Abstract
The Mu transposon system of maize is highly active, with each of the ∼50–100 copies transposing on average once each generation. The approximately one dozen distinct Mu transposons contain highly similar ∼215 bp terminal inverted repeats (TIRs) and generate 9-bp target site duplications (TSDs) upon insertion. Using a novel genome walking strategy that uses these conserved TIRs as primer binding sites, Mu insertion sites were amplified from Mu stocks and sequenced via 454 technology. 94% of ∼965,000 reads carried Mu TIRs, demonstrating the specificity of this strategy. Among these TIRs, 21 novel Mu TIRs were discovered, revealing additional complexity of the Mu transposon system. The distribution of >40,000 non-redundant Mu insertion sites was strikingly non-uniform, such that rates increased in proportion to distance from the centromere. An identified putative Mu transposase binding consensus site does not explain this non-uniformity. An integrated genetic map containing more than 10,000 genetic markers was constructed and aligned to the sequence of the maize reference genome. Recombination rates (cM/Mb) are also strikingly non-uniform, with rates increasing in proportion to distance from the centromere. Mu insertion site frequencies are strongly correlated with recombination rates. Gene density does not fully explain the chromosomal distribution of Mu insertion and recombination sites, because pronounced preferences for the distal portion of chromosome are still observed even after accounting for gene density. The similarity of the distributions of Mu insertions and meiotic recombination sites suggests that common features, such as chromatin structure, are involved in site selection for both Mu insertion and meiotic recombination. The finding that Mu insertions and meiotic recombination sites both concentrate in genomic regions marked with epigenetic marks of open chromatin provides support for the hypothesis that open chromatin enhances rates of both Mu insertion and meiotic recombination.
Author Summary
Genomic insertion sites of Mu transposons were amplified and sequenced via next generation technology, revealing more than 40,000 non-redundant Mu insertion sites that are non-uniformly distributed across the maize genome and within genes. Along chromosomes, frequencies of Mu transposon insertions are strongly correlated with recombination rates. Although both Mu and recombination occur preferentially in genes, gene density does not fully explain these patterns. Instead, the finding that Mu insertions and meiotic recombination sites both concentrate in genomic regions marked with epigenetic marks of open chromatin provides support for the hypothesis that open chromatin enhances rates of both Mu insertion and meiotic recombination.
Introduction
Gene knockouts are indispensable tools for genetic and functional genomics. The maize Mutator (Mu) transposon is the most active DNA transposon in plants [1]. In maize, a model species for which transformation can be achieved at only a low efficiency, Mu insertion mutagenesis has been an important tool for cloning genes due to its high copy numbers and high rate of germinal transposition [1],[2],[3]. In addition, because Mu elements do not exhibit a preference for transposition to nearby sites [4], as is the case for Ac/Ds transposons [5], they are ideally suited for genome-wide mutagenesis screens.
The Mutator transposon family is a two-component system. MuDR controls the transposition of itself and the other classes of the 12 nonautonomous Mu elements that have been reported so far [6]. All Mu elements share highly similar ∼215 bp terminal inverted repeats (TIRs) and upon insertion generate 9-bp target site duplications (TSDs) directly flanking Mu elements. Mu exhibits a preference for insertion in genes [7],[8],[9]. In addition, a few case studies reported a preference for insertion within 5′-UTRs or exons of genes [7],[8],[9],[10].
Although many investigations have been conducted on Mutator transposons, little is known about the genome-wide distribution of Mu insertions sites and the mechanisms by which these sites are selected. In this study, ∼965,000 Mu flanking sequences (MFSs) were obtained from 454 pyrosequencing libraries generated via Digestion-Ligation-Amplification [11], a novel approach for amplifying unknown sequences flanking known sequences. Analyses of these MFSs revealed 21 novel Mu TIR sequences and 324 genic Mu insertion hotspots that each contains ≥9 independent Mu insertions. Within genes, the Mu insertions exhibited a pronounced preference for 5′-ends with the strongest preference near transcription start sites. Additionally, regions close to the ends of chromosomes experience more Mu insertions than do peri-centromeric regions. This non-uniform pattern is similar to chromosomal distributions of recombination events and gene density. However, gene density does not fully explain the non-uniformity in genome distribution of Mu and recombination. Analyses using both cytosine methylation and histone modification data [12],[13] revealed a strong correlation between Mu insertion and cytosine methylation, H3K4me3 and H3K9ac modifications. Mu insertions and meiotic recombination sites both concentrate in genomic regions marked with epigenetic marks of open chromatin. We, therefore, hypothesize that open chromatin structure plays a key role in determining site selection of both Mu insertions and meiotic recombination events.
Results
The application of DLA-454 strategy in sequencing MFSs
DLA is a PCR-based method to amplify unknown sequences flanking known sequences [11]. DLA was adapted to sequence Mu flanking sequences using 454 pyrosequencing, a strategy that is termed DLA-454 [11]. DLA is a novel adaptor-mediated PCR-based method that uses a single-stranded oligo as an adaptor and the conserved ∼215 bp TIRs of Mu transposons as primer binding sites to amplify MFSs. In DLA-454, 6-bp barcodes [14] are inserted between the 454 primer A and a Mu-specific primer, while an adaptor primer, Nsp-P, is appended to the 454 primer B. The resulting library is sequenced using 454 primer A. By doing so, sequencing reads should begin at the barcode, followed by the Mu-specific primer and a portion of the TIR (pTIR), and end with the MFS or in cases of short MFSs the Nsp-P primer. From two technically replicated 454 GS-FLX runs, ∼964,808 reads were obtained. 99% of these sequences can be unambiguously categorized using the barcodes because the first 6 bp of each read exactly matched one of the barcode sequences. A two-step trimming strategy (Methods) was applied to remove barcodes, Mu primer, amplified Mu TIR, 454 primer B and the Nsp-P adaptor primer to obtain MFSs. Based on the results of this two-step trimming process, almost all (99.7%) reads include the Mu-specific primer and over 94% carry amplified Mu TIR sequences, demonstrating that most reads are generated from sites that contain a Mu insertion. Those trimmed MFSs (638,492) that were associated with TIR sequences were aligned to the maize B73 reference genome (B73 RefGen_v1) provided by the Maize Genome Sequencing Project (MGSP) using BLASTN (Figure S1). 58% (370,632/638,492) of the trimmed MFSs satisfied our stringent alignment cut-offs (Methods). This rate of mapping is comparable to that obtained by aligning Mo17 reads (sequenced by Joint Genome Institute using 454 pyrosequencing) to the B73 RefGen_v1 using the same criteria (data not shown). Of the aligning MFSs, 98.6% (365,600/370,632) could be uniquely mapped to a single position on the B73 RefGen_v1 and the positions of the corresponding Mu insertions determined. SNP identified between the MFSs and the sequences of the B73 RefGen_v1 were used to distinguish independent Mu insertions in different plants at the same genomic positions.
Novel Mu elements
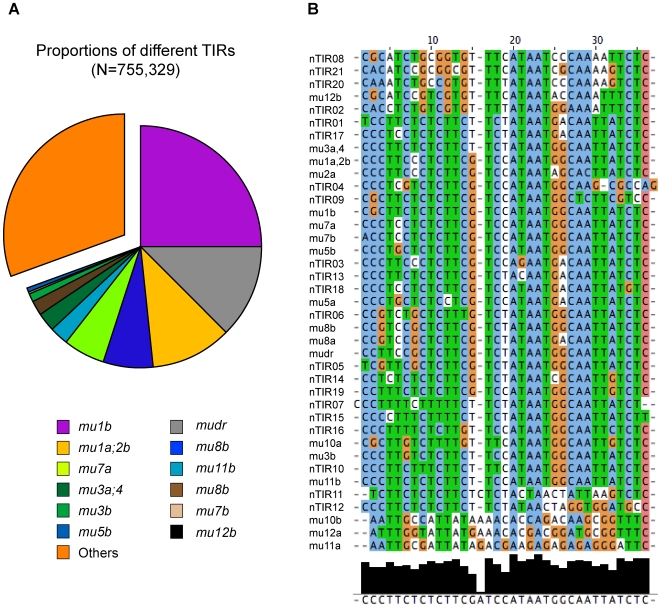
About 70% (524,696/755,329) of the 454 reads that resulted from the first trimming contained 34 bp pTIR sequences that perfectly matched known pTIRs. pTIRs from all but one of the previously described Mu elements were detected. Assuming the frequency at which pTIRs were recovered is correlated with the frequency of the corresponding classes of Mu elements in the Mu stocks, we can conclude that Mu1 and MuDR have the highest copy numbers (Figure 1A). Only a few 454 reads contained pTIRs from Mu12 and none contained Mu10 pTIRs. The two TIRs (left and right) of most Mu elements are not perfectly conserved. This allowed us to determine that TIRs from both sides of six classes of Mu elements (Mu1, Mu3, Mu4, Mu7, Mu8 and MuDR) could be successfully amplified via DLA-454. MFS from only one side of four classes of Mu elements (Mu2, Mu5, Mu11 and Mu12) were recovered in the DLA-454 data set (Figure 1).
Figure 1. Frequencies of known and novel Mu pTIRs.
(A) Proportions of different 34 bp pTIRs detected in the 454 dataset. The codes “a” and “b” designate arbitrarily defined left and right TIRs of a given Mu element. (B) Clustalw-based clustering of novel pTIRs (nTIRs), each of which was supported by at least 100 reads and exhibited a minimum edit distance of 2 from all previously described pTIRs (Methods).
Approximately, 31% of the DLA-454 reads contain pTIRs that do not perfectly match any known pTIRs. These novel sequences could be the result of sequencing errors or be evidence for the presence of novel pTIRs. We stringently required 34-bp pTIRs to have a minimum edited distance (MED) of at least 2 relative to all known pTIRs before classifying them as potentially novel pTIRs (Methods). A total of 21 novel pTIRs each of which has at least 100 supporting reads were identified (Figure 1B, Table S1). Eight of the Mu elements associated with these novel pTIRs were PCR amplified using the TIR primer in combination with primers designed based on the MFSs associated with the novel pTIR. Seven of the PCR products were successfully sequenced using Sanger technology. All seven novel pTIRs contained the expected polymorphisms relative to known pTIRs, suggesting that this data set has defined 21 novel Mu TIRs. Among the 21 novel TIRs (nTIRs), 13 were associated with multiple independent MFSs (and one, nTIR14, was associated with over 100 independent MFSs), suggesting that they are or were mobile.
Genic hotspots for Mu insertions
It has previously been established that Mu insertions exhibit a preference for typically low-copy genes as compared to non-genes [7],[8],[9]. Our first observation in support of this preference was that only ∼6% of all trimmed MFSs (600,139/638,492) contain repeat sequences as per Emrich et al., 2004 [15]. In addition, more than 98% of mappable MFSs (365,600/370,632) could be uniquely mapped to the genome even though up to 80% maize genome is repetitive [16],[17],[18]. To more directly test whether this preference of Mu elements to insert into genes holds true in our data set, we examined the numbers of Mu insertions in all of the 32,540 annotated genes in the MGSP's “filtered gene set” [18]. Even though the filtered gene set comprises only 7.5% of the genome, almost 75% of the mapped insertions are located within the 13,307 filtered genes. Similar results were obtained when these analyses were repeated with less stringently called gene sets. We therefore conclude that consistent with prior studies, Mu exhibits a strong preference for genic regions.
We then asked whether certain genes are “hotspots” for insertion. To do so, we used a simulation to determine that the probability of one or more genes acquiring nine or more insertions would be rare (p<0.05) if all genes were equally likely to acquire Mu insertions (see Methods). In the experimental data, 1% (324/32,477) of the filtered gene set had nine or more Mu insertions. Variation in gene length was not considered in this simulation because the correlation between Mu insertions and gene length is very low (r = 0.1). We used this set of genic “hotspots” to test the hypothesis that genes that experience high frequencies of Mu insertions are expressed at higher than average levels. Gene expression levels were estimated using mRNA-seq data from several tissues (Methods). Both hotspot genes (≥9 Mu insertions) and all genes that contained 1–8 Mu insertions have significantly higher levels of gene expression than genes without Mu insertions (Wilcoxon-test, all p-values<0.001, Table 1). Hotspot genes exhibit higher levels of gene expression than those genes with 1–8 Mu insertions (Wilcoxon-test, all p-values<0.001, Table 1). This relationship was observed consistently using data from each of three independent mRNA-seq experiments conducted using different tissues. Hence, we conclude that genes that experience elevated rates of Mu tend to be expressed at higher than average levels.
Table 1. Mu insertion versus gene expression.
| Category | No. Genes | Mean No. reads from various mRNA-seqs | ||
| B73_L11 | B73_L22 | F1_Seedling3 | ||
| Gene | 32,477 | 73 | 230 | 315 |
| Zero-Mu-gene4 | 19,170 | 50 | 156 | 235 |
| Mu-gene5 | 12,983 | 105* | 335* | 428* |
| Hotspot gene6 | 324 | 133* , ** | 423* , ** | 542* , ** |
Solexa mRNA-seq of L1 layer of shoot apical meristem (SAM).
Solexa mRNA-seq of L2 layer of shoot apical meristem (SAM).
Solexa mRNA-seq of 14-day seedlings from reciprocal F1 (B73×Mo17 and Mo17×B73). Data from reciprocal crosses were pooled for this analysis.
Genes without Mu insertions.
Genes with 1–8 Mu insertions.
≥9 Mu insertions per gene.
*: p-value<0.001, Wilcoxon-test of gene expression with zero-Mu-gene group.
**: p-value<0.001, Wilcoxon-test of gene expression with Mu-genes.
Mu insertions exhibit a preference for the 5′-ends of genes
Previously, several studies identified a tendency for Mu insertions to target the 5′ ends of genes. For example, Hardeman and Chandler 1989 [19] reported a preference for the first two exons of bronze1 gene and Dietrich et al. 2002 [10] reported a pronounced preference for the 5′ UTR of the glossy8 gene. This pattern has been confirmed later in multiple genes [12],[20]. To explore the distribution of Mu insertions within genes in our data set we used a set of full-length cDNAs [21] to generate a set of genes (N = 15,050) whose complete structures could be defined (the “flcDNA gene set”; Methods). After aligning the MFSs to the flcDNA gene set, the average numbers of Mu insertions per Mb were computed for each genic region (e.g., 5′ and 3′ UTRs, exons and introns) across all genes in the flcDNA gene set. A Pearson's Chi-square test (see legend of Figure S2) supported the hypothesis that frequencies of Mu insertions vary significantly across genic regions (χ 2 = 16,375, df = 7, p-value<2.2e-16). Overall, the 5′ most exons of genes had the highest frequencies of Mu insertions per Mb, particularly the 5′ UTRs and regions further upstream (Figure S2). In contrast, the 3′ portions of genes had relatively low frequencies of Mu insertions. Similar results were obtained using the MGSP's “filtered gene set”. Mu insertions occur in exons at much higher rates than in introns, which is consistent with a previous report using an engineered Mu transposon [7]. But not all exons have higher rates of insertion than introns. Indeed, the previously reported preference of Mu insertion of exons [7] can probably be explained simply by the preference of Mu to insert into 5′-most exons.
Further analyses were performed to understand the pattern of Mu insertion within genes without considering gene structure. Each gene, beginning at the transcription start site and ending at the polyA site, was split into 20 equally sized bins. The number of Mu insertions was counted in each of the 20 bins across all 15,050 genes. Figure 2A reveals a pronounced preference for insertion in the 5′-most bin and decreasing frequencies from 5′ to 3′. This pattern is observed even when using other numbers (viz., 10 and 50) of bins, indicating that Mu transposons exhibit a preference for insertion into the 5′ ends of genes.
Figure 2. The distribution of Mu insertion sites within genes.
(A) Sequences of genes (from annotated transcriptional start to poly-adenylation sites) were extracted from 15,050 full-length genes. Each gene sequence was divided into 20 equally sized bins. Because gene lengths differ, bin sizes differ from gene to gene. The x-axis lists these 20 bins 5′ to 3′. For each gene, the number of Mu insertions in each of the 20 bins was determined. Subsequently, the numbers of Mu insertions in each of the 20 bins and the lengths of each of the 20 bins were summed across the 15,050 genes. It was then possible to calculate the number of Mu insertions per Mb (y-axis) for each of the 20 bins. (B) 200-bp sequences around translation start sites (ATG, 200 bp left side and 200 bp right side) from each full-length gene were extracted and were divided into 20 bins, each of which was 20 bp in size. The x-axis lists these 20 bins 5′ to 3′. For each gene, the number of Mu insertions in each of the 20 bins was calculated. Subsequently, the numbers of Mu insertions in each of the 20 bins were summed across the 15,050 genes. The total summed length of each bin is 150,500 bp (20 bp bin length×15,050 genes). Using these data it was then possible to calculate the number of Mu insertions per Mb (y-axis) for each of the 20 bins. (C) 200-bp sequences around transcription start sites (TSS, 200 bp left side and 200 bp right side) from each full-length gene were extracted and were divided into 20 bins, each of which was 20 bp in size. The x-axis lists these 20 bins 5′ to 3′. For each gene, the number of Mu insertions in each of the 20 bins was calculated. Subsequently, the numbers of Mu insertions in each of the 20 bins were summed across the 15,050 genes. The total summed length of each bin is 150,500 bp (20 bp bin length×15,050 genes). Using these data it was then possible to calculate the number of Mu insertions per Mb (y-axis) for each of the 20 bins.
To more specifically map the positions of preferred sites for Mu insertion, 400-bp sequences (200 bp each side) surrounding the transcription start sites (TSS) and translation start sites (ATG) were extracted from each of the 15,050 genes. The extracted 400-bp sequences were each divided into 20 bins and the numbers of Mu insertions in each bin counted and plotted, revealing that Mu exhibits a preference for regions 5′ of ATGs (Figure 2B) at or slightly 5′ of the TSS (Figure 2C).
Non-uniform distribution of Mu insertions along chromosomes
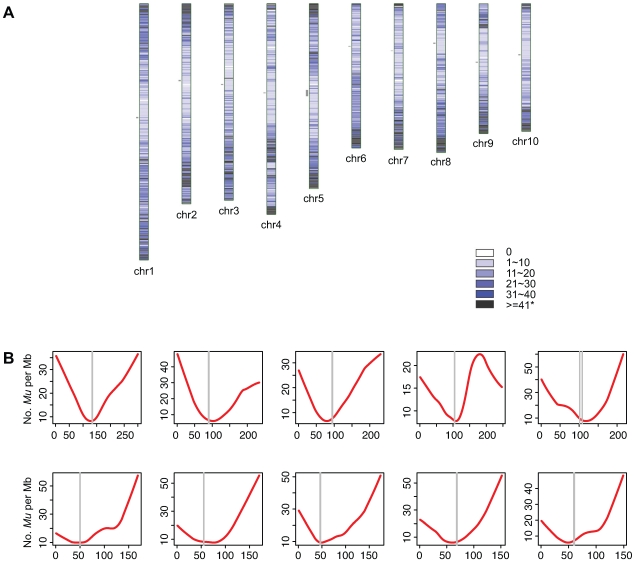
Access to the B73 reference genome allowed us to examine the distributions of Mu insertions across chromosomes. Plotting numbers of Mu insertions per Mb reveals a non-uniform distribution on each of the chromosomes (Figure 3A). Chi-square tests provided strong evidence for non-uniformity on each chromosome (Pearson's 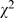 test, all p-values <2.2×10−16). To better visualize these non-uniform patterns of Mu insertion, LOWESS curves were plotted (Figure 3B) [22]. For each chromosome, a pronounced “bowl-like” trend was observed in which frequencies of Mu insertions are higher at the ends of chromosomes than in peri-centromeric regions.
test, all p-values <2.2×10−16). To better visualize these non-uniform patterns of Mu insertion, LOWESS curves were plotted (Figure 3B) [22]. For each chromosome, a pronounced “bowl-like” trend was observed in which frequencies of Mu insertions are higher at the ends of chromosomes than in peri-centromeric regions.
Figure 3. The distribution of Mu insertion sites in the maize genome.
(A) Each horizontal line on chromosomes represents a 1-Mb window. Lines are intensity- and color-coded to indicate the number of Mu insertions per Mb. Grey vertical lines represent the approximate positions of centromeres [67]. (B) The locally-weighted polynomial regression (LOWESS) curve with smooth span (f) equaling to 0.4 of the number of Mu insertions per 1-Mb window (y-axis) was plotted versus the corresponding window's coordinates (Mb, x-axis). The vertical paired grey lines represent approximate centromere positions [67]. Those patterns we observed are unlikely to be artifacts of the removal of repetitive MFS, because only a small proportion of all MFSs (1.4%) were removed based on their ability to map to multiple positions in the genome.
Because this trend is reminiscent of the distribution of meiotic recombination events in maize and other species [23],[24],[25], we were interested in examining the genome-wide distribution of meiotic recombination events per Mb and comparing these distributions to those of Mu insertion. We generated a combined genetic map containing 10,143 sequence-based genic markers (Methods). The sequences of 6,362 of these genetic markers could be uniquely aligned to the B73 RefGen_v1 and have consistent positions on both the genetic and physical maps (Methods; Figure S3). Using data from Figure S3, rates of recombination per Mb were calculated for each 1-Mb window and LOWESS curves of rates of recombination per Mb were plotted versus the physical coordinates of the B73 RefGen_v1 (Figure S4).
Each chromosome exhibits a “bowl-like” pattern of recombination per Mb similar to the frequencies of Mu insertions per Mb, which is consistent with previous cytogenetic observations [26]. The similarity between the distributions of Mu insertions and recombination events is also evident at greater granularity; viz., the numbers of Mu insertions and meiotic recombination sites in 1-Mb bins are well correlated genome-wide (r = 0.6).
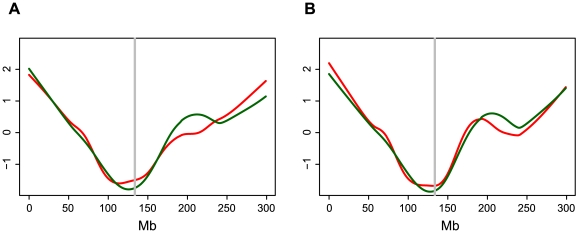
Because both meiotic crossovers and Mu insertions exhibit preferences for genes, we wondered whether the bowl-like patterns simply reflected gene density. To test this hypothesis we used the MGSP's “filtered gene set” to plot the number of genes (and bp of genic sequence) per Mb across the ten chromosomes. Similar “bowl-like” patterns were observed for the distributions of annotated genes/Mb and the proportion of genic DNA/Mb (data not shown). Even so, after expressing numbers of Mu insertions and recombination rates on a per gene or per bp of genic sequence basis, the bowl-like patterns persist (Figure 4, Figure S5, and Figure S6), demonstrating that the number of Mu insertions per gene (or per bp of genic regions) is generally greater at the ends of chromosomes than near the centromeres. Therefore, gene density can not per se fully explain the “bowl-like” patterns of Mu insertions and recombination.
Figure 4. Number of Mu insertions and recombination rate (cM) per Mb corrected by gene number and gene length on chromosome 1.
(A) Numbers of Mu insertions per gene per Mb (red line) and cM per gene per Mb (green line) are standardized as described in Methods. Locally-weighted polynomial regression (LOWESS) curves with smooth span (f) equaling to 0.4 for both standardized values were plotted against the physical coordinates of chromosome 1 (Mb, x-axis). The approximate centromere position is shown in grey [67]. (B) Numbers of Mu insertions per bp of genic sequence per Mb (red line) and cM per bp of genic sequence per Mb (green line) are standardized as described in Methods. Locally-weighted polynomial regression (LOWESS) curves with smooth span (f) equaling to 0.4 for both standardized values were plotted against the physical coordinates of chromosome 1 (Mb, x-axis). The approximate centromere position is shown in grey [67].
GC content of target sequences and a consensus sequence for Mu insertion sites
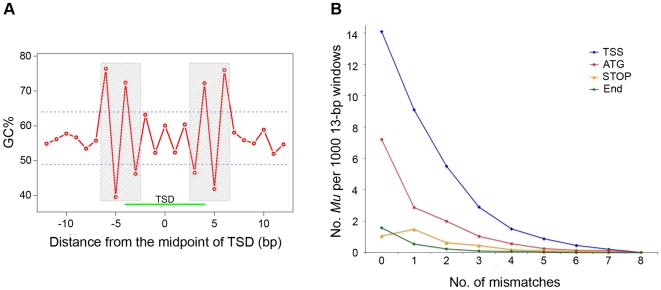
Weak consensus sequences for Mu insertion sites have been identified by several groups [7],[10],[27],[28]. To extend these results, we identified 2,217 non-redundant Mu insertion sites for which both the left and right MFSs were available and at which the expected 9 bp TSD could be detected. The mid-point of the TSD was set as position zero. The TSD is located at positions −4 to +4. GC content across the 2,217 sequences was calculated for each position (−12 through +12) separately. The null hypothesis that the GC content at each position does not differ from random can be rejected for positions −6 to −3 and +3 to +6 (p-values<0.01; Methods) (Figure 5A). The consensus sequence for position −6 to +6 is “SW::SWNNNNNWS::WS” (consistent with terminology of Dietrich et al. 2002 [10], the TSD is flanked by pairs of double colons), where S represents G or C, W represents A or T, and N can be any base. During Mu insertion the 9-bp TSD is thought to arise via the introduction and subsequent repair of staggered single-strand cuts before and after positions −4 and +4, respectively. These cuts are presumably generated by the Mu transposase. Position −6 to −3 and +3 to +6 have the pattern “SW::SW” and “WS::WS” (subsequently referred to as SWSW), respectively. We therefore hypothesize that SWSW is the preferred sequence of the Mu transposase binding/cutting site.
Figure 5. GC patterns at Mu insertion sites.
(A) More than 2,000 non-redundant Mu insertion sites for which MFSs were available on both sides and exhibiting 9-bp TSDs were collected. The mid-point of each TSD was set as position 0. Relative positions decrease to the left and increase to the right. Average GC% was calculated for each position separately across >2,000 sequences and plotted against the relative positions. Regions between the dashed lines represent the 99% confidence interval of randomly sampling 10,000 GC percentages (Methods). The shaded boxes cover positions with GC% that differ significantly from expected by chance (Methods). Boxed regions are hypothesized to be Mu transposase binding sites. (B) 101-bp sequences centered at transcription start sites (TSS), translation start sites (ATG), translational end sites (STOP) and transcriptional end sites (END) were extracted from over 15,000 full-length genes, respectively. All 13-bp windows sliding from 1 to the end of each 101-bp sequence was compared to the consensus sequence: “SW::SWNNNNNWS::WS”. The number of mismatches was computed and sequences of these windows were assigned to nine groups containing 0–8 mismatches (x-axis). 13-bp sequences of Mu insertion sites were categorized into these nine groups as well. The frequency of sequences with Mu insertions (y-axis) in each group was plotted for four sets of sequences (TSS, ATG, STOP and END) respectively.
Although SWSW is the preferred pattern, it is not required for Mu insertion, because only 61 of the 2,127 Mu insertion sites have the exact “SWSW” pattern. The 2,127 Mu insertions are a subset of the whole set of Mu insertions (N = 42,948). The remaining Mu insertion data (N = 40,821) were used to cross-validate the consensus. In this independent data set, 13-bp windows surrounding Mu insertion sites with fewer mismatches relative to the consensus were over-represented (data not shown).
To rule out the possibility that an ascertainment bias explains these results, i.e., that “SWSW” is simply enriched in those genic regions that experience the highest frequencies of Mu insertions (e.g., promoters or 5′-UTRs), similar analyses were performed on regions surrounding certain genic landmarks: viz., transcription start sites (TSS), translation start sites (ATG), translation stops sites (STOP) and transcriptional end sites (END) from the flcDNA genes (Figure 5B, Methods). Surrounding each of these genic landmarks, the frequency of Mu insertions generally decreases as the number of mismatches increases. Interestingly, even after controlling for the number of mismatches in the 13-bp window, frequencies of Mu insertions are highest in the TSS, demonstrating that the TSS is enriched for Mu insertions for reasons other than having a high frequency of the putative transposase binding sites. Hence, the distribution of the consensus sequence is not sufficient to explain the non-uniform distribution of Mu insertion sites within genes.
Epigenetic modifications are correlated with frequencies of Mu insertions
It has been hypothesized that frequencies of Mu insertions are associated with chromatin structure [7],[29]. To test this hypothesis, frequencies of Mu insertions in single-copy regions of the entire genome associated with various types of histone modifications (Table 2, Methods) were compared. The average number of Mu insertions per Mb was significantly greater for regions that contained H3K4me3 modifications than for regions that contained no H3K4me3 modification (Wilcoxon rank-sum p-value <0.0001). The same held true for H3K9ac and H3K36me3 modifications. However, for H3K27me3 modification, the situation was reversed in that the presence of H3K27me3 modifications was associated with a statistically significant decrease (Wilcoxon rank-sum p-value <0.0001) in the average number of Mu insertions per Mb.
Table 2. Mu insertions in single-copy regions with different epigenetic modifications.
| H3K4me31 | H3K9ac2 | H3K36me33 | H3K27me34 | Meth5 | MF6 | WGS-GSS7 | |
| Total length (Mb) | 34.3 | 44.2 | 31.2 | 7.7 | 15.5 | 77.7 | 11.3 |
| No. Mu | 22,871 | 24,866 | 15,552 | 599 | 156 | 21,864 | 623 |
| No. Mu/Mb | 668 | 563 | 499 | 78 | 10 | 281 | 55 |
CHIP-seq of trimethylation of lysine 4 in histone 3 [13].
CHIP-seq of acetylation of lysine 9 in histone 3 [13].
CHIP-seq of trimethylation of lysine 36 in histone 3 [13].
CHIP-seq of trimethylation of lysine 27 in histone 3 [13].
DNA methylation (McrBC sensitive, [13]).
Methylation filtration (hypomethylated, [12]).
Whole genome shotgun (WGS) - genome survey sequences (GSS) as per [66].
To check for possible interactions among histone modifications with respect to Mu insertions, a linear model with the number of Mu insertions per Mb as the response variable and the presence or absence of each histone modification and all possible interactions as explanatory variables was fit to the data. Each term in the model was significant at the 0.01 level except for two of the four three-way interactions and the four-way interaction among the four indicator variables corresponding to the four histone modifications. Thus, there is good evidence that the effects of the histone modifications on Mu insertion rates are not simply additive.
Table S2 shows that average number of Mu insertions per Mb for all 16 possible combinations of the four histone modifications. A second linear model was fit to the data used to generate Table S2. This model allowed each of the 16 possible histone modification patterns to have a different underlying Mu insertion rate. After testing for differences between each pair of patterns using the Tukey-Kramer method [30] for all pairwise comparisons, many significant differences across histone modification patterns were identified. In particular, regions with all histone modifications except H3K27me3 had a significantly higher average number of Mu per Mb than each of the other 15 patterns. Generally speaking, the H3K9ac or H3K4me3 modifications were most associated with elevated frequencies of Mu insertions among four examined histone modifications. H3K27me3 regions had relatively low frequency of Mu insertions even when other modifications were co-located. In contrast, H3K36me3 regions with H3K4me3 and/or H3K9ac co-located had much higher frequencies of Mu insertions than did H3K36me3 regions without H3K4me3 and/or H3K9ac co-located (Table S2). This illustrates one example of the type of interaction identified in our initial linear model analysis.
We also examined the relationship between DNA methylation and Mu insertion rates. This was done by identifying single-copy regions that either have evidence of containing DNA methylation (McrBC sensitive) or evidence of being hypomethylated (methylation filtration data). Regions with evidence of being methylated and being hypomethylated have frequencies of Mu insertions per Mb 5× lower or 5× higher than the WGS-GSS control, indicating that at least in single-copy regions methylation is strongly negatively correlated with frequencies of Mu insertions.
Discussion
The stocks used in this study exhibited a high rate of Mu element transposition based on standard Robertson's seedling test assays [31]. Consequently, most Mu insertions investigated in this study were recently generated. Most of them are far less likely to have been subjected to selection than are less active elements. Hence, their distributions are expected to better reflect insertion site preferences than do the distributions of “ancient” transposons detected during genome sequencing projects.
Although there are some exceptions (viz., the long arms of Chr 2 and 5), overall the distributions of Mu insertions and meiotic recombination are similar, suggesting that common features may be involved in site selection for both kinds of events. Both types of events cluster in genes (this study and [32]), but we have demonstrated that gene density is not sufficient to explain the non-uniform distributions of Mu insertions and recombination events along chromosomes.
Both types of events share a preference for GC-rich regions. Meiotic recombination events cluster in GC-rich regions in humans and yeast [33],[34]. Similarly, our data are consistent with a prior report [7] that Mu insertion sites exhibit a bias toward GC-rich regions. The average GC content of the 100-bp intervals surrounding Mu insertion sites is 56% in our data set versus the average 47% GC in the filtered gene set. In addition, like the P elements of Drosophila [35],[36] to which they have been compared mechanistically [37], Mu transposons exhibit a strong preference for 5′-ends of genes, which are typically enriched for GC relative to 3′ ends [38].
What is not clear from these data is whether the high GC content of preferred Mu insertion sites is a cause or an effect of Mu insertion site preference. The finding that the preferred Mu insertion site consensus sequence does not exhibit a strong GC signal (50% GC) suggests that although Mu transposon exhibits a preference for regions that happen to have a high GC content, they do so for reasons other than the GC content of these regions. The overall frequency of Mu insertions in a given sequence is related to its similarity to the preferential consensus sequence (SW::SWNNNNNWS::WS). However, in different genic contexts (e.g., 5′-UTRs, 3′-UTRs) this preferential sequence has dramatically different impacts on Mu insertion frequency, strongly indicating that Mu insertion site selection is more dependent on genic context than on DNA sequences per se.
If DNA sequences are not a major factor in site selection for Mu insertions, what are the causal factor(s)? Frequencies of Mu insertions and meiotic recombination are both low in pericentromeric regions, which are rich in heterochromatin, suggesting an association with chromatin structure. Indeed, our genome-wide results demonstrate that various types of epigenetic modifications are differentially correlated with frequencies of Mu insertions. In addition, the 5′-ends of genes, which are preferred sites for both Mu insertion in maize and meiotic recombination in yeast and maize [25],[33],[34],[35],[36] have distinctive epigenetic modifications. We demonstrated that Mu insertions particularly favor regions surrounding the TSS, which also exhibit strong signal for both H3K4me3 and H3K9ac [13],[39] and exhibit low levels of cytosine methylation [13],[40],[41]. In addition, H3K4me3 modifications which cluster at DSB hotspots in yeast [42] were highly correlated with Mu insertions in this study. Both of these epigenetic modifications are associated with open chromatin structure [39],[40],[43],[44],[45],[46]. Hence, it is likely that chromatin structure plays a key, and perhaps even a causal role, in site selection for both Mu insertion and meiotic recombination. A number of other transposons (e.g., Ac/Ds, P elements, MITEs, and Tos17) exhibit preferences for low-copy genic regions [3],[29],[35],[47],[48],[49],[50],[51], which cluster in the euchromatin. It is therefore possible that these transposons may share a common mechanism of insertion site selection. To further test the hypothesis that chromatin structure is a common feature driving transposon insertion site preferences, the co-localization of new transposon insertion sites and/or transposase binding site with epigenetic marks could be assayed in mutants such as mop1 [52] that would be expected to alter the distribution of epigenetic marks.
Methods
Genetic stocks
The Mu activity of Mu stocks was determined using Robertson's standard seedling test assays [31] that detects the effects of Mu transposition via the appearance of new mutations. The progeny of crosses between Mu active lines and various inbreds and hybrids were self-pollinated to produce the Mu stocks used in this study. The resulting kernels were planted and leaves harvested for genomic DNA isolation. Mu flanking sequences were amplified from these genomic DNAs via DLA [11].
Genetic mapping was conducted using (N≤357) of the IBM (Intermated B73×Mo17) Recombinant Inbred Lines (RILs) as shown in Figure S7 and listed in Table S3. The IBM RILs are the standard mapping population for the maize genetics community.
DLA for amplification, primer design, and barcode design
DLA-454 was conducting following the protocol in [11]. As compared to Liu et al., additional barcode primers were used:
AaMu
5′ GCCTCCCTCGCGCCATCAGTCTGAGGCCTCYATTTCGTCGAATC
AbMu
5′ GCCTCCCTCGCGCCATCAGGTTAGCGCCTCYATTTCGTCGAATC
AcMu
5′ GCCTCCCTCGCGCCATCAGGGTACTGCCTCYATTTCGTCGAATC
AdMu
5′ GCCTCCCTCGCGCCATCAGCATGTGGCCTCYATTTCGTCGAATC
AhMu
5′ GCCTCCCTCGCGCCATCAGATTCTGGCCTCYATTTCGTCGAATC
Data processing of Mu 454 reads
Categorization of reads based on barcodes and trimming
Raw 454 reads were categorized by their barcodes. SeqClean (http://compbio.dfci.harvard.edu/tgi/software/) was used to trim barcodes, primers and partial Mu TIR sequences. A two-step trimming strategy was applied. First, the Mu primer and adaptor primer (default overlapping requirement, ≥80% identity with primers) were removed. Trimmed sequences with the size ≥60bp were subjected to a second round trimming to remove Mu TIR regions (≥30 bp overlapping, ≥80% identity with known, novel and putative pTIRs (34 bp)).
Mapping MFS to the B73 reference genome (B73 RefGen_v1)
Trimmed MFSs with size ≥40 bp were aligned to the B73 RefGen_v1 using BLASTN. Only if there was a single best BLAST alignment (lowest E-value hit) with ≥95% identity and ≥40 bp covering the 5′ region of the query MFS, was a MFS considered to be unambiguously mapped onto the genome, which allowed the corresponding Mu insertion site to be determined. The requirement for a 5′ match was established because the exact Mu insertion sites were inferred by aligning the first 5′ bases of MFSs to the reference B73 genome. Failure of MFSs to align to the B73 RefGen_v1 could be due to DNA sequence polymorphisms between B73 and the Mu stocks and/or DNA sequencing errors and/or incomplete trimming.
Identification of SNPs between MFSs and the B73 RefGen_v1
SNPs were identified within unique alignments of MFSs and the B73 RefGen_v1. For individual B73 genomic regions where multiple MFSs were mapped, MOSAIK, a reference-guided read aligner, was used to conduct realignment of all sequences in the region (≥95% identity) (http://bioinformatics.bc.edu/marthlab/Mosaik) and SNPs were identified using an updated version of PolyBayes [53] (http://bioinformatics.bc.edu/marthlab/PolyBayes). SNP calls required at least two 454 reads of support and the proportion of reads with a given SNP among the total reads that cover a SNP site must have been ≥20%. The probability of called SNPs being correct (as calculated by PolyBayes) was larger than 0.9. Among the ∼1.5 Mb of the B73 RefGen_v1 with at least 2× MFSs coverage, 19,252 SNPs were identified. These SNPs were used to distinguish among unique Mu insertions at the same genomic positions.
Determination of non-redundant Mu insertions
To conservatively correct for errors in trimming MFS 454 reads, insertion sites with the same orientation that clustered within 3 bp of each other were regarded as the same site. Those MFS with different orientations could be the result of the same or different insertion events. Therefore, if the distance between two sites was 8-bp, indicating a 9-bp target site duplication (TSD), they were counted as paired insertions. Otherwise, they were regarded as different insertions.
MFSs were further distinguished using TIR and SNP data if multiple reads were aligned to the same insertion sites. Different types of TIRs indicate independent insertions from different genetic lines. To avoid inappropriately separating MFSs at the same position based on sequence errors in TIRs, additional criteria were used. MFSs were treated as independent insertion events if either of the criteria below was satisfied:
Both TIRs and SNPs provide evidence of independence.
A TIR was supported by at least two reads and the total number of reads containing this TIR is not less than 20% of the total reads at this site.
Identification of novel Mu TIRs
An edit distance between two sequences is the minimum number of base modifications, including base substitution, deletion and insertion, required to change one sequence into the other sequence [14]. The first 34-bp sequence was extracted from each of 454 reads after the first step of trimming. Edit distances were calculated between the 34-bp sequence and all known 34-bp pTIRs. The smallest edit distance was obtained for this 34-bp sequence versus all known pTIRs. This procedure was repeated for additional 34-bp windows obtained by starting at nucleotide positions 2 to 7. Among seven smallest edit distances from seven repeats of each read, the smallest value was considered the minimum edit distance (MED) between possible pTIRs and known pTIRs. If the MED was between 2 and 10, the corresponding 34-bp sequence was extracted and treated as the putative pTIR. In total, 153,136 reads with unknown pTIRs were obtained. After removing redundancy, 292 non-redundant putative pTIRs that had at least 100 reads of support were obtained and imported to Sequencher 4.7 for sequence clustering. Finally, 21 novel pTIRs were obtained.
Calculation of a cut-off to identify Mu insertion hotspot genes
A total of 31,838 unique Mu insertions were mapped onto 32,477 annotated genes (including 500 bp extensions upstream and downstream from the transcription start and poly-adenylation sites). We randomly assigned each of the 31,838 Mu insertions to one of the 32,477 genes. When making each random assignment, each gene was equally likely to receive an insertion. This simulates a scenario in which no genes are preferential targets for insertion. After all assignments were made, we noted the maximum number of insertions received by any one gene. The entire process was repeated 10,000 times to obtain 10,000 maximum numbers of insertions. For 95% of the 10,000 simulation runs, the maximum number of insertions in any one gene was less than 9. Thus, we declared genes with 9 or more insertions to be hotspot genes.
Approximately 1% [4],[54] or fewer [37] of gametes carry deletions adjacent to a given Mu insertion. Such events could potentially cause mis-classifications of the same insertion as two (or more) independent events. To determine if this rare event could account for the observed hotspots. All of the Mu insertions at eight hotspot genes were manually checked for independence using data from the pTIRs, SNPs in the MFSs and TSDs. These analyses found no evidence that Mu insertion hotspots artifacts caused by Mu adjacent deletions.
mRNA-seq of B73 SAMs
Shoot apical meristems (SAMs) L1 and L2 were harvested from 14-day B73 seedlings. SAMs were fixed, embedded in paraffin, sectioned and tissues collected via LCM as described previously [55]. L1 and L2 were collected separately from the same SAMs. RNA extraction, amplification and synthesis of double-stranded cDNA were conducted according to previous procedures [55].
mRNA-seq of B73×Mo17 hybrid seedlings
RNAs of B73×Mo17 hybrids (F1) were extracted from 14-day-old F1 seedlings. RNAs were purified using DNaseI treatment followed by cleanup with the RNeasy Plant Mini Kit (Qiagen, Valencia, CA) as per manufacturer instructions. Sequencing library construction was constructed using the Illumina mRNA-Seq sample preparation kit.
Alignments of mRNA–seq reads
Illumina reads were aligned to the B73 RefGen_v1 [18] with the short read aligner NOVOALIGN (http://www.novocraft.com). One and two mismatches per read were allowed in B73 and F1 RNA-seq data, respectively. Only reads that uniquely mapped to the B73 RefGen_v1 were considered for further analysis.
Generation of the full-length gene set
A total of 22,490 full-length cDNAs [21] were mapped to the B73 RefGen_v1 using GMAP [56]. Alignments were accepted only if a cDNA mapped to a single location with ≥95% identity for all exons, ≥90% cDNA coverage and ≤60 bp tail lengths. Gene structure was defined by the original annotation in combination with the GMAP splice-alignment results.
Statistical test of significant GC content at Mu insertion sites
2,127 highly reliable non-redundant Mu insertion sites with both sides of MFSs and 9 bp TSDs were used to study the sequence characteristics of Mu insertion sites (e.g. GC content). All sequences were aligned at the mid-point of the TSD, which was set as position 0. Relative positions decreased to the left and increased to the right. Average GC content was calculated at each specific position separately across 2,127 sequences. To obtain the probability of observed GC% at each given position, one base was randomly extracted from each of the 2,127 Mu insertion sites. An insertion site for purposes of this analysis is a 400 bp region centered on the 0 position of the TSD. Mean GC% was computed across all extracted nucleotides (2,127 bases). This procedure was repeated 10,000 times to obtain 10,000 GC% values. 99% of these 10,000 GC% values were between the dashed lines on Figure 4A. GC percentages falling outside this range were considered significant.
Consensus analysis of Mu insertion sites in different genic regions
101-bp sequences centered at transcription start sites (TSS), translation start sites (ATG), translation stops sites (STOP) and transcriptional end sites (END) were extracted from each of the 15,050 flcDNA genes. The number of mismatches of 13-bp-window sequence compared with “SW::SWNNNNNWS::WS” was computed and sequences of all sliding windows (1-bp stepping) from 101-bp sequences were categorized to nine groups with 0–8 mismatches, respectively. 13-bp of Mu insertion sites presumably centered at the midpoints of TSDs were categorized to these nine groups as well. The frequency of Mu insertions of each group was plotted (Figure 5B).
Recombination and Mu insertion
Construction of an integrated genetic map
An integrated genetic map of maize was constructed using 10,188 markers generated by multiple mapping projects (Missouri Mapping Project [57],[58], Genoplante [59], and our ISU mapping project ([60],[61] and unpublished work). All of these markers had been used to genotype overlapping sets of recombinant inbred lines (RILs) from the IBM population (Figure S7). As part of this study some of the ISU IDP markers were used to genotype additional IBM RILs (Figure S8). All downloaded and newly generated genotyping scores were analyzed using the MultiPoint mapping software (Initial threshold recombination rate: 0.05; Final threshold recombination rate: 0.35; Kosambi function for genetic distances calculation) [62],[63]. The genetic positions of 10,143 (∼99%) mapped markers (Table S4) were quality checked with previous map builds to repair inconsistencies.
Mapping genetic markers to physical positions
Most of the genetic markers included in the integrated genetic map were developed from known sequences. The source sequences (genomic DNAs and ESTs) of these mapped markers were trimmed using Seqclean against the UniCore vector database (dated: April 22nd, 2008). Sequences of 9,127 markers were successfully mapped to the B73 RefGen_v1 genome reference sequence via BLASTN (≥99% identity, ≤20 bp tails) and GMAP (≤10 kb intron span, ≥95% identity of all exons, ≥90% coverage, ≤5% tails or ≤60 bp tails) for the genomic DNAs and ESTs, respectively. Only sequences that mapped to a single location in the genome were used for subsequent analyses (N = 7,185). For the purposes of this analysis, we assumed that the structure of the B73 reference genome was correct. Hence, 823 markers that aligned to the B73 RefGen_v1 were removed because they had genetic positions that were inconsistent with the reference genome sequence. It is, however, expected that in subsequent analyses these currently excluded genetic markers may be useful in improving the structure of the B73 reference genome. Using the pipeline described above, 6,362 of the markers could be anchored to the B73 RefGen_v1.
Estimation of genetic positions
The 6,362 genetic markers that were anchored to the reference genome were used to draw physical-genetic curves for each chromosome by fitting a General Additive Model (GAM) using the gam function of R [64],[65]. The fitted curve was then used to estimate genetic positions based on physical positions for estimating recombination rates.
Estimation of recombination rates (cM/Mb)
Each chromosome was divided into non-overlapping 1-Mb bins. The genetic positions of the starting and ending positions (expressed in cM) of bins were estimated based on the fitted physical-genetic curves. The physical positions of the starting and ending positions of bins (expressed in Mb) were determined based on the B73 reference genome. The estimated recombination rate of a bin (cM/Mb) is calculated by dividing the genetic length of the bin expressed in cM by the physical length of the bin expressed in Mb (physical length).
Correction of recombination rates and Mu insertion frequencies by gene numbers and genic lengths
Numbers of Mu insertions and genetic length (cM) were determined in each of non-overlapping 1-Mb bins in each chromosome of the reference genome. The number of genes and the total genic length (bp) were calculated for the each 1-Mb bin as well. Numbers of Mu insertions per gene (or bp) per Mb and cM per gene (or bp) per Mb were then computed as the gene numbers (or genic lengths) corrected Mu insertion frequencies and recombination rates, respectively.
Standardization of recombination rates and Mu insertion frequencies
To visualize recombination rates and Mu insertion frequencies on the same plot, both values were standardized. Values were standardized using the formula: 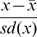 , where
, where  is either recombination rate (cM/Mb) or Mu insertion frequency (number of Mu insertions per Mb).
is either recombination rate (cM/Mb) or Mu insertion frequency (number of Mu insertions per Mb). 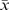 represents the mean of all
represents the mean of all  values in the same chromosome;
values in the same chromosome; 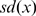 represents the standard deviation of these
represents the standard deviation of these  values. Gene and genic length corrected recombination rates and Mu insertion frequencies were standardized using the same method.
values. Gene and genic length corrected recombination rates and Mu insertion frequencies were standardized using the same method.
Epigenetic modifications and Mu insertion
The positions of histone modifications (H3K4me3, H3K9ac, H3K36me3 and H3K27me3) and cytosine methylation relative to BAC sequences from [13] were downloaded from http://www.ncbi.nlm.nih.gov/geo/ and mapped to the B73 RefGen_v1 (100% identity, 100% coverage). The 80–90% of sequences from each data set that uniquely mapped to the genome were used for further analysis. Whole genome shotgun genomic survey sequences (WGS-GSS, N = 17,232) and methylation filtration (MF) sequences (N = 349,950) [12] (http://magi.plantgenomics.iastate.edu/) [66] were mapped to the B73 RefGen_v1 (≥98% identity, ≥95% coverage). Only reads with a single best alignment (lowest e-value) were considered for the further analysis, resulting in the mapping of 81% (13,938/17,232) and 87% (304,490/349,950) of the WGS-GSS and MF reads, respectively.
Accession numbers
Sequence read archive accession no: SRX007377, SRX007378.
Supporting Information
Clustering of 454 MFSs via alignment to the B73 RefGen_v1. Reads were categorized by their barcodes. A two-step trimming strategy was applied to remove barcodes, primers and amplified TIR sequences. Trimmed MFSs were mapped to the B73 RefGen_v1. Alignments were required to exhibit ≥95% identity, ≥40 bp overlap and have no 5′ tails that failed to align to the B73 RefGen_v1. Only reads with a single best hit (lowest e-value) were used for further analyses (Methods). Finally, redundancy was removed to obtain a non-redundant set of Mu insertions.
(1.50 MB TIF)
Distribution of Mu insertion sites in different gene regions. The numbers of Mu insertions were calculated for eight different genic regions (100 bp upstream of the TSS: purple; the 5′ UTR: orange; 5′-most exons, all internal exons [as a group], and 3′-most exons: blue; all introns [as a group]: black lines) of the ∼12,000 full-length genes (excluding single-exon genes from the 15,050 full-length gene set). These regions are listed on the x-axis. For each gene, the number of Mu insertions in each of the eight genic regions was determined. Subsequently, the numbers of Mu insertions in each of the eight genic regions and the lengths of each of eight genic regions were summed across the 15,050 genes. It was then possible to calculate the number of Mu insertions per Mb (y-axis) for each of the eight genic regions. A Pearson's Chi-square test was used to test the null hypothesis that the probability of an insertion in each genic region is proportional to its total length. This null hypothesis was rejected (p-value <2.2e-16), providing strong evidence that frequencies of Mu insertion vary across genic regions.
(0.40 MB EPS)
Genetic-physical map of 6,362 genetic markers. The genetic position (cM) of each marker was plotted against its physical coordinates on the 10 chromosomes of the B73 reference genome (Mb) (Methods). Approximate centromere positions (Wolfgruber et al., [67]) are flanked by pairs of vertical grey lines. Those chromosomal regions with a paucity of polymorphic genetic markers are, based on comparative genomic hybridization (CGH) data (from Springer et al., PLoS Genetics 2009 [doi:10.1371/journal.pgen.1000734]), highly conserved between B73 and Mo17. The log intensity ratio of Mo17 to B73 (log2(Mo17/B73), y-axis) for each CGH probe is plotted versus its physical position on the B73 RefGen_v1 (Mb). CGH probes with statistically significant values of log2(Mo17/B73) (q-value<0.05) are indicated in red (Mo17>B73) and blue (B73>Mo17), while non-significant probes are indicated in green (B73 = Mo17).
(2.51 MB TIF)
The distribution of recombination events on reference chromosomes. The recombination rate was estimated for every non-overlapping 1-Mb window across the genome (Methods). The locally-weighted polynomial regression (LOWESS) curve with smooth span (f) equaling to 0.4 was plotted (green line) using the recombination rate (cM) per 1-Mb window (y-axis) versus the corresponding window's coordinates on the B73 reference chromosome (Mb, x-axis). Approximate centromere positions from Wolfgruber et al. [67] are shown in grey.
(0.33 MB EPS)
Numbers of Mu insertions and recombination rates per Mb corrected by numbers of genes on reference chromosomes 2–10. Numbers of Mu insertions per gene per Mb (red lines) and cM per gene per Mb (green lines), respectively were standardized as described in Methods. The locally-weighted polynomial regression (LOWESS) curves with smooth span (f) equaling to 0.4 of both standardized values were plotted against the physical coordinates (Mb, x-axis) on reference chromosomes 2–10. Approximate centromere positions from Wolfgruber et al. [67] are shown in grey.
(0.38 MB EPS)
Numbers of Mu insertions and recombination rates per Mb corrected by bp of genic sequences on reference chromosomes 2–10. Numbers of Mu insertions per bp of genic sequence per Mb (red lines) and cM per bp of genic sequence per Mb (green lines), respectively were standardized as described in Methods. The locally-weighted polynomial regression (LOWESS) curves with smooth span (f) equaling to 0.4 of both standardized values were plotted against the physical coordinates (Mb, x-axis) on reference chromosomes 2–10. Approximate centromere positions from Wolfgruber et al. [67] are shown in grey.
(0.38 MB EPS)
IBM RILs used to generate various genetic maps. Venn diagram shows numbers of IBM RILs used in each of several genetic mapping studies (Table S3). Data from these mapping projects, supplemented with additional data (Figure S8), were used to construct the integrated genetic map presented in Table S4.
(0.48 MB EPS)
Markers used to construct the integrated genetic map. An integrated genetic map of maize was constructed based on genotyping data from 10,143 markers from multiple mapping projects (Missouri Mapping Project (MMP) (Coe et al., Plant Phys 2002, Cone et al., Plant Phys 2002), Genoplante (Falque et al., Genetics 2005), ISU-IDP/TIDP (Map 7) (unpublished), ISU SNP (Liu et al., Genetics 2010)). Some IDP markers were used to genotype additional IBM RILs as part of this study. This flowchart provides types and numbers of markers used to genotype RILs. See also Figure S7 and Tables S3 and S4.
(0.56 MB TIF)
Novel pTIRs.
(0.10 MB DOC)
Frequencies of Mu insertions in different combinations of four epigenetic modifications in histone 3.
(0.05 MB DOC)
List of 357 RILs in the integrated genetic map.
(0.38 MB DOC)
10,143 genetic markers in the integrated map.
(9.95 MB XLS)
Acknowledgments
We thank Ms. Natasha Zazubovich (McGill University) for constructing the genetic map; Drs. Abraham Korol, Yefim I. Ronin, and David I. Mester (University of Haifa) for assistance using the MultiPoint mapping software; Ms. Jia-Ling Pik for experimental wet lab assistance; Dr. Kazuhiro Ohtsu and graduate student Yi Jia for sharing unpublished RNA-seq data; Drs. Alain Murigneux, Laurent Decousset, and Jorge Duarte for discussions regarding the relationship between physical and genetic distances; Drs. Jeffery Essner, Thomas Peterson, Hui-Hsien Chou, Xiaoqiu Huang, An-Ping Hsia, and graduate student Ruth Swanson-Wagner for useful discussions. In addition we thank the following consortia for sharing data prior to publication: the Maize Genome Sequencing Project for sharing genome sequences and annotation, the Maize Centromere Project for sharing the positions of centromeres, Maize Transposable Element Consortium for sharing the transposon data, and Dr. Dan Rokhsar of the DOE's Joint Genome Institute for sharing unpublished whole genome shotgun sequences of the Mo17 genome.
Footnotes
The authors have declared that no competing interests exist.
This research was supported by a grant from the National Research Initiative of the USDA Cooperative State Research, Education, and Extension Service, grant no. 2005-35301-15715 to P. S. Schnable. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lisch D. Mutator transposons. Trends Plant Sci. 2002;7:498–504. doi: 10.1016/s1360-1385(02)02347-6. [DOI] [PubMed] [Google Scholar]
- 2.McCarty DR, Settles AM, Suzuki M, Tan BC, Latshaw S, et al. Steady-state transposon mutagenesis in inbred maize. Plant J. 2005;44:52–61. doi: 10.1111/j.1365-313X.2005.02509.x. [DOI] [PubMed] [Google Scholar]
- 3.Candela H, Hake S. The art and design of genetic screens: maize. Nat Rev Genet. 2008;9:192–203. doi: 10.1038/nrg2291. [DOI] [PubMed] [Google Scholar]
- 4.Lisch D, Chomet P, Freeling M. Genetic characterization of the Mutator system in maize: behavior and regulation of Mu transposons in a minimal line. Genetics. 1995;139:1777–1796. doi: 10.1093/genetics/139.4.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brutnell TP. Transposon tagging in maize. Funct Integr Genomics. 2002;2:4–12. doi: 10.1007/s10142-001-0044-0. [DOI] [PubMed] [Google Scholar]
- 6.Walbot V, Rudenko GN. MuDR/Mu transposons of maize. Washington, D. C.: Amer. Soc. Microbiology; 2002. [Google Scholar]
- 7.Fernandes J, Dong Q, Schneider B, Morrow DJ, Nan GL, et al. Genome-wide mutagenesis of Zea mays L. using RescueMu transposons. Genome Biol. 2004;5:R82. doi: 10.1186/gb-2004-5-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cresse AD, Hulbert SH, Brown WE, Lucas JR, Bennetzen JL. Mu1-related transposable elements of maize preferentially insert into low copy number DNA. Genetics. 1995;140:315–324. doi: 10.1093/genetics/140.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Settles AM, Latshaw S, McCarty DR. Molecular analysis of high-copy insertion sites in maize. Nucleic Acids Res. 2004;32:e54. doi: 10.1093/nar/gnh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dietrich CR, Cui F, Packila ML, Li J, Ashlock DA, et al. Maize Mu transposons are targeted to the 5′ untranslated region of the gl8 gene and sequences flanking Mu target-site duplications exhibit nonrandom nucleotide composition throughout the genome. Genetics. 2002;160:697–716. doi: 10.1093/genetics/160.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu S, Dietrich CR, Schnable PS. DLA-Based Strategies for Cloning Insertion Mutants: Cloning the gl4 Locus of Maize using Mu Transposon Tagged Alleles. Genetics in press. 2009 doi: 10.1534/genetics.109.108936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmer LE, Rabinowicz PD, O'Shaughnessy AL, Balija VS, Nascimento LU, et al. Maize genome sequencing by methylation filtration. Science. 2003;302:2115–2117. doi: 10.1126/science.1091265. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Elling AA, Li X, Li N, Peng Z, et al. Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in Maize. Plant Cell. 2009 doi: 10.1105/tpc.109.065714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu F, Guo L, Wen TJ, Liu F, Ashlock DA, et al. DNA sequence-based “bar codes” for tracking the origins of expressed sequence tags from a maize cDNA library constructed using multiple mRNA sources. Plant Physiol. 2003;133:475–481. doi: 10.1104/pp.103.025015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emrich SJ, Aluru S, Fu Y, Wen TJ, Narayanan M, et al. A strategy for assembling the maize (Zea mays L.) genome. Bioinformatics. 2004;20:140–147. doi: 10.1093/bioinformatics/bth017. [DOI] [PubMed] [Google Scholar]
- 16.SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 17.Hake S, Walbot V. The genome of Zea mays, its organization and homology to related grasses. Chromosoma. 1980;79:251–270. [Google Scholar]
- 18.Schnable PS, Ware D, Fulton RS, Stein JC, Wei F. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326 doi: 10.1126/science.1178534. doi:1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 19.Hardeman KJ, Chandler VL. Characterization of bz1 mutants isolated from mutator stocks with high and low numbers of Mu1 elements. Dev Genet. 1989;10:460–472. doi: 10.1002/dvg.1020100607. [DOI] [PubMed] [Google Scholar]
- 20.May BP, Liu H, Vollbrecht E, Senior L, Rabinowicz PD, et al. Maize-targeted mutagenesis: A knockout resource for maize. Proc Natl Acad Sci U S A. 2003;100:11541–11546. doi: 10.1073/pnas.1831119100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soderlund C, Descour A, Kudrna D, Bomhoff M, Boyd L, et al. Sequencing, Mapping and Analysis of 27,455 Maize Full-length cDNAs. PLoS Genet. 2009;5:e740. doi: 10.1371/journal.pgen.1000740. doi: 10.1371/journal.pgen.1000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cleveland WS. Robust locally weighted regression and smoothing scatterplots. Journal of the American Statistical Association. 1979;74:829–836. [Google Scholar]
- 23.Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 24.Akhunov ED, Goodyear AW, Geng S, Qi LL, Echalier B, et al. The organization and rate of evolution of wheat genomes are correlated with recombination rates along chromosome arms. Genome Res. 2003;13:753–763. doi: 10.1101/gr.808603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fengler K, Allen SM, Li B, Rafalski A. Distribution of genes, recombination, and repetitive elements in the maize genome. the Plant Genome. 2007;47:S83–S95. [Google Scholar]
- 26.Wang CJ, Harper L, Cande WZ. High-resolution single-copy gene fluorescence in situ hybridization and its use in the construction of a cytogenetic map of maize chromosome 9. Plant Cell. 2006;18:529–544. doi: 10.1105/tpc.105.037838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanley S, Edwards D, Stevenson D, Haines S, Hegarty M, et al. Identification of transposon-tagged genes by the random sequencing of Mutator-tagged DNA fragments from Zea mays. Plant J. 2000;23:557–566. doi: 10.1046/j.1365-313x.2000.00830.x. [DOI] [PubMed] [Google Scholar]
- 28.Raizada MN, Walbot V. The late developmental pattern of Mu transposon excision is conferred by a cauliflower mosaic virus 35S -driven MURA cDNA in transgenic maize. Plant Cell. 2000;12:5–21. doi: 10.1105/tpc.12.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennetzen JL. Transposable element contributions to plant gene and genome evolution. Plant Mol Biol. 2000;42:251–269. [PubMed] [Google Scholar]
- 30.Kramer CY. Extension of multiple range tests to group means with unequal numbers of replications. Biometrics. 1956;12:307–310. [Google Scholar]
- 31.Robertson DS. Characterization of a Mutator system in maize. Mutation Research. 1978;51:21–28. [Google Scholar]
- 32.Dooner HK, Hsia AP, Schnable PS. Homologous recombination in maize. Springer; 2009. pp. 377–402. [Google Scholar]
- 33.Fullerton SM, Bernardo Carvalho A, Clark AG. Local rates of recombination are positively correlated with GC content in the human genome. Mol Biol Evol. 2001;18:1139–1142. doi: 10.1093/oxfordjournals.molbev.a003886. [DOI] [PubMed] [Google Scholar]
- 34.Gerton JL, DeRisi J, Shroff R, Lichten M, Brown PO, et al. Inaugural article: global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao GC, Rehm EJ, Rubin GM. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2000;97:3347–3351. doi: 10.1073/pnas.050017397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spradling AC, Stern DM, Kiss I, Roote J, Laverty T, et al. Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc Natl Acad Sci U S A. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Wen TJ, Schnable PS. Role of RAD51 in the repair of MuDR-induced double-strand breaks in maize (Zea mays L.). Genetics. 2008;178:57–66. doi: 10.1534/genetics.107.080374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong GK, Wang J, Tao L, Tan J, Zhang J, et al. Compositional gradients in Gramineae genes. Genome Res. 2002;12:851–856. doi: 10.1101/gr.189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009;10:R62. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borde V, Robine N, Lin W, Bonfils S, Geli V, et al. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 44.Yan C, Boyd DD. Histone H3 acetylation and H3 K4 methylation define distinct chromatin regions permissive for transgene expression. Mol Cell Biol. 2006;26:6357–6371. doi: 10.1128/MCB.00311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, Wang X, He K, Ma Y, Su N, et al. High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell. 2008;20:259–276. doi: 10.1105/tpc.107.056879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamazaki M, Tsugawa H, Miyao A, Yano M, Wu J, et al. The rice retrotransposon Tos17 prefers low-copy-number sequences as integration targets. Mol Genet Genomics. 2001;265:336–344. doi: 10.1007/s004380000421. [DOI] [PubMed] [Google Scholar]
- 48.Feschotte C, Jiang N, Wessler SR. Plant transposable elements: where genetics meets genomics. Nat Rev Genet. 2002;3:329–341. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- 49.Kolkman JM, Conrad LJ, Farmer PR, Hardeman K, Ahern KR, et al. Distribution of Activator (Ac) throughout the maize genome for use in regional mutagenesis. Genetics. 2005;169:981–995. doi: 10.1534/genetics.104.033738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, et al. Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell. 2003;15:1771–1780. doi: 10.1105/tpc.012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang N, Bao Z, Zhang X, Hirochika H, Eddy SR, et al. An active DNA transposon family in rice. Nature. 2003;421:163–167. doi: 10.1038/nature01214. [DOI] [PubMed] [Google Scholar]
- 52.Alleman M, Sidorenko L, McGinnis K, Seshadri V, Dorweiler JE, et al. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. doi: 10.1038/nature04884. [DOI] [PubMed] [Google Scholar]
- 53.Marth GT, Korf I, Yandell MD, Yeh RT, Gu Z, et al. A general approach to single-nucleotide polymorphism discovery. Nat Genet. 1999;23:452–456. doi: 10.1038/70570. [DOI] [PubMed] [Google Scholar]
- 54.Das L, Martienssen R. Site-selected transposon mutagenesis at the hcf106 locus in maize. Plant Cell. 1995;7:287–294. doi: 10.1105/tpc.7.3.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohtsu K, Smith MB, Emrich SJ, Borsuk LA, Zhou R, et al. Global gene expression analysis of the shoot apical meristem of maize (Zea mays L.). Plant J. 2007;52:391–404. doi: 10.1111/j.1365-313X.2007.03244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 57.Coe E, Cone K, McMullen M, Chen SS, Davis G, et al. Access to the maize genome: an integrated physical and genetic map. Plant Physiol. 2002;128:9–12. doi: 10.1104/pp.128.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cone KC, McMullen MD, Bi IV, Davis GL, Yim YS, et al. Genetic, physical, and informatics resources for maize. On the road to an integrated map. Plant Physiol. 2002;130:1598–1605. doi: 10.1104/pp.012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Falque M, Decousset L, Dervins D, Jacob AM, Joets J, et al. Linkage mapping of 1454 new maize candidate gene Loci. Genetics. 2005;170:1957–1966. doi: 10.1534/genetics.104.040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fu Y, Wen TJ, Ronin YI, Chen HD, Guo L, et al. Genetic dissection of intermated recombinant inbred lines using a new genetic map of maize. Genetics. 2006;174:1671–1683. doi: 10.1534/genetics.106.060376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu S, Chen HD, Makarevitch I, Shirmer R, Emrich SJ, et al. High-Throughput Genetic Mapping of Mutants via Quantitative SNP-typing. Genetics in press. 2010 doi: 10.1534/genetics.109.107557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mester D, Ronin Y, Minkov D, Nevo E, Korol A. Constructing large-scale genetic maps using an evolutionary strategy algorithm. Genetics. 2003;165:2269–2282. doi: 10.1093/genetics/165.4.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mester DI, Ronin YI, Nevo E, Korol AB. Fast and high precision algorithms for optimization in large-scale genomic problems. Comput Biol Chem. 2004;28:281–290. doi: 10.1016/j.compbiolchem.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 64.Wood SN. Fast stable direct fitting and smoothness selection for generalized additive models. Journal of the Royal Statistical Society: Series B. 2008;70:495–518. [Google Scholar]
- 65.Wood SN. mgcv:GAMs and Generalized Ridge Regression for R. R News. 2001;1:20–25. [Google Scholar]
- 66.Fu Y, Emrich SJ, Guo L, Wen TJ, Ashlock DA, et al. Quality assessment of maize assembled genomic islands (MAGIs) and large-scale experimental verification of predicted genes. Proc Natl Acad Sci U S A. 2005;102:12282–12287. doi: 10.1073/pnas.0503394102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wolfgruber TK, Sharma A, Schneider KL, Albert PS, Koo D, et al. Maize centromere structure and evolution: sequence analysis of centromeres 2 and 5 reveals dynamic loci shaped primarily by retrotransposons. PLoS Genet. 2009;5:e743. doi: 10.1371/journal.pgen.1000743. doi: 10.1371/journal.pgen.1000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clustering of 454 MFSs via alignment to the B73 RefGen_v1. Reads were categorized by their barcodes. A two-step trimming strategy was applied to remove barcodes, primers and amplified TIR sequences. Trimmed MFSs were mapped to the B73 RefGen_v1. Alignments were required to exhibit ≥95% identity, ≥40 bp overlap and have no 5′ tails that failed to align to the B73 RefGen_v1. Only reads with a single best hit (lowest e-value) were used for further analyses (Methods). Finally, redundancy was removed to obtain a non-redundant set of Mu insertions.
(1.50 MB TIF)
Distribution of Mu insertion sites in different gene regions. The numbers of Mu insertions were calculated for eight different genic regions (100 bp upstream of the TSS: purple; the 5′ UTR: orange; 5′-most exons, all internal exons [as a group], and 3′-most exons: blue; all introns [as a group]: black lines) of the ∼12,000 full-length genes (excluding single-exon genes from the 15,050 full-length gene set). These regions are listed on the x-axis. For each gene, the number of Mu insertions in each of the eight genic regions was determined. Subsequently, the numbers of Mu insertions in each of the eight genic regions and the lengths of each of eight genic regions were summed across the 15,050 genes. It was then possible to calculate the number of Mu insertions per Mb (y-axis) for each of the eight genic regions. A Pearson's Chi-square test was used to test the null hypothesis that the probability of an insertion in each genic region is proportional to its total length. This null hypothesis was rejected (p-value <2.2e-16), providing strong evidence that frequencies of Mu insertion vary across genic regions.
(0.40 MB EPS)
Genetic-physical map of 6,362 genetic markers. The genetic position (cM) of each marker was plotted against its physical coordinates on the 10 chromosomes of the B73 reference genome (Mb) (Methods). Approximate centromere positions (Wolfgruber et al., [67]) are flanked by pairs of vertical grey lines. Those chromosomal regions with a paucity of polymorphic genetic markers are, based on comparative genomic hybridization (CGH) data (from Springer et al., PLoS Genetics 2009 [doi:10.1371/journal.pgen.1000734]), highly conserved between B73 and Mo17. The log intensity ratio of Mo17 to B73 (log2(Mo17/B73), y-axis) for each CGH probe is plotted versus its physical position on the B73 RefGen_v1 (Mb). CGH probes with statistically significant values of log2(Mo17/B73) (q-value<0.05) are indicated in red (Mo17>B73) and blue (B73>Mo17), while non-significant probes are indicated in green (B73 = Mo17).
(2.51 MB TIF)
The distribution of recombination events on reference chromosomes. The recombination rate was estimated for every non-overlapping 1-Mb window across the genome (Methods). The locally-weighted polynomial regression (LOWESS) curve with smooth span (f) equaling to 0.4 was plotted (green line) using the recombination rate (cM) per 1-Mb window (y-axis) versus the corresponding window's coordinates on the B73 reference chromosome (Mb, x-axis). Approximate centromere positions from Wolfgruber et al. [67] are shown in grey.
(0.33 MB EPS)
Numbers of Mu insertions and recombination rates per Mb corrected by numbers of genes on reference chromosomes 2–10. Numbers of Mu insertions per gene per Mb (red lines) and cM per gene per Mb (green lines), respectively were standardized as described in Methods. The locally-weighted polynomial regression (LOWESS) curves with smooth span (f) equaling to 0.4 of both standardized values were plotted against the physical coordinates (Mb, x-axis) on reference chromosomes 2–10. Approximate centromere positions from Wolfgruber et al. [67] are shown in grey.
(0.38 MB EPS)
Numbers of Mu insertions and recombination rates per Mb corrected by bp of genic sequences on reference chromosomes 2–10. Numbers of Mu insertions per bp of genic sequence per Mb (red lines) and cM per bp of genic sequence per Mb (green lines), respectively were standardized as described in Methods. The locally-weighted polynomial regression (LOWESS) curves with smooth span (f) equaling to 0.4 of both standardized values were plotted against the physical coordinates (Mb, x-axis) on reference chromosomes 2–10. Approximate centromere positions from Wolfgruber et al. [67] are shown in grey.
(0.38 MB EPS)
IBM RILs used to generate various genetic maps. Venn diagram shows numbers of IBM RILs used in each of several genetic mapping studies (Table S3). Data from these mapping projects, supplemented with additional data (Figure S8), were used to construct the integrated genetic map presented in Table S4.
(0.48 MB EPS)
Markers used to construct the integrated genetic map. An integrated genetic map of maize was constructed based on genotyping data from 10,143 markers from multiple mapping projects (Missouri Mapping Project (MMP) (Coe et al., Plant Phys 2002, Cone et al., Plant Phys 2002), Genoplante (Falque et al., Genetics 2005), ISU-IDP/TIDP (Map 7) (unpublished), ISU SNP (Liu et al., Genetics 2010)). Some IDP markers were used to genotype additional IBM RILs as part of this study. This flowchart provides types and numbers of markers used to genotype RILs. See also Figure S7 and Tables S3 and S4.
(0.56 MB TIF)
Novel pTIRs.
(0.10 MB DOC)
Frequencies of Mu insertions in different combinations of four epigenetic modifications in histone 3.
(0.05 MB DOC)
List of 357 RILs in the integrated genetic map.
(0.38 MB DOC)
10,143 genetic markers in the integrated map.
(9.95 MB XLS)