Abstract
To identify the physiological functions of the retinoid-related orphan receptor γ (RORγ), a member of the nuclear receptor superfamily, mice deficient in RORγ function were generated by targeted disruption. RORγ−/− mice lack peripheral and mesenteric lymph nodes and Peyer's patches, indicating that RORγ expression is indispensable for lymph node organogenesis. Although the spleen is enlarged, its architecture is normal. The number of peripheral blood CD3+ and CD4+ lymphocytes is reduced 6- and 10-fold, respectively, whereas the number of circulating B cells is normal. The thymus of RORγ−/− mice contains 74.4% ± 8.9% fewer thymocytes than that of wild-type mice. Flow cytometric analysis showed a decrease in the CD4+CD8+ subpopulation. Terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) staining demonstrated a 4-fold increase in apoptotic cells in the cortex of the thymus of RORγ−/− mice. The latter was supported by the observed increase in annexin V-positive cells. RORγ−/− thymocytes placed in culture exhibit a dramatic increase in the rate of “spontaneous” apoptosis. This increase is largely associated with CD4+CD8+ thymocytes and may, at least in part, be related to the greatly reduced level of expression of the anti-apoptotic gene Bcl-XL. Flow cytometric analysis demonstrated a 6-fold rise in the percentage of cells in the S phase of the cell cycle among thymocytes from RORγ−/− mice. Our observations indicate that RORγ is essential for lymphoid organogenesis and plays an important regulatory role in thymopoiesis. Our findings support a model in which RORγ negatively controls apoptosis in thymocytes.
The nuclear hormone receptor superfamily consists of structurally related, ligand-dependent transcription factors and a large number of orphan receptors for which the ligand has not yet been identified (1, 2). Nuclear receptors share a common modular structure composed of several domains that have functions in DNA binding, ligand binding, nuclear localization, dimerization, repression, and transactivation (3). Typically, the transactivation activity of nuclear receptors is controlled by small lipophilic molecules that bind to the receptor, thereby causing a conformational change in the receptor. This change in conformation causes dissociation of corepressor complexes and promotes interaction of the receptor with coactivators (4). The latter leads then to increased gene expression and consequently modulation of many physiological processes.
The retinoid-related orphan receptors RORα, -β, and -γ constitute a subfamily of nuclear orphan receptors (5–11). Each of these receptors binds as a monomer to response elements (ROREs) consisting of the consensus core motif AGGTCA preceded by an A/T-rich region (7, 12). RORs have been reported to play critical roles in a wide variety of biological processes (13, 14). RORγ is highly expressed in thymus, kidney, liver, muscle, and brown fat but not in white fat tissue (6, 7, 10, 15). In the thymus, two isoforms, γ1 and γ2 (also named RORγT), have been identified (15). The γ2 differs from the γ1 isoform in that it lacks the amino terminus of γ1. The expression of the γ2 isoform is highly restricted to the double-positive thymocytes, suggesting a regulatory role for RORγ2 in these cells (15, 16). Recently, overexpression of RORγ in T-cell hybridomas has been shown to inhibit T cell antigen receptor (TCR)-activation-induced apoptosis by repressing the induction of Fas ligand (FasL) (15) (M.S., S.K., and A.M.J., unpublished observations).
To investigate the biological role(s) of RORγ in vivo, we used homologous recombination in embryonic stem cells to generate mice in which the RORγ gene was disrupted. In this study, we show that RORγ−/− mice lack lymph nodes and Peyer's patches, suggesting that RORγ is essential for their development. In addition, we demonstrate that RORγ plays a critical role in thymopoiesis and T cell homeostasis. The rapid induction of apoptosis observed in RORγ−/− thymocytes is consistent with the concept that RORγ functions as a negative regulator of apoptosis in these cells.
Materials and Methods
Generation of RORγ-Deficient Mice.
The RORγ genomic clone used to create the targeting vector pPNT-ΔRORγ was derived from the mouse strain 129/Sv and described previously (17). The pPNT-ΔRORγ targeting vector was designed to replace the region between introns 2 and 6 by the 1.8-kb neomycin cassette, containing the phosphoglycerate kinase (PGK) poly(A)+ signal and the neomycin phosphotransferase (NEO) selectable marker under the control of a PGK promoter, as shown in Fig. 1. The cassette was flanked by a 5.0- and 1.8-kb region of the RORγ gene. pPNT-ΔRORγ was linearized by NotI and electroporated into 129/Sv embryonic stem cells. Clones with a disrupted RORγ allele were isolated after G418 selection. Gene-targeted embryonic stem cells were microinjected into blastocysts from C57BL/6 mice, and the blastocysts were then implanted in pseudopregnant B6D2 mice. Offspring demonstrating agouti coat color were analyzed for the presence of the disrupted RORγ gene. The chimeric mice were crossed with B6D2 or C57BL/6 mice to identify transmitting chimeras and to obtain mice heterozygous for the mutant allele. Heterozygous mice were intercrossed to obtain animals homozygous for the mutant allele and to obtain wt littermate controls. This gene knock-out was performed at the University of North Carolina Transgenic Facility, headed by Beverly Koller.
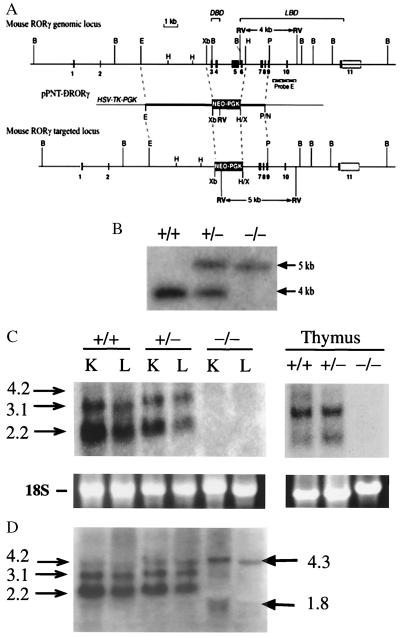
Figure 1.
Targeting the RORγ locus. (A) Schematic representation of the mouse RORγ gene locus, the pPNT-ΔRORγ targeting vector, and the recombination at the RORγ locus. In the targeted locus the region from exon 3 through exon 6 is deleted. DBD, DNA-binding domain; LBD, ligand-binding domain. (B) Diagnostic Southern blot analysis. Genomic DNA was cut by EcoRV, electrophoresed, and hybridized to the 3′-flanking probe E (indicated in A), which detected fragments of the expected size of 4.0 kb for wild-type and 5.0 kb for the mutant allele. Lanes indicate digested DNA from wild-type (wt; +/+), heterozygous (+/−), and homozygous mutant (−/−) mice. (C and D) Northern blot analysis was performed with RNA isolated from liver (L), kidney (K), and thymus of wt, RORγ+/−, and RORγ−/− mice by using either a probe encompassing the deleted region encoded by exons 3–6 (C) or a full-length RORγ probe (D).
Genotyping and Southern Blot Analysis.
Total DNA was prepared from embryonic stem cells and tail biopsies as described previously (18). DNA (8 μg) was digested with EcoRV and separated by electrophoresis in a 0.8% agarose gel containing 40 mM Tris acetate, 2 mM EDTA, pH 8.5 (TAE) buffer. After transfer to an Immobilon-Ny+ membrane (Millipore), DNA was hybridized to [α-32P]dCTP-labeled probe E (Fig. 1).
Northern Blot Analysis.
RNA from liver, kidney, and thymus was isolated by using Tri-Reagent (Sigma) according to the manufacturer's protocol. Total RNA (30 μg) was separated by electrophoresis on a formaldehyde/1.2% agarose gel, blotted to an Immobilon-Ny+ membrane, and hybridized to 32P-labeled RORγ cDNAs as described previously (7).
Cells and Tissue Culture.
Thymocytes, splenocytes, and peripheral blood cells from 8-week-old mice were isolated as described (19) and used immediately for analysis. Thymocytes (1–2 × 106 cells per ml) were also cultured for various times in RPMI medium 1640 containing 10% FBS in the presence or absence of 0.1 μM dexamethasone before analysis.
Flow Cytometry.
Single-cell suspensions were incubated with Cy-Chrome (Cy)-, fluorescein isothiocyanate (FITC)-, or phycoerythrin (PE)-conjugated CD3, CD4, CD8α, TCRβ, or B220 antibodies (PharMingen). For cell cycle analysis, cells were fixed in 70% ethanol and resuspended in PBS containing propidium iodide (50 μg/ml). Analysis of FITC-annexin V binding was carried out with the ApoAlert Annexin V kit (CLONTECH) according to the manufacturer's instructions. Flow cytometric analyses of labeled cells were performed with a FACSort EpicsXL (Beckman–Coulter) flow cytometer and accompanying FACS Convert, CellQuest (Becton Dickinson), and MacCycle (Phoenix Flow Systems) software programs.
Terminal Deoxynucleotidyltransferase-Mediated dUTP Nick End Labeling (TUNEL) Staining.
For detection of DNA fragmentation in situ, paraffin-embedded sections were analyzed by the TUNEL method using an In Situ Cell Death Detection kit (Roche) according to the manufacturer's protocol. Labeling by TMR red-labeled dUTP was examined in a Zeiss Axioplan fluorescence microscope.
RNase Protection Assay (RPA).
Total RNA (20 μg) isolated from the thymocytes of RORγ+/− or RORγ−/− mice was analyzed by RPA using the RiboQuant MultiProbe RNase protection system mAPO-2 (PharMingen). RPA was performed according to the manufacturer's protocol.
Results
To investigate the biological role(s) of RORγ in vivo, we used homologous recombination in embryonic stem cells to generate mice in which the RORγ gene was disrupted. A schematic representation of the targeting vector is shown in Fig. 1A. The targeting vector was created such that after homologous recombination the 1.8-kb neomycin cassette would be inserted between introns 2 and 6, resulting in the deletion of exons 3–6. These exons encode the region of RORγ between amino acids Gln-25 and Lys-309, containing the entire DNA-binding domain, the hinge domain, and part of the ligand-binding domain (7). This deletion is thus predicted to disrupt the DNA-binding ability as well as the transactivating activity of both RORγ1 and -γ2. The disrupted RORγ allele was ultimately transmitted through the germ line. Cross-breeding of RORγ+/− mice produced litters with all three possible genotypes. The genotypes exhibited the expected Mendelian distribution. Genotypes were determined by Southern blot analysis of genomic DNA digested with EcoRV. Hybridization to a 3′-flanking probe detected fragments of the expected size of 4 kb for wt and 5 kb for the mutant allele (Fig. 1B). Additional diagnostic Southern blot analysis using BamHI restriction or a NEO probe confirmed the correct targeting at the RORγ locus (not shown).
Expression of RORγ mRNA was examined by Northern blot analysis in samples from thymus, kidney, and liver. The RORγ probe, encoding the region from exon 3 to 6 deleted during homologous recombination, hybridized strongly to a 2.2- and 3.1-kb mRNA and weakly to a 4.2-kb RNA in samples prepared from wt and RORγ+/− mice. As we reported previously, the different sizes of RNA are generated by the use of alternative polyadenylation signals (17). In contrast, this probe did not show any signal in RNA samples prepared from RORγ−/− mice, confirming the disruption of the RORγ gene (Fig. 1C). A probe including the entire coding region of RORγ hybridized to RNAs 4.3 and 1.8 kb in size. These RNAs are likely generated by alternative splicing. Since the codons of exons 2 and 7 are not in the same reading frame, the protein product generated by such RNAs should be unrelated to RORγ. Immunohistochemical analysis using an RORγ-specific antibody showed nuclear staining in sections of thymus from wt mice, whereas RORγ protein was undetectable in thymus from RORγ−/− mice (not shown).
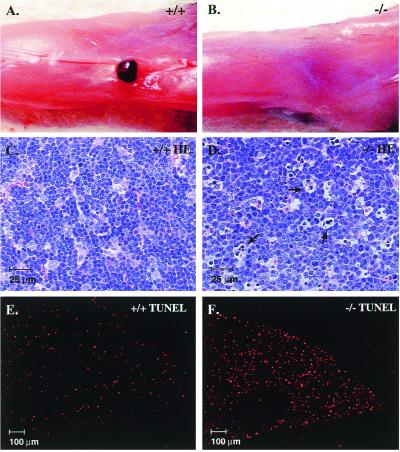
The general appearance of RORγ−/− mice is normal, and mice are healthy during early stages of life. Necropsy studies showed that RORγ−/− mice lacked lymph nodes. Peripheral (e.g., popliteal, inguinal, cervical), para-aortic, and mesenteric lymph nodes as well as Peyer's patches were absent (Fig. 2 A and B and data not shown), indicating that lymph node development is completely arrested. In contrast, lymphatic vessels seemed to be normal. These observations suggest that RORγ plays a critical regulatory role in the organogenesis of lymph nodes and Peyer's patches. Although the spleen from RORγ−/− mice was 2-fold larger, its structure was normal and its germinal centers and follicular dendritic cell network were intact. The spleen contained almost 3 times more lymphocytes (Table 1), and the number of B lymphocytes (B220+ cells) was increased relative to T lymphocytes (CD3+ cells). Analysis of CD4+ and CD8+ cells in spleen showed that the percentage of CD4+ cells was somewhat reduced, whereas that of CD8+ cells remained unchanged. The number of peripheral blood T lymphocytes in RORγ−/− mice was decreased about 6-fold, whereas the number of B lymphocytes did not change significantly (Table 1). The T lymphopenia was more pronounced in the CD4+ compartment than in the CD8+ compartment, with a 10-fold reduction in circulating CD4+ T cells compared with only a 3-fold reduction in circulating CD8+ T cells. Because RORγ is most highly expressed in the thymus, we were particularly interested in examining this tissue for phenotypic changes. The average weight of the thymus from RORγ−/− mice was reduced by 15–20% compared with those from wt mice. The total number of thymocytes retrieved from the thymus of RORγ−/− mice was consistently 74.4% ± 8.9% less than that from wt mice (Table 1), suggesting that disruption of the RORγ gene affects thymopoiesis.
Figure 2.
RORγ−/− mice lack peripheral lymphoid organs and manifest increased apoptosis in the thymus. (A and B) India ink was injected into the hind footpads and the popliteal fossae were dissected. A lymph node is clearly visible in wt mice (A), whereas RORγ−/− mice did not contain detectable lymph node tissue (B). (C–F) Sections of thymus from RORγ+/+ (C and E) and RORγ−/− (D and F) mice stained by hematoxylin/eosin (C and D) or by TUNEL staining using TMR red-dUTP (E and F). Arrows indicate apoptotic cells.
Table 1.
Comparison of various lymphocyte populations in spleen, peripheral blood, and thymus from RORγ+/+ and RORγ−/− mice
| Tissue | Weight, mg
|
Total cell number
|
CD3+/B220+,
%
|
CD4+/CD8+
ratio
|
||||
|---|---|---|---|---|---|---|---|---|
| RORγ+/+ (n = 4) | RORγ−/− (n = 5) | RORγ+/+ (n = 4) | RORγ−/− (n = 5) | RORγ+/+ (n = 3) | RORγ−/− (n = 3) | RORγ+/+ (n = 3) | RORγ−/− (n = 3) | |
| Spleen | 56.6 ± 9.3 | 122.5 ± 16.0 | 48.3 ± 20.4* | 141.9 ± 76.7* | 50.0 ± 5.7/30.9 ± 5.6 | 44.1 ± 7.6/40.3 ± 9.0 | 2.43 ± 0.12 | 1.72 ± 0.46 |
| Peripheral blood | NA | NA | 4.41 ± 2.46† | 2.09 ± 0.80† | 27.9 ± 7.62/35.0 ± 13.4 | 10.3 ± 1.80/74.0 ± 2.42 | 1.28 ± 0.17 | 0.74 ± 0.45 |
| Thymus | 76.4 ± 21.0 | 62.4 ± 13.0 | 156.3 ± 38.5* | 40.1 ± 15.7* | ND | ND | See Fig. 3 and Table 2 | |
The number of mice studied in each group is indicated by the number in parentheses. CD3+/B220+ indicates the percent of CD3+ or B220+ cells. Data are mean ± SD. NA, not applicable; ND, not determined.
Total lymphocytes (× 106 cells per tissue).
White blood cells (× 103 cells per μl).
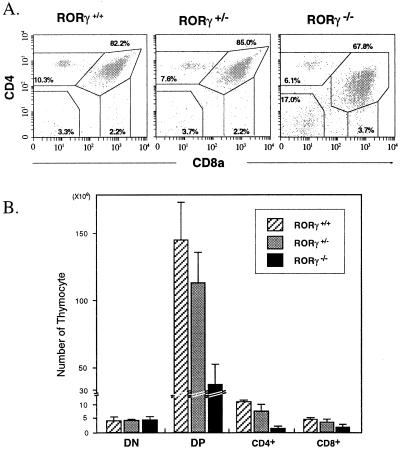
The generation of various T cells in the thymus is a complex biological process that combines proliferation, differentiation, apoptosis, selection, and commitment to different lineages (20–22). Stem cells differentiate into immature CD4−CD8− double-negative (DN) thymocytes that differentiate into CD4+CD8+ double-positive (DP) thymocytes by a multistep process and subsequently undergo positive and negative selection. The surviving DP cells differentiate further into single-positive (SP) CD4+ helper and CD8+ cytotoxic lineages. To study further the impact of the disruption of the RORγ gene on thymopoiesis, we examined its effect on the various CD4/CD8 subpopulations by flow cytometry using fluorescent antibodies against these cell surface markers (Fig. 3A). Comparison of the CD4/CD8 subpopulations between wt and RORγ−/− mice showed that in isolates from RORγ−/− mice the percentage of DN cells was greatly enhanced and that of SP CD4+ and DP thymocytes was reduced (Fig. 3). CD5 and TCRβ flow cytometry confirmed that the DP compartment in RORγ−/− mice indeed consisted of DP cells (not shown). The number of SP CD8+ cells in RORγ−/− mice was more difficult to determine because the population of DP cells exhibited reduced levels of CD4 and therefore began to overlap with the SP CD8+ population. Although the percentage of DN cells in RORγ−/− mice increased, the total number of DN cells per thymus did not differ significantly from that in wt mice (Fig. 3B; Table 2). In contrast, the number of DP, SP CD4+, and SP CD8+ cells was reduced by 74.6% ± 9.7%, 87.4% ± 4.4%, and 55.4% ± 21.4%, respectively. In heterozygous mice, the number of DP, SP CD4+, and SP CD8+ cells was only moderately affected (Table 2). Our results demonstrate that disruption of RORγ function affects in a major way the number of DP, helper, and cytotoxic thymocytes.
Figure 3.
Flow cytometric analysis of CD4 and CD8 expression. (A) Isolated thymocytes from RORγ+/+, RORγ+/−, and RORγ−/− mice stained with anti-CD4-PE and anti-CD8-FITC cell surface markers and then analyzed by flow cytometry. Numbers within each of the four outlined areas indicate the percentage of the respective subpopulation. The results shown are representative for four independent analyses. (B) Comparison of the total cell number in each CD4/CD8 subpopulation.
Table 2.
Comparison of CD4/CD8 subpopulations in the thymus from RORγ+/+, RORγ+/−, and RORγ−/− mice
| Source of thymocytes | Average total thymocyte no. (×106) | DN
|
DP
|
SP
(CD4)
|
SP (CD8)
|
||||
|---|---|---|---|---|---|---|---|---|---|
| % | × 106 | % | × 106 | % | × 106 | % | × 106 | ||
| RORγ+/+ (n = 3) | 169.0 ± 25.2 | 2.6 ± 1.0 | 4.2 ± 0.9 | 86.4 ± 2.0 | 146.6 ± 20.4 | 6.6 ± 0.7 | 10.9 ± 0.4 | 2.7 ± 0.5 | 4.5 ± 0.4 |
| RORγ+/− (n = 3) | 124.5 ± 25.7 | 3.4 ± 0.5 | 4.3 ± 0.3 | 86.6 ± 1.3 | 114.0 ± 16.8 | 5.8 ± 1.4 | 7.5 ± 1.7 | 2.6 ± 0.3 | 3.5 ± 0.7 |
| RORγ−/− (n = 3) | 42.2 ± 11.5 | 10.9 ± 3.7 | 4.5 ± 0.9 | 78.5 ± 6.6 | 36.1 ± 11.2 | 3.3 ± 1.8 | 1.4 ± 0.5 | 3.8 ± 0.5 | 1.8 ± 0.7 |
The number of mice studied in each group is indicated by the number in parentheses. Data are mean ± SEM.
Examination of hematoxylin/eosin-stained sections showed a significant increase in the presence of apoptotic nuclei in the cortex of thymuses from RORγ−/− mice (Fig. 2 C and D). This increase in apoptosis was confirmed by TUNEL staining. Sections prepared from the thymus of RORγ−/− mice consistently contained 4 times as many TUNEL-positive cells compared with those from wt mice of the same litter (Fig. 2 E and F). TUNEL-positive cells were associated with the cortex region of the thymus and not with the medulla. The number of apoptotic cells stained by TUNEL in the thymus from heterozygous mice (RORγ+/−) was comparable to that from wt mice (not shown).
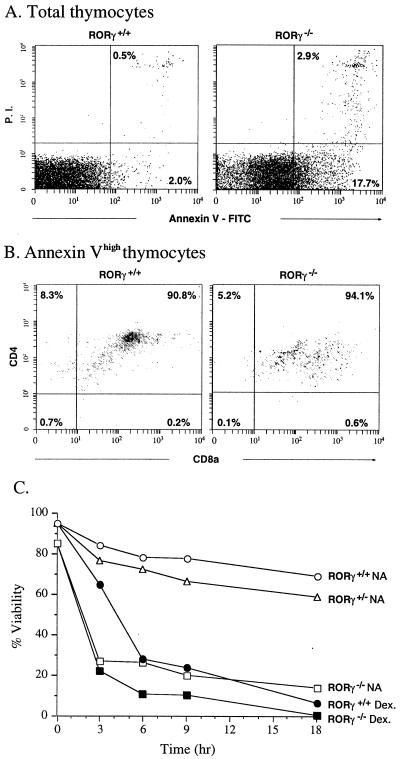
An early event in apoptosis is the loss of membrane asymmetry and the subsequent exposure of phosphatidylserine at the cell surface, which can be detected by binding of FITC-labeled annexin V (23). Analysis of isolated thymocytes by flow cytometry showed that almost the entire RORγ−/− thymocyte population labeled more intensely with annexin V. The percentage of highly labeled annexin V thymocytes (early and late apoptotic cells) from RORγ−/− mice was about 20–25% compared with 2–4% for wt mice (Fig. 4A). It is likely that this 4-fold increase in apoptosis is at least partly responsible for the decrease in the total number of thymocytes in RORγ−/− mice. To determine with which subpopulation of cells the increase in apoptosis was associated, we determined the percentage of SP CD4+, DP, SP CD8+, and DN thymocytes among the annexin Vhigh population. As shown in Fig. 4B, 90.8% and 94.1%, respectively, of the annexin Vhigh thymocyte population in RORγ+/+ and RORγ−/− mice was composed of DP cells. These results suggest that most of the enhanced apoptosis in RORγ−/− mice is associated with DP thymocytes. Isolated RORγ−/− thymocytes placed in culture underwent “spontaneous” apoptosis at a dramatically enhanced rate compared with wt or RORγ+/− thymocytes (Fig. 4C). The rate of apoptosis was almost identical to that of wt cells treated with dexamethasone, a known inducer of apoptosis in thymocytes (24). Treatment of RORγ−/− thymocytes with dexamethasone only slightly enhanced apoptosis. Our results indicate that RORγ−/− thymocytes in culture have a greatly enhanced probability for undergoing apoptosis and suggest that RORγ functions as a negative regulator of apoptosis.
Figure 4.
Apoptosis is largely associated with DP thymocytes. (A) Thymocytes were isolated and treated with annexin V-FITC, anti-CD4-PE and anti-CD8-Cy. Total thymocytes were first analyzed by flow cytometry for annexin V binding. The lower and upper right quadrants indicate early and late apoptotic cells, respectively. (B) Thymocytes highly labeled by annexin V (early and late apoptotic cells) were separated further by flow cytometry on the basis of anti-CD4 and anti-CD8 labeling. (C) The rate of apoptosis in cultured ROR−/− thymocytes is greatly increased. Thymocytes were cultured in RPMI medium 1640 plus 10% FBS in the presence (Dex.) or absence (NA) of 0.1 μM dexamethasone. At the indicated times, samples were removed and analyzed for annexin V binding by flow cytometry as in A. The percentage of annexin Vlow cells (% viability) was calculated and plotted as a function of time in culture.
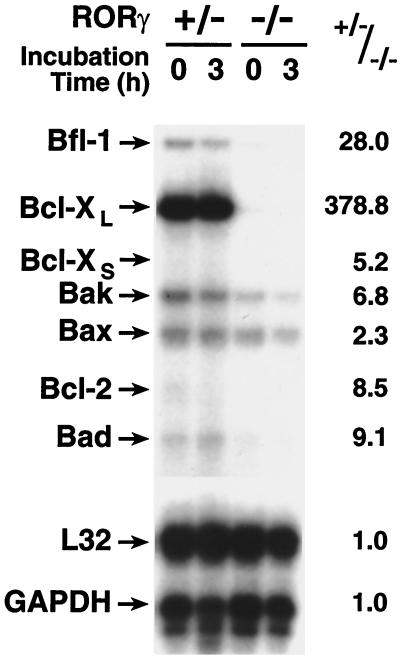
Programmed cell death often involves members of the Bcl-2 family, which includes both anti- and pro-apoptotic proteins (25, 26). To determine whether the increase in apoptosis in RORγ−/− mice was accompanied by any changes in the expression of pro- or anti-apoptotic genes, we analyzed RNA of isolated thymocytes from RORγ+/− and RORγ−/− mice by RPAs. As shown in Fig. 5, although the mRNA level of several anti- and pro-apoptotic genes decreased, the level of the anti-apoptotic gene Bcl-XL was most affected and consistently decreased more than 300-fold in the thymocytes from RORγ−/− mice compared with that of RORγ+/− mice. This difference is much larger than one would expect on the basis of the observed changes in CD4/CD8 subpopulations. During the 3-hr incubation in culture little change in the expression levels of anti- as well as pro-apoptotic mRNAs was observed. These results indicate that decreased expression of Bcl-XL as well as the decrease in the ratio between anti-/pro-apoptotic gene expression relates to the increased sensitivity of RORγ−/− thymocytes to undergo “spontaneous” apoptosis. Likely, additional biochemical events are involved in triggering this cell death.
Figure 5.
Effect of RORγ disruption on the expression of anti- and pro-apoptotic genes. RNA was prepared from freshly isolated thymocytes from RORγ+/− or RORγ−/− mice, and thymocytes were cultured for 3 hr. RNA was then analyzed by RPA. The positions of mRNAs encoding Bcl-2, Bfl-1, Bcl-XL, Bak, Bax, Bad, L32, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are indicated. The results are representative for two independent experiments. The level of each RNA relative to the level of GAPDH was determined, and the ratio between the level of each RNA in RORγ+/− versus RORγ−/− thymocytes was calculated. The ratios are shown on the right.
The homeostasis between various subpopulations in the thymus depends on a balance between proliferation, differentiation, and apoptosis. These subpopulations have very different proliferative capacities (22, 27). Cell cycle analysis showed that RORγ−/− mice exhibited a dramatic increase in the percentage of thymocytes in the S phase of the cell cycle (Table 3). About 4.4% ± 0.7% of thymocytes from wt mice were in S phase, in agreement with previous reports, compared with about 25.7% ± 5.7% of the cells from RORγ−/− mice. This increase in the percentage of cycling cells may be in part a reflection of the increase in the percentage of rapidly cycling DN thymocytes.
Table 3.
Cell cycle analysis of thymocytes from RORγ+/+ and RORγ−/− mice
| Source of thymocytes | %
|
||
|---|---|---|---|
| G1/G0 | S | G2/M | |
| RORγ+/+ (n = 3) | 85.6 ± 2.6 | 4.4 ± 0.7 | 9.9 ± 2.2 |
| RORγ−/− (n = 3) | 64.5 ± 5.7 | 25.7 ± 5.5 | 9.6 ± 0.5 |
Isolated thymocytes from RORγ+/+ and RORγ−/− mice were stained with propidium iodide and then analyzed by flow cytometry. The number of mice studied in each group is indicated by the number in parentheses. Data are mean ± SEM.
Discussion
Characterization of RORγ−/− mice identified several important functions for RORγ. RORγ−/− mice lack lymph nodes, including peripheral and mesenteric lymph nodes, as well as Peyer's patches. These observations suggest a critical role for RORγ in the control of lymph node and Peyer's patch development. Recent studies have reported the importance of several proteins, including members of the tumor necrosis factor family and their receptors, in lymph node genesis (28, 29). Mice deficient in lymphotoxin (LT) α (30) and LTβ-R (31) lack all lymph nodes and Peyer's patches, whereas mice deficient in LTβ (32) lack Peyer's patches and peripheral lymph nodes but still have mesenteric lymph nodes. RANK−/− mice lack peripheral lymph nodes but retain mesenteric and Peyer's patches (33). In contrast to RORγ−/− mice, mice lacking LTα, LTβ or LTβ-R also manifest defects in splenic architecture. Id2, a member of the helix–loop–helix family of transcription factors, also plays a key role in the development of peripheral lymphoid organs. The phenotype of Id2−/− mice resembles that of RORγ−/− mice. Like RORγ−/− mice, Id2−/− mice lack lymph nodes and Peyer's patches, and have a normal splenic architecture. These observations are in agreement with previous reports indicating that organogenesis among secondary lymphoid organs is regulated differently. The lack of Peyer's patches and lymph nodes in RORγ−/− mice could be due to defects at different stages of lymph node genesis, including differentiation of progenitor cells, control of homing activity of lymphocytes, or the regulation of LTs. The population of CD4+CD3−IL-7Rα+ progenitor cells appears to play a key role in the formation of lymph nodes and Peyer's patches and are absent in Id2−/− mice (34). As does Id2, RORγ may positively regulate the formation of these cells. Future studies have to determine whether RORγ acts upstream or downstream of Id2, whether one protein regulates the expression of the other, or whether the Id2 and RORγ signaling pathways are involved in crosstalk.
Our study also identified an important regulatory role for RORγ in thymopoiesis and T cell homeostasis. We demonstrated that the total numbers of thymocytes and circulating T cells in RORγ−/− mice are dramatically reduced. TUNEL staining showed increased apoptosis in the cortex of the thymus, and RORγ−/− thymocytes placed in culture rapidly undergo apoptosis. This increase in apoptosis is largely associated with DP thymocytes and may be at least partially responsible for the reduction in the number of DP, SP CD4+, and SP CD8+ cells. No apparent difference was found in the number of DN cells, suggesting that disruption of RORγ function may not affect very early stages of thymopoiesis. Our observations are consistent with the hypothesis that RORγ plays a role in the negative regulation of apoptosis and appears to promote cell survival. Previous studies have shown that the expression of the RORγ2 isoform is highly restricted to DP thymocytes (15, 16). This expression pattern correlates with the increased apoptosis observed in DP thymocytes and supports the concept that the γ2 is the most likely isoform involved in the control of apoptosis in these cells. Although the γ1 isoform is also expressed in the thymus, it may have other regulatory functions.
The precise mechanism by which RORγ regulates apoptosis is not yet understood. RORγ could inhibit signaling pathways that promote apoptosis and/or stimulate signals that support cell survival. RPAs demonstrated that particularly the expression of the anti-apoptotic Bcl-XL gene is decreased in RORγ−/− thymocytes. Bcl-XL is highly regulated during T cell development and maximally expressed in DP thymocytes and shown to inhibit multiple forms of apoptosis (35). Because of the extent of the decrease in Bcl-XL, its down-regulation is likely only partially due to the observed decrease in DP thymocytes. The reduction in Bcl-XL expression by RORγ could be mediated by a direct or indirect mechanism. RORγ may influence Bcl-XL transcription by binding directly to response elements in its promoter or affect its transcription through crosstalk with other transcriptional factors. Alternatively, RORγ may affect its expression by an indirect mechanism through regulation of other genes involved in the control of Bcl-XL, such as certain cytokines or their receptors. Since little change was observed in Bcl-XL expression during 3 hr of culture, down-regulation of Bcl-XL may increase the sensitivity of RORγ−/− thymocytes to undergo “spontaneous” apoptosis but may not be sufficient to initiate cell death. Likely other biochemical events such as a conformational change and translocation of Bax from the cytosol to the mitochondria may be involved in the induction of this apoptosis (36).
Recently, RORγ2 has been reported to inhibit TCR-induced apoptosis in T cell hybridomas by repressing the induction of FasL (15) (M.S., S.K., and A.M.J., unpublished observations). RORγ could have a similar effect in thymocytes. However, the role of Fas/FasL-mediated apoptosis in thymocyte development remains unclear. Although several reports have indicated that Fas/FasL interaction is not involved in negative selection (37), modulation of apoptosis by Fas has been described by others (38, 39). RPAs and Northern blot analyses showed no difference in the expression of FasL mRNA between wt and RORγ−/− mice (not shown). These results suggest that enhanced thymocyte apoptosis in RORγ−/− mice does not relate to increased FasL expression.
In summary, our results demonstrate that RORγ is essential for lymphoid organogenesis. In addition, our study demonstrates that RORγ plays a critical role in thymopoiesis and T cell homeostasis. Our results support a model in which RORγ functions as a suppressor of apoptosis. Therefore, defects in either the expression of RORγ or the RORγ signaling pathway may lead to impairments in the control of apoptosis, maturation of thymocytes, and T cell homeostasis. Identification of RORγ target genes will facilitate the elucidation of the molecular mechanisms that regulate thymopoiesis and the development of lymph node and Peyer's patches.
Acknowledgments
We thank Dr. Beverly Koller and Anne Latour (Transgenic Facility, University of North Carolina) for their outstanding assistance and guidance in generating the RORγ−/− mice, Aron Boney and Prithu Metty (Duke University) for their technical assistance, Dr. Carl Bortner (National Institute of Environmental Health Sciences) for his advice with the flow cytometric analysis, and Dr. Donna Newman (National Institute of Environmental Health Sciences) for her comments on the manuscript. This study was supported by the National Institutes of Health Grant RO1 AI47604 and by the Japan Society for the Promotion of Science.
Abbreviations
- ROR
retinoid-related orphan receptor
- TUNEL
terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling
- TCR
T cell antigen receptor
- wt
wild-type
- RPA
RNase protection assay
- DN
double-negative
- DP
double-positive
- SP
single-positive
References
- 1.Willy P J, Mangelsdorf D J. In: Hormones and Signaling. O'Malley B W, editor. Vol. 1. San Diego: Academic; 1998. pp. 308–358. [Google Scholar]
- 2.Kumar R, Thompson E B. Steroids. 1999;64:310–319. doi: 10.1016/s0039-128x(99)00014-8. [DOI] [PubMed] [Google Scholar]
- 3.McKenna N J, Xu J, Nawaz Z, Tsai S Y, Tsai M J, O'Malley B W. J Steroid Biochem Mol Biol. 1999;69:3–12. doi: 10.1016/s0960-0760(98)00144-7. [DOI] [PubMed] [Google Scholar]
- 4.Xu L, Glass C K, Rosenfeld M G. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 5.Giguere V, Tini M, Flock G, Ong E, Evans R M, Otulakowski G. Genes Dev. 1994;8:538–553. doi: 10.1101/gad.8.5.538. [DOI] [PubMed] [Google Scholar]
- 6.Hirose T, Smith R J, Jetten A M. Biochem Biophys Res Commun. 1994;205:1976–1983. doi: 10.1006/bbrc.1994.2902. [DOI] [PubMed] [Google Scholar]
- 7.Medvedev A, Yan Z H, Hirose T, Giguere V, Jetten A M. Gene. 1996;181:199–206. doi: 10.1016/s0378-1119(96)00504-5. [DOI] [PubMed] [Google Scholar]
- 8.Carlberg C, Hooft van Huijsduijnen R, Staple J K, DeLamarter J F, Becker-Andre M. Mol Endocrinol. 1994;8:757–770. doi: 10.1210/mend.8.6.7935491. [DOI] [PubMed] [Google Scholar]
- 9.Becker-Andre M, Andre E, DeLamarter J F. Biochem Biophys Res Commun. 1993;194:1371–1379. doi: 10.1006/bbrc.1993.1976. [DOI] [PubMed] [Google Scholar]
- 10.Ortiz M A, Piedrafita F J, Pfahl M, Maki R. Mol Endocrinol. 1995;9:1679–1691. doi: 10.1210/mend.9.12.8614404. [DOI] [PubMed] [Google Scholar]
- 11.Wiesenberg I, Missbach M, Kahlen J P, Schrader M, Carlberg C. Nucleic Acids Res. 1995;23:327–333. doi: 10.1093/nar/23.3.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giguere V, McBroom L D, Flock G. Mol Cell Biol. 1995;15:2517–2526. doi: 10.1128/mcb.15.5.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dussault I, Fawcett D, Matthyssen A, Bader J A, Giguere V. Mech Dev. 1998;70:147–153. doi: 10.1016/s0925-4773(97)00187-1. [DOI] [PubMed] [Google Scholar]
- 14.Andre E, Conquet F, Steinmayr M, Stratton S C, Porciatti V, Becker-Andre M. EMBO J. 1998;17:3867–3877. doi: 10.1093/emboj/17.14.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Y W, Deftos M L, Ojala E W, Bevan M J. Immunity. 1998;9:797–806. doi: 10.1016/s1074-7613(00)80645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villey I, de Chasseval R, de Villartay J P. Eur J Immunol. 1999;29:4072–4080. doi: 10.1002/(SICI)1521-4141(199912)29:12<4072::AID-IMMU4072>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 17.Medvedev A, Chistokhina A, Hirose T, Jetten A M. Genomics. 1997;46:93–102. doi: 10.1006/geno.1997.4980. [DOI] [PubMed] [Google Scholar]
- 18.Medvedev A, Saunders N A, Matsuura H, Chistokhina A, Jetten A M. J Biol Chem. 1999;274:3887–3896. doi: 10.1074/jbc.274.6.3887. [DOI] [PubMed] [Google Scholar]
- 19.Naka T, Matsumoto T, Narazaki M, Fujimoto M, Morita Y, Ohsawa Y, Saito H, Nagasawa T, Uchiyama Y, Kishimoto T. Proc Natl Acad Sci USA. 1998;95:15577–15582. doi: 10.1073/pnas.95.26.15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellmeier W, Sawada S, Littman D R. Annu Rev Immunol. 1999;17:523–554. doi: 10.1146/annurev.immunol.17.1.523. [DOI] [PubMed] [Google Scholar]
- 21.Killeen N, Irving B A, Pippig S, Zingler K. Curr Opin Immunol. 1998;10:360–367. doi: 10.1016/s0952-7915(98)80176-3. [DOI] [PubMed] [Google Scholar]
- 22.Zuniga-Pflucker J C, Lenardo M J. Curr Opin Immunol. 1996;8:215–224. doi: 10.1016/s0952-7915(96)80060-4. [DOI] [PubMed] [Google Scholar]
- 23.Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. J Immunol Methods. 1995;184:39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
- 24.Schwartzman R A, Cidlowski J A. Int Arch Allergy Immunol. 1994;105:347–354. doi: 10.1159/000236781. [DOI] [PubMed] [Google Scholar]
- 25.Reed J C. Oncogene. 1998;17:3225–3236. doi: 10.1038/sj.onc.1202591. [DOI] [PubMed] [Google Scholar]
- 26.Gross A, McDonnell J M, Korsmeyer S J. Genes Dev. 1999;13:1899–1911. doi: 10.1101/gad.13.15.1899. [DOI] [PubMed] [Google Scholar]
- 27.Kisielow P, von Boehmer H. Adv Immunol. 1995;58:87–209. doi: 10.1016/s0065-2776(08)60620-3. [DOI] [PubMed] [Google Scholar]
- 28.Fu Y X, Chaplin D D. Annu Rev Immunol. 1999;17:399–433. doi: 10.1146/annurev.immunol.17.1.399. [DOI] [PubMed] [Google Scholar]
- 29.Ruddle N H. Immunol Res. 1999;19:119–125. doi: 10.1007/BF02786481. [DOI] [PubMed] [Google Scholar]
- 30.De Togni P, Goellner J, Ruddle N H, Streeter P R, Fick A, Mariathasan S, Smith S C, Carlson R, Shornick L P, Strauss-Schoenberger J, et al. Science. 1994;264:703–707. doi: 10.1126/science.8171322. [DOI] [PubMed] [Google Scholar]
- 31.Futterer A, Mink K, Luz A, Kosco-Vilbois M H, Pfeffer K. Immunity. 1998;9:59–70. doi: 10.1016/s1074-7613(00)80588-9. [DOI] [PubMed] [Google Scholar]
- 32.Koni P A, Sacca R, Lawton P, Browning J L, Ruddle N H, Flavell R A. Immunity. 1997;6:491–500. doi: 10.1016/s1074-7613(00)80292-7. [DOI] [PubMed] [Google Scholar]
- 33.Dougall W C, Glaccum M, Charrier K, Rohrbach K, Brasel K, De Smedt T, Daro E, Smith J, Tometsko M E, Maliszewski C R, et al. Genes Dev. 1999;13:2412–2424. doi: 10.1101/gad.13.18.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, Gruss P. Nature (London) 1999;397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- 35.Grillot D A, Merino R, Nunez G. J Exp Med. 1995;182:1973–1983. doi: 10.1084/jem.182.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khaled A R, Kim K, Hofmeister R, Muegge K, Durum S K. Proc Natl Acad Sci USA. 1999;96:14476–14481. doi: 10.1073/pnas.96.25.14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singer G G, Abbas A K. Immunity. 1994;1:365–371. doi: 10.1016/1074-7613(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 38.Fleck M, Zhou T, Tatsuta T, Yang P, Wang Z, Mountz J D. J Immunol. 1998;160:3766–3775. [PubMed] [Google Scholar]
- 39.Kishimoto H, Surh C D, Sprent J. J Exp Med. 1998;187:1427–1438. doi: 10.1084/jem.187.9.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]