endogenous MAPK1, did not result in a significant difference in luciferase expression compared to controls when a wild-type GATE element reporter construct is used (Figure 5H). However, the DNA-binding-deficient mutant of MAPK1 led to substantially up-regulated reporter expression when co-transfected with MAPK1-targeted siRNA. In contrast, kinase-dead mutants of MAPK1 efficiently repressed reporter expression. Neither wild-type nor mutant proteins showed any effect on the activity of the mutant GATE element reporter when overexpressed (Figure 5H). These results clearly demonstrated that MAPK1 specifically and directly represses expression of the luciferase reporter genes driven by canonical GATE element via its DNA-binding domain in vivo.
To further confirm the transcriptional repressor activity of MAPK1 against chromosomal genes, we monitored gene expression level of two known IFNγ-induced genes, IRF9 and OAS1, by overexpressing different mutant forms of MAPK1 in HeLa cells. We first determined that siRNA-mediated knockdown of endogenous MAPK1 significantly de-repressed expression of IRF9 and OAS1 (Figure 5I). However, in cells that lack endogenous MAPK1, overexpression of kinase-dead MAPK1 repressed expression of IRF9 and OAS1 as efficiently as overexpression of wild-type MAPK1, whereas overexpression of DNA-binding-deficient MAPK1 did not show any significant effects (Figure 5I). These results suggest that MAPK1 plays an important role in regulating expression of IFNγ-induced genes via its DNA-binding activity.
The above data suggest that low expression of IFNγ-induced genes might be maintained by the occupancy of MAPK1 on the promoters. Therefore, we predicted that promoter occupancy of these genes by MAPK1 might inversely correlate with induction of gene expression in response to IFNγ application. Using a combination of quantitative ChIP and qRT-PCR, we measured the dynamics of promoter occupancy by MAPK1 and gene expression of IRF9 and OAS1. During the course of IFNγ treatment we observed that MAPK1 was rapidly depleted from the promoters of IRF9 and OAS1 within the first four hours and the MAPK1 occupancy reached its lowest level between 6 and 8 hours post-treatment. Interestingly, promoter occupancy by MAPK1 gradually rose and almost fully recovered to its original level at 48 hours post-treatment. As predicted, the mRNA level of both IRF9 and OAS1 shows a near-perfect inverse correlation to promoter occupancy by MAPK1 (Figure 5J).
DISCUSSION
The identification of many sequence-specific PDIs for both conventional TFs and uDBPs raises an interesting question; that is whether these uDBPs bind to different target sequences than do annotated TFs. While some proteins in the same functional class were found to have preferred DNA-binding profiles selective to that protein family, the overlap in the DNA motifs recognized by the TFs and uDBPs is remarkable and substantial (Figure S18), which suggests a complex landscape for human PDI networks and possible crosstalk between TFs and uDBPs. As an example, we found that MAPK1 regulates expression of IFNγ-induced genes via binding to GATE element, which has also been shown to be bound by C/EBP-β (Roy et al., 2000).
Our study suggests that a crosstalk between C/EBP-β and the DNA-binding and kinase activities of MAPK1 results in a negative feedback loop to tightly control the temporal expression pattern of IRF9 and OAS1 upon IFNγ induction. Previously, Kalvakolanu and colleagues showed that upon IFNγ induction C/EBP-β is phosphorylated by MAPK1/2 to activate expression of the GATE-driven genes (Roy et al., 2002). However, this model does not explain up-regulation of the GATE-driven genes when only MAPK1 is knocked down in cells (Huang et al., 2008) or the suppression of IRF9 and OAS1 8 hours post IFNγ-treatment (Figure 5J). Based on the newly discovered DNA-binding activity of MAPK1, a plausible explanation is that expression of the GATE-driven genes is dictated by competitive binding of C/EBP-β and MAPK1 to GATE element. In untreated cells, GATE is directly bound by MAPK1 via its DNA-binding domain and transcription of the downstream genes is inhibited, which explains the up-regulation of those IFN-response genes when MAPK1 is knocked down (Huang et al., 2008). When cells are treated with IFNγ, C/EBP-β is rapidly induced and phosphorylated by MAPK1/2, which are activated by the MEKK1/MEK1 pathway (Roy et al., 2002). The activated C/EBP-β in the nucleus then rapidly competes off MAPK1 bound to GATE, resulting in a rapid activation of the GATE-driven genes and a sharp decline of MAPK1 occupancy at GATE (Figure 5J). As this proceeds, the concentration of nuclear MAPK1 gradually increases to a level that it starts to compete off bound C/EBP-β and therefore posts a negative feedback to eventually shut down expression of these genes. Taken together, we believe that the crosstalk between the two independent MAPK1 activities and C/EBP-β partially explains the dynamics of IFNγ-induced gene expression.
A significant advantage of the presented protein-centered approach is that the binding specificity of a given DNA motif can be simultaneously measured for thousands of proteins in a single assay. In our studies, we made careful choice for the biologically meaningful DNA motifs that are either highly conserved during evolution or highly enriched in the regulatory regions of co-expressed genes. Therefore, by exploring the DNA space predicted to be enriched for cis-regulatory elements, we have established possible connections to their upstream effectors. Indeed, the fact that virtually all of the DNA motifs tested in this study bound selectively to proteins on the array supports this notion. Furthermore, our approach can examine a large variety of protein families, providing an opportunity to discover novel DNA-binding proteins. It is expected that combined with DNA-centered approaches, such as protein-binding DNA microarrays and one-hybrid analysis, we will be able to precisely determine DNA binding consensus sequences for many uDBPs.
EXPERIMENTAL PROCEDURES
Probe Preparation
Double-stranded DNA probes were generated according to a protocol described previously (Ho et al., 2006).
Human ORF Cloning
Using the Gateway recombinant cloning system (Invitrogen, CA), human ORFs were shuttled from the selected entry clones of the Ultimate Human ORF Collection (Invitrogen, CA) or from the entry clones generated in our own laboratories to a yeast high-copy expression vector (pEGH-A) that produces GST-His6 fusion proteins under the control of the galactose-inducible GAL1 promoter. Plasmids were rescued into E. coli and verified by restriction endonuclease digestion. Plasmids with inserts of correct size were transformed into yeast for protein purification.
Protein Purification
Human proteins were purified as GST-His6 fusion proteins from yeast using a high-throughput protein purification protocol as described previously (Zhu et al., 2001).
Protein Microarrays
Purified human proteins were arrayed in a 384-well format and printed on FAST slides (Whatman, Germany) in duplicate. The protein microarrays were probed with Cy5-labeled DNA motifs using a protocol similar to that previously described (Ho et al., 2006): A protein chip was blocked for 3 h with 3% BSA in hybridization buffer (25 mM HEPES at pH 8.0, with 50 mM KGlu, 0.1% Triton X-100, 8 mM MgAC2, 3 mM DTT, 4 μM poly (dA-dT), and 10% glycerol) and then incubated with a Cy5-labeled DNA motif at a final concentration of 40 nM in hybridization buffer at 4° C overnight. The chip was washed once in cold hybridization buffer without poly (dA-dT) for 5 min and spun to dryness. The slides were finally scanned with a GenePix 4000 scanner (MDS Analytical Technologies, CA) and the binding signals were acquired using the GenePix software.
EMSAs
Each binding reaction was carried out with 100 fmol of biotinylated dsDNA probe and 2 pmol of purified protein in 20 μl of binding buffer (25 mM HEPES at pH 8.0 with 50 mM KGlu, 0.1% Triton X-100, 2 mM MgAC2, 3 mM DTT, and 5% glycerol). Twenty-five pmol (a 250-fold excess) of unlabeled (cold) DNA motifs were added in the competition assays. Reactions were carried out for 30 min at room temperature, followed by overnight incubation at 4° C. Reaction mixtures were loaded onto 5% TBE polyacrylamide gels and separated at 100 V on ice until the dye front migrated two-thirds of the way to the bottom of the gel. Nucleic acids were transferred to nylon membranes and visualized with the LightShift EMSA Kit (Pierce, USA) according to the manufacturer’s recommendations. All the expression clones for proteins used in EMSA were verified by DNA sequencing.
Luciferase Assays
Four tandem repeats of the DNA motif and the GATE element (Weihua et al., 1997) were subcloned into pTK-Luc vector (McKnight et al., 1981) and pGL3 vector (Promega, USA), respectively. DNA was transfected using the FugeneHD reagent (Roche, Switzerland). For the 4 x DNA-motif, GT1-7 cells were co-transfected with 3 constructs: pTK-Luc, pCAGIG expressing MAPK1, and pRL-TK (Promega, USA). For the GATE element, three hours after the transfection of pGL3 construct, siRNA against 3′UTR ofMAPK1 was tranfected using TransPass R1 reagent (NEB, USA). Cells were harvested 48 hrs post-transfection for luciferase reporter assay using the Dual-Luciferase reporter assay system (Promega, USA). The luciferase activity was normalized by the internal control pRL-TK Renilla luciferase activity. All assays were performed in three separate experiments done in triplicate.
Chromatin Immunoprecipitation (ChIP)
ChIP was carried out on HeLa cells using a mouse anti-MAPK1 antibody (Millipore, USA) or a rabbit anti-PCK2 antibody (Santa Cruz, USA) according to a protocol described previously (Nelson et al., 2006), except that the protein A-Sepharose was replaced with salmon sperm DNA/protein A-agarose (Millipore, USA). Normal mouse or rabbit IgG was used for mock IP as a negative control.
Site-Directed Mutagenesis
Site-directed Mutagenesis was carried out followed a protocol described previously (Jensen and Weilguny, 2005) using the QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene, USA).
Computational Analysis
The tissue specific motifs were identified using algorithms previously described (Yu et al., 2006), and see Supplemental Method for details. The procedures of protein chip data analysis include image scan, background correction, within-chip normalization, identification of positive hits, and non-specific binding filtering. Normalization and identification of positive hits were performed using the algorithms described in Supplemental Method in detail. DNA-binding logos were discovered using AlignACE (Roth et al., 1998). The DNA-binding logos were aligned using the ungapped Smith-Waterman algorithm (Smith and Waterman, 1981). The clustering tree of the TF logos was built using Neighbor-joint algorithm. The tree was visualized using MEGA4 (Tamura et al., 2007). Potential DNA motifs in the promoter regions were identified using MDscan (Liu et al., 2002). The distance between the DNA-binding profiles of any two proteins in the phylogenetic tree is defined in Supplemental Method. The initial phylogenetic tree was constructed based on the distance information using the minimum evolution method in MEGA4. The length of the branches was log-transformed. The curved layout was built manually. The length of the branches was in some cases slightly altered when the curved layout was constructed, and therefore the length was not precisely proportional to the actual distances between binding profiles. P value of GO analysis was calculated using one-sided Fisher exact test corrected for multiple testing using the minimum P method of Westfall and Young (Westfall, 1993) as provided in Ontologizer (Bauer et al., 2008). ChIP-chip data was analyzed using Cisgenome (Ji et al., 2008).
Supplementary Material
Acknowledgments
We thank Drs. J. Boeke, P. Cole, J. Nathans, G. Seydoux, T. Shimogori, S. Chen, D. Griffin, S. Taverna, J. Pomerantz, and D. Zack for their comments and suggestions. We also thank Drs. K. Dalby, D. Kalvakolanu, and R. Weiner for providing reagents, and D. McClellan for editorial assistance. This work was supported by the National Institutes of Health (GM076102 to H.Z., J.Q., RR020839 to H.Z., NEI Vision Core Grant to J.Q.), a W. M. Keck Foundation Distinguished Young Investigator in Medical Research Award to S.B., a grant from the Ruth and Milton Steinbach Fund to S.B., and a generous gift from Mr. and Mrs. Robert and Clarice Smith.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL DATA
Supplemental Data include Supplemental Experimental Procedures, 20 figures, 13 tables, and Supplemental References.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S, Grossmann S, Vingron M, Robinson PN. Ontologizer 2.0--a multifunctional tool for GO term enrichment analysis and data exploration. Bioinformatics. 2008;24:1650–1651. doi: 10.1093/bioinformatics/btn250. [DOI] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggon TJ, Shan WS, Santagata S, Myers SC, Shapiro L. Implication of tubby proteins as transcription factors by structure-based functional analysis. Science. 1999;286:2119–2125. doi: 10.1126/science.286.5447.2119. [DOI] [PubMed] [Google Scholar]
- Diella F, Gould CM, Chica C, Via A, Gibson TJ. Phospho.ELM: a database of phosphorylation sites--update 2008. Nucleic Acids Res. 2008;36:D240–244. doi: 10.1093/nar/gkm772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elemento O, Slonim N, Tavazoie S. A universal framework for regulatory element discovery across all genomes and data types. Mol Cell. 2007;28:337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elemento O, Tavazoie S. Fast and systematic genome-wide discovery of conserved regulatory elements using a non-alignment based approach. Genome Biol. 2005;6:R18. doi: 10.1186/gb-2005-6-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DA, Zhu H, Zhu X, Royce T, Gerstein M, Snyder M. Regulation of gene expression by a metabolic enzyme. Science. 2004;306:482–484. doi: 10.1126/science.1096773. [DOI] [PubMed] [Google Scholar]
- Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SW, Jona G, Chen CT, Johnston M, Snyder M. Linking DNA-binding proteins to their recognition sequences by using protein microarrays. Proc Natl Acad Sci U S A. 2006;103:9940–9945. doi: 10.1073/pnas.0509185103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Liu LY, Li ZF, Wang P, Ni L, Song LP, Xu DH, Song TS. Effects of small interfering RNAs targeting MAPK1 on gene expression profile in HeLa cells as revealed by microarray analysis. Cell Biol Int. 2008;32:1081–1090. doi: 10.1016/j.cellbi.2008.04.019. [DOI] [PubMed] [Google Scholar]
- Jensen PH, Weilguny D. Combination primer polymerase chain reaction for multi-site mutagenesis of close proximity sites. J Biomol Tech. 2005;16:336–340. [PMC free article] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos ET, Wang JY. Cell cycle-regulated binding of c-Abl tyrosine kinase to DNA. Science. 1992;256:382–385. doi: 10.1126/science.256.5055.382. [DOI] [PubMed] [Google Scholar]
- Kummerfeld SK, Teichmann SA. DBD: a transcription factor prediction database. Nucleic Acids Res. 2006;34:D74–81. doi: 10.1093/nar/gkj131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Matrubutham U, Parvizi B, Yen J, Duan D, Mirchandani J, Hashima S, Nguyen U, Ubil E, Loewenheim J, et al. ORFDB: an information resource linking scientific content to a high-quality Open Reading Frame (ORF) collection. Nucleic Acids Res. 2004;32:D595–599. doi: 10.1093/nar/gkh118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Brutlag DL, Liu JS. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat Biotechnol. 2002;20:835–839. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- McKnight SL, Gavis ER, Kingsbury R, Axel R. Analysis of transcriptional regulatory signals of the HSV thymidine kinase gene: identification of an upstream control region. Cell. 1981;25:385–398. doi: 10.1016/0092-8674(81)90057-x. [DOI] [PubMed] [Google Scholar]
- Messina DN, Glasscock J, Gish W, Lovett M. An ORFeome-based analysis of human transcription factor genes and the construction of a microarray to interrogate their expression. Genome Res. 2004;14:2041–2047. doi: 10.1101/gr.2584104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova GV, Pezza RJ, Vanevski F, Ploquin M, Masson JY, Camerini-Otero RD. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat Struct Mol Biol. 2005;12:449–453. doi: 10.1038/nsmb923. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313:533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- Robinson MJ, Harkins PC, Zhang J, Baer R, Haycock JW, Cobb MH, Goldsmith EJ. Mutation of position 52 in ERK2 creates a nonproductive binding mode for adenosine 5′-triphosphate. Biochemistry. 1996;35:5641–5646. doi: 10.1021/bi952723e. [DOI] [PubMed] [Google Scholar]
- Roth FP, Hughes JD, Estep PW, Church GM. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat Biotechnol. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- Roy SK, Hu J, Meng Q, Xia Y, Shapiro PS, Reddy SP, Platanias LC, Lindner DJ, Johnson PF, Pritchard C, et al. MEKK1 plays a critical role in activating the transcription factor C/EBP-beta-dependent gene expression in response to IFN-gamma. Proc Natl Acad Sci U S A. 2002;99:7945–7950. doi: 10.1073/pnas.122075799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SK, Wachira SJ, Weihua X, Hu J, Kalvakolanu DV. CCAAT/enhancer-binding protein-beta regulates interferon-induced transcription through a novel element. J Biol Chem. 2000;275:12626–12632. doi: 10.1074/jbc.275.17.12626. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Teichmann SA, Babu MM. Gene regulatory network growth by duplication. Nat Genet. 2004;36:492–496. doi: 10.1038/ng1340. [DOI] [PubMed] [Google Scholar]
- Tokai-Nishizumi N, Ohsugi M, Suzuki E, Yamamoto T. The chromokinesin Kid is required for maintenance of proper metaphase spindle size. Mol Biol Cell. 2005;16:5455–5463. doi: 10.1091/mbc.E05-03-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihua X, Kolla V, Kalvakolanu DV. Interferon gamma-induced transcription of the murine ISGF3gamma (p48) gene is mediated by novel factors. Proc Natl Acad Sci U S A. 1997;94:103–108. doi: 10.1073/pnas.94.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PaYS. Resampling-Based Multiple Testing: Examples and Methods for p-value Adjustment. New York: Wiley; 1993. [Google Scholar]
- Wingender E, Dietze P, Karas H, Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, Lander ES. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc Natl Acad Sci U S A. 2007;104:7145–7150. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Lin J, Zack DJ, Qian J. Computational analysis of tissue-specific combinatorial gene regulation: predicting interaction between transcription factors in human tissues. Nucleic Acids Res. 2006;34:4925–4936. doi: 10.1093/nar/gkl595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol Cell. 2003;12:1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.