Abstract
Fenofibric acid (FA), the active moiety of fenofibrate, is an agonist of the peroxisome proliferator-activated nuclear receptor α that modulates triglyceride and cholesterol profiles. Lipid response to fenofibrate and FA serum concentrations is highly variable. Although FA is reported to be almost exclusively inactivated by UDP-glucuronosyltransferases (UGTs) into FA-glucuronide (FA-G), the contribution of UGT isoenzymes has never been systematically assessed. Heterologously expressed human UGT1A and UGT2B and their coding variants were tested for FA glucuronidation using liquid chromatography/mass spectrometry. Recombinant UGT2B7 presented the highest Vmax/Km value (2.10 μl/min/mg), 16-fold higher than the activity of other reactive UGTs, namely, UGT1A3, UGT1A6, and UGT1A9 (0.13, 0.09, and 0.02 μl/min/mg, respectively). UGT2B7.1 (His268) and UGT2B7.2 (Tyr268) enzyme activity was similar, whereas UGT1A3.2 (R11A47), UGT1A3.3 (Trp11), and UGT1A9.3 (Thr33) showed 61 to 96% reduced Vmax/Km values compared with the respective (1) reference proteins. FA-G formation by a human liver bank (n = 48) varied by 10-fold, but the rate of formation was not associated with common genetic variations in UGT1A3, UGT1A6, UGT1A9, and UGT2B7. Correlation with activities for the probe substrates zidovudine (UGT2B7; r2 = 0.75), mycophenolic acid (UGT1A9; r2 = 0.42), fulvestrant (UGT1A3; r2 = 0.36), but not serotonin (UGT1A6; r2 = 0.06) indicated a primary role for UGT2B7 and lesser roles of UGT1A9 and UGT1A3 in hepatic FA glucuronidation. This was confirmed by a strong correlation of FA-G formation with UGT2B7 protein content and inhibition by fluconazole, a known UGT2B7 selective inhibitor. Additional studies are required to identify genetic factors contributing to the observed FA glucuronidation variability.
Fenofibrate is a widely prescribed drug approved for triglyceride-lowering in patients with mixed dyslipidemia (Guay, 2002). Fenofibrate is mostly known to reduce serum triglycerides (by 35–50%) and to elevate serum high-density lipoprotein-cholesterol (by 15–25%) (Staels et al., 1998). Lipid response to fenofibrate is highly variable.
Fenofibric acid (FA), the active moiety of fenofibrate, is responsible for the primary pharmacodynamic effects of the drug, including decrease of triglycerides, cholesterol, and very low-density lipoprotein levels, as well as increase of high-density lipoprotein-cholesterol (Guay, 2002). These effects are modulated at the transcription level through the activation of peroxisome proliferator-activated receptor α. FA serum concentrations have been associated with lipid response, and they are characterized by high interindividual variability (Straka et al., 2007).
Most common adverse effects associated with fenofibrate treatment are dose-related increases of serum levels of liver-specific enzymes (3–13%), as well as headache (3%), abdominal pain (5%), and respiratory disorders (6%) (Steinmetz et al., 1996). Although infrequent, rhabdomyolysis is the most threatening adverse reaction. Fibrates are also frequently prescribed together with hydroxymethylglutaryl-coenzyme A reductase inhibitors (statins) to treat patients with mixed hyperlipidemia, but this combination has been reported to increase risk of myopathy, including rhabdomyolysis (Murdock et al., 1999). Fenofibrate and some statins are known to share common drug-metabolizing pathways. However, the underlying mechanisms and factors involved in such an adverse effect remain unclear.
Potential causes of variability in response to fenofibrate therapy include genetic variations in drug-metabolizing pathways and transport. FA is known to be exclusively metabolized by the phase II enzymes UDP-glucuronosyltransferases (UGTs) (Prueksaritanont et al., 2002), and no other phase II or cytochrome P450 drug-metabolizing enzymes have been implicated (Prueksaritanont et al., 2005). UGTs are responsible for clearance of various endobiotics, and this pathway is also critical for numerous drugs. Among known human UGTs, only six were previously evaluated for FA glucuronidation, of which UGT1A1, UGT1A3, UGT1A9, and UGT2B7 were able to glucuronidate FA (Prueksaritanont et al., 2002). However, the influence of common genetic variants on FA glucuronidation has yet to be addressed.
The aims of this study were 1) to perform a systematic study to identify human UGTs involved in FA glucuronidation using in vitro recombinant human UGTs and human liver microsome (HLM) samples, and 2) to explore the influence of common heterologous UGT variant allozymes and perform correlative studies concerning FA-glucuronide (FA-G) formation by HLMs. A specific and sensitive high-performance liquid chromatography (HPLC) coupled with tandem mass spectrometry (MS/MS) method was developed to achieve these goals.
Materials and Methods
Chemicals and Reagents.
FA was obtained from EDQM (Strasbourg, France). Preparation of FA-G standard is described below. All the other chemicals and reagents were of the highest grade commercially available.
Genomic DNA and Liver Samples.
Human genomic DNA and liver samples were obtained as described previously (Hesse et al., 2004); approvals from human research institutional review boards were obtained. Available subject information included gender, age, race, ethnicity, and histories of smoking and alcohol use. The enzymatic quality of liver samples was confirmed with at least 10 other measures of glucuronidation activity with this sample set as previously stated in other studies (Hesse et al., 2004; Girard et al., 2006). UGT2B7 protein content measurements have been reported previously (Court et al., 2003).
Genotyping.
UGT1A1 −53TA6>7, −3156G>A, and −3279T>G genotypes for all 48 subjects were determined in a previous study (Girard et al., 2005), as for those of UGT1A9 exon 1 polymorphisms at codons 3 (Tyr3) and 33 (Thr33); intron polymorphisms at positions I 143C>T, I 152G>A, I 201A>C, I 219T>A, I 313A>C, and I 399C>T (relative to the end of UGT1A9 exon 1); and 13 promoter polymorphisms from −87 to −5366 (Girard et al., 2004, 2006). UGT1A3 genotypes (12 polymorphisms in the first exon and 7 promoter polymorphisms from −66 to −758 relative to the ATG) were also available (Caillier et al., 2007). Finally, UGT2B7 and UGT1 3′UTR variants 1813T>C, 1941G>C, and 2042G>C were assessed using the strategies described by Lévesque et al. (2008).
Human UGT1A and UGT2B Variant Allozymes.
Microsomal proteins were prepared as described previously (Villeneuve et al., 2003; Thibaudeau et al., 2006; Caillier et al., 2007). To ascertain the level of UGT protein content in the stably transfected UGT1A- and UGT2B-human embryonic kidney (HEK) 293 clones, a semiquantitative immunoblot analysis method was used as previously reported (Gagné et al., 2002; Villeneuve et al., 2003) and using the anti-human UGT1A common carboxyl terminus region (amino acids 312–531) antiserum RC-71 and the anti-human UGT2B antibody (EL-93) (Guillemette et al., 1997). To normalize for sample loading, blots were probed with anti-calnexin antibody (Stressgen Biotechnologies, Victoria, BC, Canada) to detect this endoplasmic reticulum-resident protein. Bands were visualized using enhanced chemiluminescence (GE Healthcare, Little Chalfont, Buckinghamshire, UK) and quantified by Bioimage Visage 110s from Genomic Solutions Inc. (Ann Arbor, MI). Ratios between UGT and calnexin signals were calculated for each UGT. Because of the use of two distinct unpurified antibodies, a direct comparison of expression levels between UGT1 and UGT2 could not be established. In addition, preparations of UGT1A and UGT2B were obtained from commercial sources to compare with UGT-HEK293 (BD Gentest, Woburn, MA; PanVera Corp., Madison, WI). The level of UGT protein expression was determined using the same semiquantitative immunoblot analysis method. In addition, based on a previous report indicating higher activity of UGT1A10 in homogenates compared with microsomes, UGT1A10 cell homogenates were also prepared by resuspending pelleted cells in phosphate buffer solution with 0.5 mM dithiothreitol and then stored at −80°C until use.
Glucuronidation Activity and Inhibition Assays.
To assess which UGT(s) might be involved in the formation of FA-G, initial experiments with UGT1A and UGT2B microsomes, including commercial preparations consisting of 4-h incubation at 37°C with 0.2 mM FA, 10 to 50 μg of UGT membrane protein, 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 2 mM UDP-glucuronic acid, 20 μg/ml alamethicin, 10 μg/ml phosphatidylcholine, 5 μg/ml pepstatin, and 0.5 μg/ml leupeptin in a standard procedure, were performed in our laboratory (Gagné et al., 2002). Assays with liver samples and UGT1A10 homogenates, as well as inhibition assays, were performed in the same conditions. Inhibition assays were performed with varying concentrations of fluconazole (0–4 mM) and fixed FA (125 μM) concentration; 3′-azido-2′,3′-dideoxythymidine (1.1 mM) was used as positive control of UGT2B7 activity. The assays were terminated by adding 100 μl of methanol + 0.02% formic acid and were centrifuged at 14,000g for 10 min before analysis. Before kinetics experiments with UGTs with a significant glucuronidation activity for FA, experiments were designed to assess the linearity of the glucuronidation reaction. Determination of kinetics parameters (Vmax and apparent Km) was then performed for UGT1A3, UGT1A6, UGT1A9, and UGT2B7, and microsomes were incubated 1 h in the presence of substrate concentrations ranging from 5 to 2000 μM. Kinetic parameters were also assessed for the following allelic variant proteins: UGT1A3.1, UGT1A3.2 (R11A47), UGT1A3.3 (Arg11), UGT1A9.1, UGT1A9.2 (Tyr3), UGT1A9.3 (Thr33), UGT2B7.1, and UGT2B7.2 (Tyr268). Absolute glucuronidation activities were adjusted by UGT protein relative levels and expressed as relative glucuronidation activities. UGT protein levels were determined by Western blotting following a previously published procedure (Gagné et al., 2002). Visualization and expression level quantification were performed as previously published (Villeneuve et al., 2003). Statistical evaluation of best-fit model was used to select the enzyme kinetic model and confirmed by a visual inspection of fitted functions (V as a function of [S]) and Eadie-Hofstee plots (V as a function of V/[S]). Kinetic parameter calculations were performed with Sigma Plot 8.0 software assisted by Enzyme Kinetics 1.1 software (SPSS Inc., Chicago, IL). Values are expressed as the mean of at least two experiments performed in triplicate.
Mass Spectrometry Analysis.
Detection of FA-G was performed by HPLC coupled with MS/MS. The analysis system consisted of an HPLC module (Agilent 1200 series; Agilent Technologies, Ville St-Laurent, QC, Canada) and a triple-quadrupole mass spectrometer (API 3200; MDS Sciex, Concord, ON, Canada). Ten-microliter samples, maintained at 4°C, were injected on a 100 × 4.6-mm (4.0-μm diameter) Synergic RP-Hydro C-18 reversed-phase column (Phenomenex, Torrance, CA). The mobile phase consisted of solution A (H2O + 2% formic acid) and solution B (MeOH + 2% formic acid) using the following gradient: 30% A (0–2 min), 20% A (2–3.1 min), 5% A (3.1–4.1 min), and 30% A (4.1–7 min). The flow rate was 0.9 ml/min. MS detection of FA-G was followed in the multiple reactions monitoring, in negative ion mode with mass fragmentation of 492.9 → 230.8 (FA-G) and 498.9 → 230.8 (d6 FA-G used as internal standard). Intra- and interday precisions and accuracy validation assays were performed in the glucuronidation assay buffer spiked with three concentrations of FA-G (3, 500, and 800 ng/ml). The intraday assay was performed by analyzing three aliquots of each concentration, and interday data were realized by repeating the analysis on three different days.
Preparation of FA-G Standard.
FA-G was obtained from enzymatic assays using liver microsomes. In brief, media from enzymatic assays were diluted with 1% formic acid and loaded on Strata X cartridges (60 mg; Phenomenex) preconditioned with methanol followed by 1% formic acid. After loading the sample, the cartridge was washed with ultrapure water and chloroform to remove the unconjugated FA. FA-G was eluted with methanol, evaporated under nitrogen, and diluted in methanol and 0.02% formic acid, and the purity of the compound was confirmed by HPLC/MS. An aliquot was treated with β-glucuronidase, and the residue was quantified with a calibration curve of FA. The concentration of FA obtained from the aliquot digested by β-glucuronidase was then converted into a concentration of FA-G.
Statistical Analyses.
All the statistical analyses were done using JMP 4.0.2 program (SAS Institute, Cary, NC). Correlation analyses between activity values were done using Spearman's r. The r values greater than 0.50 and p values less than 0.05 were considered significant. One-way analysis of variance and a comparison for each pair using Student's t test were used to determine the relationship between genotypes and glucuronosyltransferase activities. Normal distribution of expression and activity values was assessed with the Shapiro-Wilk W test. Raw data that were not normally distributed were transformed with a logarithm function to achieve a normal distribution. For all the analyses, a p value less than 0.05 was considered significant.
Results
Detection of FA-G.
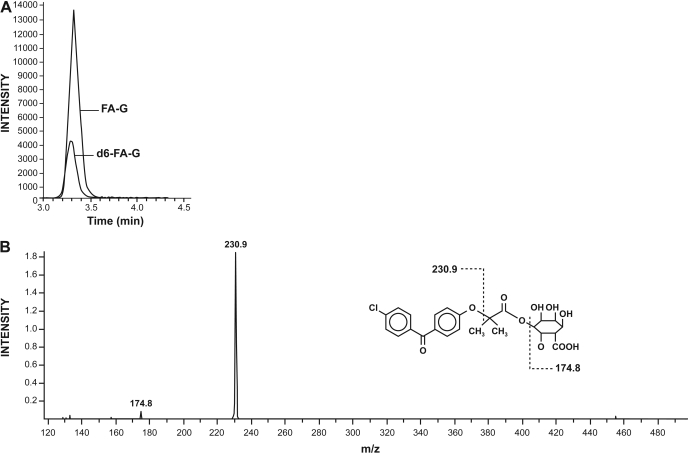
A representative chromatogram and MS/MS spectra of FA-G are presented in Fig. 1. Retention time of FA-G was 3.32 min. The limit of quantification was 1 ng/ml using a signal-to-noise ratio of 10:1. The inter- and intraday precision (CV%) and accuracy (bias%) for the measurement of FA-G are presented. Signals were found to be linear from 1 to 1000 ng/ml for FA-G. All of the precision values (CV) were less than 10%, whereas accuracy was included in a range of 10% (Table 1).
Fig. 1.
Detection of FA-G by HPLC coupled with MS/MS. A, representative chromatogram of FA-G from an incubated sample. Retention time of both the FA-G and internal standard (d6 FA-G) was 3.32 min. B, MS/MS scan confirms the glucuronide nature of the observed FA-G peak.
TABLE 1.
LC/MS/MS-based assay validation results for FA-G quantification
| FA-G Working Solution | Intra-assay |
Interassay |
||
|---|---|---|---|---|
| Accuracy (Bias) | Precision (CV) | Accuracy (Bias) | Precision (CV) | |
| ng/ml | % | % | % | % |
| 3 | 4.4 | 5.3 | 3.3 | 6.2 |
| 500 | 2.6 | 2.1 | 3.3 | 1.7 |
| 800 | 6.0 | 5.2 | 5.9 | 3.7 |
In Vitro Glucuronidation Assays with Recombinant Human UGT Enzymes.
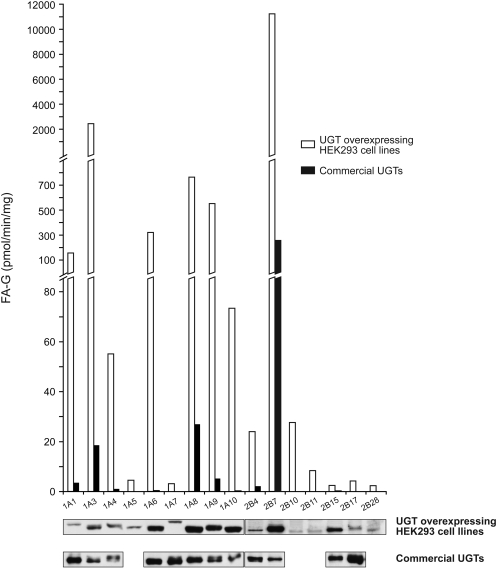
Microsomes from recombinant UGT expressed in HEK293 cells were initially incubated with 200 μM FA for 4 h to detect formation of FA-G. UGT1A3, UGT1A6, UGT1A9, and UGT2B7 generated a significant amount of FA-G (>200 pmol/min/mg protein/UGT content), whereas other liver-expressed UGTs showed lower or undetectable activity (Fig. 2). From UGTs not expressed in liver, UGT1A8 was the most reactive (∼600 pmol/min/mg/UGT). UGT1A10 homogenates were also tested based on recent observations of higher activity for 4-OH-tamoxifen, endoxifen, 2-amino-1-methyl-6-phenylimidazo[4,5-f]pyridine, and N-hydroxy-2-amino-1-methyl-6-phenylimidazo[4,5-f]pyridine compared with microsomes (Dellinger et al., 2007; Sun et al., 2007). However, for UGT1A10, FA-G formation was 1.6-fold higher in microsomes than the activity observed with homogenates (data not shown). Our results were confirmed concomitantly performing assays with commercial preparations of UGTs (Supersomes), although they were less active than HEK293 recombinant UGTs in the assay conditions used (Fig. 2).
Fig. 2.
FA glucuronidation by recombinant UGT expressed in HEK293 cells compared with commercial UGT Supersomes. The screening experiment with UGT1A and UGT2B microsomes included UGT-expressing HEK293 cells and commercial preparations and consisted of 4-h incubation at 37°C. Western blot of UGTs (UGT-overexpressing HEK293 cell lines and UGT Supersomes) is shown below. (UGT1A1 microsomes, UGT1A10 Supersomes, and UGT2B7 microsomes and Supersomes were arbitrarily set to 1.0, respectively, for UGT1A and UGT2B family members.) UGT2A family members were not tested.
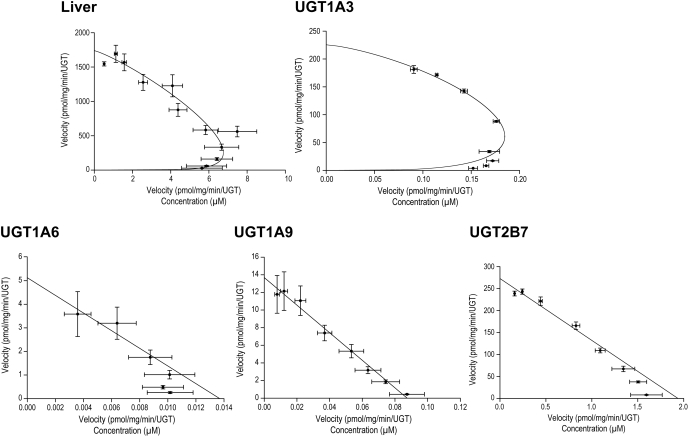
For subsequent studies, kinetic analyses were performed only for the most reactive UGTs expressed in the liver, namely, UGT1A3, UGT1A6, UGT1A9, and UGT2B7 (Tables 2 and 3). A first observation is that the kinetic profiles diverge from one enzyme to another. UGT1A6, UGT1A9, and UGT2B7 displayed a Michaelis-Menten profile, whereas UGT1A3 displayed a sigmoid profile (Hill's) (Fig. 3). Kinetics for HLM reveals a Hill's profile with an S50 of 295 ± 68 μm and a Vmax of 1.8 ± 0.2 nmol/min/mg. Of these four enzymes, UGT2B7, UGT1A9, and UGT1A6 could be categorized as high-affinity enzymes with apparent Km values ranging from 131 to 222 μM, whereas UGT1A3 exhibited the lowest affinity for FA (Km of 724 μM). UGT2B7 showed the highest catalytic efficiency for FA-G formation, at least 16-fold higher than the other active UGTs, with a Vmax/Km value of 2.1 μl/min/mg. UGT1A3, UGT1A9, and UGT1A6 had Vmax/Km values of 0.13, 0.09, and 0.02 μl/min/mg, respectively.
TABLE 2.
Kinetic parameters for the glucuronidation of FA by human UGT2B7, UGT1A6, and UGT1A9 enzymes
Results are expressed as mean ± S.D. of two independent experiments performed in triplicate. Observed Vmax values were adjusted to relative protein expression; absolute values were 243 ± 3 for UGT2B7.1, 4.9 ± 4 for UGT1A6.1, and 16.2 ± 9 for UGT1A9.1.
| Allozymes | Apparent Km | Observed Vmax | -Fold versus *1 | Vmax/Km |
|---|---|---|---|---|
| μM | pmol/min/mg | μl/min/mg | ||
| UGT2B7 | ||||
| *1 | 126 ± 51 | 243 ± 3 | 2.1 ± 0.8 | |
| *2 (Tyr268) | 131 ± 76 | 136 ± 21 | 1.2 ± 0.5 | |
| UGT1A6 | ||||
| *1 | 222 ± 155 | 3.7 ± 3 | 0.02 ± 0.001 | |
| UGT1A9 | ||||
| *1 | 131 ± 45 | 12.0 ± 7 | 0.09 ± 0.02 | |
| *2 (Tyr3) | 121 ± 2 | 9.3 ± 1 | 0.08 ± 0.007 | |
| *3 (Thr33) | 271 ± 36 | 1.1 ± 0.07 | ↓ 10.9* | 0.004 ± 0.0002 |
p value <0.05.
TABLE 3.
Kinetic parameters for the glucuronidation of FA by human UGT1A3 enzyme
Results are expressed as mean ± S.D. of two independent experiments performed in triplicate. Observed Vmax values were adjusted to relative protein expression; absolute values were 244 ± 23 for UGT1A3.1.
| Allozymes | S50 | Observed Vmax | -Fold versus *1 | n | Clmax |
|---|---|---|---|---|---|
| μM | pmol/min/mg | μl/min/mg | |||
| UGT1A3 | |||||
| *1 | 724 ± 239 | 184 ± 18 | 1.35 ± 0.21 | 0.13 ± 0.03 | |
| *2 (R11A47) | 609 ± 43 | 62 ± 8 | ↓ 3.0* | 1.30 ± 0.28 | 0.05 ± 0.01 |
| *3 (Trp11) | 700 ± 113 | 44 ± 3 | ↓ 4.2* | 1.25 ± 0.07 | 0.030 ± 0.003 |
p value <0.05.
Fig. 3.
Eadie-Hofstee plots of recombinant UGT. Whereas UGT1A6, UGT1A9, and UGT2B7 displayed a Michaelis-Menten profile, UGT1A3 was characterized by a sigmoid profile (Hill's), as for liver microsomes.
Glucuronidation by Recombinant Human UGT Variant Allozymes.
Variant UGT allozymes with common nonsynonymous polymorphisms were investigated in their ability to catalyze the formation of FA-G (Tables 2 and 3). UGT2B7.1 (His268 or *1 allele) and UGT2B7.2 (Tyr268 or *2 allele) allozymes have similar apparent Km values and no statistically different velocities, although a lower Vmax value is observed for *2 (243 pmol/min/mg for *1 versus 136 pmol/min/mg for *2). Compared with UGT1A9.1, rates of FA-G formation were unaffected for UGT1A9.2 protein, with a tyrosine residue at codon 3, but were significantly reduced for UGT1A9.3 (Thr33) allozyme by 11-fold (1.1 versus 12 pmol/min/mg for UGT1A9.1; p * 0.0045). Consequently, the Vmax/Km value of UGT1A9.3 (0.004 μl/min/mg) allele was 23-fold lower than that of the UGT1A9.1 protein (0.09 μl/min/mg) (p * 0.0049). The two of the most frequent UGT1A3 variants were also tested: UGT1A3.2 (R11A47; frequency of 36% in whites) and UGT1A3.3 (Arg11; frequency of 6% in whites) (Caillier et al., 2007; Menard et al., 2009). A substantial decrease in the formation of FA-G was observed for both variants UGT1A3.2 and UGT1A3.3 compared with the reference UGT1A3.1 protein (by 61 and 76%, respectively; p * 0.005), mainly explained by decreased velocities.
FA-G Formation in a Liver Bank and Its Relationship to Probe Substrates, UGT Expression, and Common Genetic Variations.
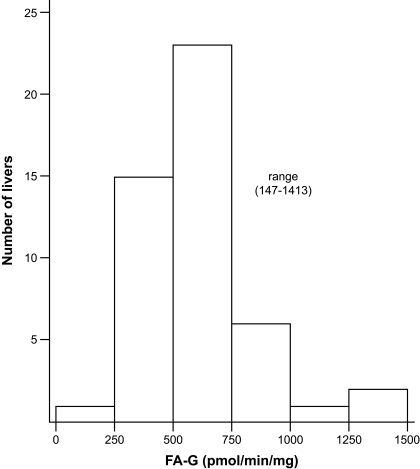
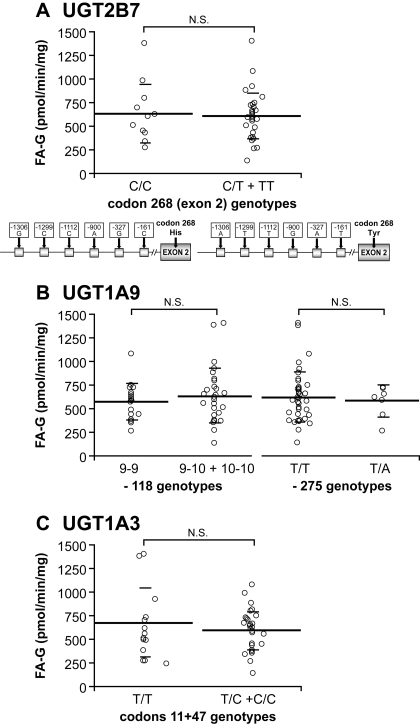
UGT activities for FA were measured, and those for common UGT marker substrates (zidovudine, mycophenolic acid, fulvestrant, and serotonin) were available for a set of HLMs (n * 48) (Court et al., 2003; Girard et al., 2004; Krishnaswamy et al., 2004). Rates of formation of FA-G ranged from 147 to 1413 pmol/min/mg and varied 10-fold (Fig. 4). Correlative studies with probe substrates revealed a significant correlation between formation of FA-G and glucuronidation of zidovudine (UGT2B7; r2 * 0.75), mycophenolic acid (UGT1A9; r2 * 0.42), and fulvestrant (UGT1A3; r2 * 0.36) (p ≤ 0.01), supporting the contribution of these UGTs in the hepatic glucuronidation of FA (Table 4). Inhibition assays with fluconazole, a known UGT2B7 selective inhibitor (Uchaipichat et al., 2006), indicate an IC50 of 36.4 μm on FA glucuronidation by liver microsomes and a maximum of 60% inhibition of FA glucuronidation at the highest concentration of fluconazole. FA-G formation was highly correlated with UGT2B7 protein content (r2 * 0.736; p < 0.0001). No significant relationship between the presence of genetic variations in the most reactive UGTs and FA-G formation was evidenced in the human liver bank (Fig. 5).
Fig. 4.
FA glucuronidation by microsomes from 48 human liver samples.
TABLE 4.
Correlation between formation of FA-G and 3′-azido-2′,3′-dideoxythymidine (AZT), MPA, fulvestrant, and serotonin
| Substrate | Hepatic UGT | r2 | p |
|---|---|---|---|
| AZT | 2B7 | 0.751 | <0.0001 |
| MPA | 1A9 | 0.422 | 0.003 |
| Fulvestrant | 1A3 | 0.357 | 0.013 |
| Serotonin | 1A6 | 0.055 | 0.712 |
Fig. 5.
Analyses of rates of FA-G formation by UGT genotypes. Genetic variants in UGT2B7 (A), UGT1A9 (B), and UGT1A3 (C; carriers of codon 11 and/or codon 47 variations were grouped together) were not associated with significant differences in FA glucuronidation. N.S., not significant.
Discussion
Fenofibrate is approved for triglyceride-lowering in subjects with types IV and V hyperlipoproteinemia. Although fibrates have been associated with muscle toxicity (Magarian et al., 1991), an effect that is more pronounced in patients also treated with a statin (Pierce et al., 1990; Miller and Spence, 1998), fenofibrate remains the preferred fibrate in patients who require combined therapy (Rosenson, 2004). The extended use of fenofibrate emphasizes the importance of understanding its metabolism to explain and eventually prevent some drug-drug interactions and adverse reactions. Our results suggest that FA glucuronidation is mediated by multiple hepatic UGTs. Although UGT2B7 has a predominant role, UGT1A9 and UGT1A3 also contribute to FA-G formation (Fig. 6). The contribution of specific genetic polymorphisms in UGT2B7, UGT1A9, or UGT1A3 to variability in glucuronidation of FA by a human liver bank could not be confirmed.
Fig. 6.
Putative in vivo metabolism of FA. Based on the present in vitro studies, UGT2B7 appears as the primary hepatic enzyme responsible for FA glucuronidation along with two other hepatic UGTs, UGT1A9 (high-affinity enzyme) and UGT1A3 (*low-affinity enzyme).
Our observations, indicating that whereas UGT2B7 and UGT1A9 showed a high affinity for FA, UGT1A3 and UGT1A6 represent low-affinity enzymes for this substrate, are in agreement with a previous study concerning FA glucuronidation. The group of Prueksaritanont et al. (2002) tested six UGTs, and their data support a role for UGT2B7, UGT1A9, and UGT1A3 in FA glucuronidation. A more systematic characterization of enzyme kinetics presented here corroborated these results, including assays with commercial preparations of UGT enzymes. In contrast to a previous study that showed recombinant UGT1A10 homogenates have greater activity compared with UGT1A10 microsomes for a variety of substrates, including tamoxifen (Dellinger et al., 2007; Sun et al., 2007), we observed that microsomes are more active for fenofibrate than homogenates, suggesting that such differences could be substrate-specific. A higher activity with microsomes compared with Supersomes was also observed, as noted in other studies (Benoit-Biancamano et al., 2009). Thus, a primary role of UGT2B7 followed by UGT1A9 and possibly UGT1A3 in the hepatic clearance of FA is predicted and further supported by correlative studies using liver microsomes and UGT probe substrates. The substantial inhibition of hepatic glucuronidation with the UGT2B7 selective inhibitor fluconazole also supports a major contribution of UGT2B7 in the liver glucuronidation of FA. The role of UGT1A3 could also be relevant for FA glucuronidation in liver because both HLMs and UGT1A3 show sigmoidal kinetics, and the S50 value for UGT1A3 is only approximately 2-fold higher than the value for HLMs. Furthermore, UGT1A3 mRNA expression in the liver was reported to be higher than UGT1A6 and UGT1A9 mRNA, both having similar level of mRNA expression (Zhang et al., 2007). It is possible that the lower affinity of UGT1A3 for FA may be partially compensated for by its significant expression in the liver, which could also explain the kinetic profile similar to UGT1A3 observed for HLMs. UGT2B7 mRNA expression was also shown to be higher than that of UGT1A6 (Somers et al., 2007), suggesting it may also play an important role in FA metabolism.
High variability in the formation of FA-G was observed in the bank of 48 HLMs. Genetic polymorphisms that affect the metabolic activity or expression of UGT biotransformation enzymes may be important contributors to interindividual differences in FA pharmacokinetics. In line with this hypothesis, we explored the relationship between known polymorphisms of UGT2B7, UGT1A9, and UGT1A3 and the hepatic formation of FA-G using a bank of human liver microsomal fractions. The UGT2B7*2 (Tyr268) allele carried by 54% of whites (Lampe et al., 2000) was first studied. Previously reported data suggest a possible substrate-specific interaction for the mutation at codon 268. For example, a decreased activity for UGT2B7.2 (Tyr268) compared with the UGT2B7.1 (His268) allozyme for lithocholic acid, hyodeoxycholic acid, estradiol, and zidovudine (Gall et al., 1999; Barbier et al., 2000) increased activity for UGT2B7.2 compared with the UGT2B7.1 allozyme for 4-hydroxy-catecholestrogen (4-OHCE) (Thibaudeau et al., 2006) and showed similar activity for the two allozymes for 2-OHCE, 4-OHCE, androsterone, menthol, opioids, propranolol, and epirubicin (Cheng et al., 1998; Coffman et al., 1998; Bhasker et al., 2000; Innocenti et al., 2001). On the other hand, the UGT2B7*2 variant has no significant influence on the conjugation of mycophenolic acid (MPA), morphine, valproic acid (Court et al., 2003; Bernard et al., 2006; Chung et al., 2008), and FA, as described herein. However, the coding variation is in linkage disequilibrium with a series of promoter variations that were associated with a 2-fold reduced transcriptional activity compared with the reference allele (Duguay et al., 2004), thus leaving open the possibility of interactions that may be observed under other conditions. There was no evidence of a relationship between UGT2B7 variations and rates of FA-G formation in the tested liver samples. In vitro correlation studies with rates of formation of FA-G were evidenced on stratification by UGT1A3, UGT1A6, or UGT1A9 genetic status for several known regulatory and coding functional variations. However, given the limited number of human liver bank samples, the contribution of genetic factors to the variability of FA glucuronidation cannot be ruled out based on our study.
Based on this study, we conclude that glucuronidation of FA is mainly mediated by a few UGT enzymes, primarily UGT2B7, but also by UGT1A9, which is a high-affinity and a low-capacity enzyme, and by UGT1A3, which is a low-affinity high-capacity enzyme. Additional studies either with in vivo experiments or in vitro studies with a sufficiently large number of subjects are required to determine whether the genetic status for the genes encoding these UGTs influence the metabolism of FA.
Acknowledgments.
We thank Lyne Villeneuve for technical assistance in the realization of this work and Mario Harvey for critical reading. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This work was supported in part by the National Institutes of Health National Institute of General Medical Sciences [Grant GM-061834]; and the Canadian Institutes of Health Research [Grant MOP-42392]. J.T. was the recipient of a studentship award from the Fonds de la Recherche en Santé du Québec. M.-O.B.-B. was supported by a Canada Graduate Scholarship Doctoral Research Award from the Canadian Institutes for Health Research and by the Canadian Federation of University Women (CFUW) Dr. Marion Elder Grant Fellowship, funded by CFUW Wolfville.
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.109.029058
- FA
- fenofibric acid
- UGT
- UDP-glucuronosyltransferase
- HLM
- human liver microsome
- FA-G
- fenofibric acid-glucuronide
- HPLC
- high-performance liquid chromatography
- MS/MS
- tandem mass spectrometry
- HEK
- human embryonic kidney
- OHCE
- hydroxy-catecholestrogen
- CV
- coefficient of variation
- MPA
- mycophenolic acid.
References
- Barbier O, Turgeon D, Girard C, Green MD, Tephly TR, Hum DW, Bélanger A. (2000) 3′-Azido-3′-deoxythimidine (AZT) is glucuronidated by human UDP-glucuronosyltransferase 2B7 (UGT2B7). Drug Metab Dispos 28: 497–502 [PubMed] [Google Scholar]
- Benoit-Biancamano MO, Connelly J, Villeneuve L, Caron P, Guillemette C. (2009) Deferiprone glucuronidation by human tissues and recombinant UDP glucuronosyltransferase 1A6: an in vitro investigation of genetic and splice variants. Drug Metab Dispos 37: 322–329 [DOI] [PubMed] [Google Scholar]
- Bernard O, Tojcic J, Journault K, Perusse L, Guillemette C. (2006) Influence of nonsynonymous polymorphisms of UGT1A8 and UGT2B7 metabolizing enzymes on the formation of phenolic and acyl glucuronides of mycophenolic acid. Drug Metab Dispos 34: 1539–1545 [DOI] [PubMed] [Google Scholar]
- Bhasker CR, McKinnon W, Stone A, Lo AC, Kubota T, Ishizaki T, Miners JO. (2000) Genetic polymorphism of UDP-glucuronosyltransferase 2B7 (UGT2B7) at amino acid 268: ethnic diversity of alleles and potential clinical significance. Pharmacogenetics 10: 679–685 [DOI] [PubMed] [Google Scholar]
- Caillier B, Lépine J, Tojcic J, Ménard V, Perusse L, Bélanger A, Barbier O, Guillemette C. (2007) A pharmacogenomics study of the human estrogen glucuronosyltransferase UGT1A3. Pharmacogenet Genomics 17: 481–495 [DOI] [PubMed] [Google Scholar]
- Cheng Z, Rios GR, King CD, Coffman BL, Green MD, Mojarrabi B, Mackenzie PI, Tephly TR. (1998) Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7. Toxicol Sci 45: 52–57 [DOI] [PubMed] [Google Scholar]
- Chung JY, Cho JY, Yu KS, Kim JR, Lim KS, Sohn DR, Shin SG, Jang IJ. (2008) Pharmacokinetic and pharmacodynamic interaction of lorazepam and valproic acid in relation to UGT2B7 genetic polymorphism in healthy subjects. Clin Pharmacol Ther 83: 595–600 [DOI] [PubMed] [Google Scholar]
- Coffman BL, King CD, Rios GR, Tephly TR. (1998) The glucuronidation of opioids, other xenobiotics, and androgens by human UGT2B7Y(268) and UGT2B7H(268). Drug Metab Dispos 26: 73–77 [PubMed] [Google Scholar]
- Court MH, Krishnaswamy S, Hao Q, Duan SX, Patten CJ, Von Moltke LL, Greenblatt DJ. (2003) Evaluation of 3′-azido-3′-deoxythymidine, morphine, and codeine as probe substrates for UDP-glucuronosyltransferase 2B7 (UGT2B7) in human liver microsomes: specificity and influence of the UGT2B7*2 polymorphism. Drug Metab Dispos 31: 1125–1133 [DOI] [PubMed] [Google Scholar]
- Dellinger RW, Chen G, Blevins-Primeau AS, Krzeminski J, Amin S, Lazarus P. (2007) Glucuronidation of PhIP and N-OH-PhIP by UDP-glucuronosyltransferase 1A10. Carcinogenesis 28: 2412–2418 [DOI] [PubMed] [Google Scholar]
- Duguay Y, Báár C, Skorpen F, Guillemette C. (2004) A novel functional polymorphism in the uridine diphosphate-glucuronosyltransferase 2B7 promoter with significant impact on promoter activity. Clin Pharmacol Ther 75: 223–233 [DOI] [PubMed] [Google Scholar]
- Gagné JF, Montminy V, Belanger P, Journault K, Gaucher G, Guillemette C. (2002) Common human UGT1A polymorphisms and the altered metabolism of irinotecan active metabolite 7-ethyl-10-hydroxycamptothecin (SN-38). Mol Pharmacol 62: 608–617 [DOI] [PubMed] [Google Scholar]
- Gall WE, Zawada G, Mojarrabi B, Tephly TR, Green MD, Coffman BL, Mackenzie PI, Radominska-Pandya A. (1999) Differential glucuronidation of bile acids, androgens and estrogens by human UGT1A3 and 2B7. J Steroid Biochem Mol Biol 70: 101–108 [DOI] [PubMed] [Google Scholar]
- Girard H, Court MH, Bernard O, Fortier LC, Villeneuve L, Hao Q, Greenblatt DJ, von Moltke LL, Perussed L, Guillemette C. (2004) Identification of common polymorphisms in the promoter of the UGT1A9 gene: evidence that UGT1A9 protein and activity levels are strongly genetically controlled in the liver. Pharmacogenetics 14: 501–515 [DOI] [PubMed] [Google Scholar]
- Girard H, Thibaudeau J, Court MH, Fortier LC, Villeneuve L, Caron P, Hao Q, von Moltke LL, Greenblatt DJ, Guillemette C. (2005) UGT1A1 polymorphisms are important determinants of dietary carcinogen detoxification in the liver. Hepatology 42: 448–457 [DOI] [PubMed] [Google Scholar]
- Girard H, Villeneuve L, Court MH, Fortier LC, Caron P, Hao Q, von Moltke LL, Greenblatt DJ, Guillemette C. (2006) The novel UGT1A9 intronic I399 polymorphism appears as a predictor of 7-ethyl-10-hydroxycamptothecin glucuronidation levels in the liver. Drug Metab Dispos 34: 1220–1228 [DOI] [PubMed] [Google Scholar]
- Guay DR. (2002) Update on fenofibrate. Cardiovasc Drug Rev 20: 281–302 [DOI] [PubMed] [Google Scholar]
- Guillemette C, Lévesque E, Beaulieu M, Turgeon D, Hum DW, Bélanger A. (1997) Differential regulation of two uridine diphospho-glucuronosyltransferases, UGT2B15 and UGT2B17, in human prostate LNCaP cells. Endocrinology 138: 2998–3005 [DOI] [PubMed] [Google Scholar]
- Hesse LM, He P, Krishnaswamy S, Hao Q, Hogan K, von Moltke LL, Greenblatt DJ, Court MH. (2004) Pharmacogenetic determinants of interindividual variability in bupropion hydroxylation by cytochrome P450 2B6 in human liver microsomes. Pharmacogenetics 14: 225–238 [DOI] [PubMed] [Google Scholar]
- Innocenti F, Iyer L, Ramírez J, Green MD, Ratain MJ. (2001) Epirubicin glucuronidation is catalyzed by human UDP-glucuronosyltransferase 2B7. Drug Metab Dispos 29: 686–692 [PubMed] [Google Scholar]
- Krishnaswamy S, Hao Q, Von Moltke LL, Greenblatt DJ, Court MH. (2004) Evaluation of 5-hydroxytryptophol and other endogenous serotonin (5-hydroxytryptamine) analogs as substrates for UDP-glucuronosyltransferase 1A6. Drug Metab Dispos 32: 862–869 [DOI] [PubMed] [Google Scholar]
- Lampe JW, Bigler J, Bush AC, Potter JD. (2000) Prevalence of polymorphisms in the human UDP-glucuronosyltransferase 2B family: UGT2B4(D458E), UGT2B7(H268Y), and UGT2B15(D85Y). Cancer Epidemiol Biomarkers Prev 9: 329–333 [PubMed] [Google Scholar]
- Lévesque E, Benoit-Biancamano MO, Delage R, Couture F, Guillemette C. (2008) Pharmacokinetics of mycophenolate mofetil and its glucuronide metabolites in healthy volunteers. Pharmacogenomics 9: 869–879 [DOI] [PubMed] [Google Scholar]
- Magarian GJ, Lucas LM, Colley C. (1991) Gemfibrozil-induced myopathy. Arch Intern Med 151: 1873–1874 [PubMed] [Google Scholar]
- Ménard V, Girard H, Harvey M, Pérusse L, Guillemette C. (2009) Analysis of inherited genetic variations at the UGT1 locus in the French-Canadian population. Hum Mutat 30: 677–687 [DOI] [PubMed] [Google Scholar]
- Miller DB, Spence JD. (1998) Clinical pharmacokinetics of fibric acid derivatives (fibrates). Clin Pharmacokinet 34: 155–162 [DOI] [PubMed] [Google Scholar]
- Murdock DK, Murdock AK, Murdock RW, Olson KJ, Frane AM, Kersten ME, Joyce DM, Gantner SE. (1999) Long-term safety and efficacy of combination gemfibrozil and HMG-CoA reductase inhibitors for the treatment of mixed lipid disorders. Am Heart J 138: 151–155 [DOI] [PubMed] [Google Scholar]
- Pierce LR, Wysowski DK, Gross TP. (1990) Myopathy and rhabdomyolysis associated with lovastatin-gemfibrozil combination therapy. JAMA 264: 71–75 [PubMed] [Google Scholar]
- Prueksaritanont T, Richards KM, Qiu Y, Strong-Basalyga K, Miller A, Li C, Eisenhandler R, Carlini EJ. (2005) Comparative effects of fibrates on drug metabolizing enzymes in human hepatocytes. Pharm Res 22: 71–78 [DOI] [PubMed] [Google Scholar]
- Prueksaritanont T, Tang C, Qiu Y, Mu L, Subramanian R, Lin JH. (2002) Effects of fibrates on metabolism of statins in human hepatocytes. Drug Metab Dispos 30: 1280–1287 [DOI] [PubMed] [Google Scholar]
- Rosenson RS. (2004) Current overview of statin-induced myopathy. Am J Med 116: 408–416 [DOI] [PubMed] [Google Scholar]
- Somers GI, Lindsay N, Lowdon BM, Jones AE, Freathy C, Ho S, Woodrooffe AJ, Bayliss MK, Manchee GR. (2007) A comparison of the expression and metabolizing activities of phase I and II enzymes in freshly isolated human lung parenchymal cells and cryopreserved human hepatocytes. Drug Metab Dispos 35: 1797–1805 [DOI] [PubMed] [Google Scholar]
- Staels B, Dallongeville J, Auwerx J, Schoonjans K, Leitersdorf E, Fruchart JC. (1998) Mechanism of action of fibrates on lipid and lipoprotein metabolism. Circulation 98: 2088–2093 [DOI] [PubMed] [Google Scholar]
- Steinmetz A, Schwartz T, Hehnke U, Kaffarnik H. (1996) Multicenter comparison of micronized fenofibrate and simvastatin in patients with primary type IIA or IIB hyperlipoproteinemia. J Cardiovasc Pharmacol 27: 563–570 [DOI] [PubMed] [Google Scholar]
- Straka RJ, Burkhardt RT, Fisher JE. (2007) Determination of fenofibric acid concentrations by HPLC after anion exchange solid-phase extraction from human serum. Ther Drug Monit 29: 197–202 [DOI] [PubMed] [Google Scholar]
- Sun D, Sharma AK, Dellinger RW, Blevins-Primeau AS, Balliet RM, Chen G, Boyiri T, Amin S, Lazarus P. (2007) Glucuronidation of active tamoxifen metabolites by the human UDP glucuronosyltransferases. Drug Metab Dispos 35: 2006–2014 [DOI] [PubMed] [Google Scholar]
- Thibaudeau J, Lépine J, Tojcic J, Duguay Y, Pelletier G, Plante M, Brisson J, Têtu B, Jacob S, Perusse L, et al. ( 2006) Characterization of common UGT1A8, UGT1A9, and UGT2B7 variants with different capacities to inactivate mutagenic 4-hydroxylated metabolites of estradiol and estrone. Cancer Res 66: 125–133 [DOI] [PubMed] [Google Scholar]
- Uchaipichat V, Winner LK, Mackenzie PI, Elliot DJ, Williams JA, Miners JO. (2006) Quantitative prediction of in vivo inhibitory interactions involving glucuronidated drugs from in vitro data: the effect of fluconazole on zidovudine glucuronidation. Br J Clin Pharmacol 61: 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve L, Girard H, Fortier LC, Gagné JF, Guillemette C. (2003) Novel functional polymorphisms in the UGT1A7 and UGT1A9 glucuronidating enzymes in Caucasian and African-American subjects and their impact on the metabolism of 7-ethyl-10-hydroxycamptothecin and flavopiridol anticancer drugs. J Pharmacol Exp Ther 307: 117–128 [DOI] [PubMed] [Google Scholar]
- Zhang W, Liu W, Innocenti F, Ratain MJ. (2007) Searching for tissue-specific expression pattern-linked nucleotides of UGT1A isoforms. PLoS ONE 2: e396. [DOI] [PMC free article] [PubMed] [Google Scholar]