Abstract
Dynamin-related protein (Drp) 1 is a key regulator of mitochondrial fission and is composed of GTP-binding, Middle, insert B, and C-terminal GTPase effector (GED) domains. Drp1 associates with mitochondrial fission sites and promotes membrane constriction through its intrinsic GTPase activity. The mechanisms that regulate Drp1 activity remain poorly understood but are likely to involve reversible post-translational modifications, such as conjugation of small ubiquitin-like modifier (SUMO) proteins. Through a detailed analysis, we find that Drp1 interacts with the SUMO-conjugating enzyme Ubc9 via multiple regions and demonstrate that Drp1 is a direct target of SUMO modification by all three SUMO isoforms. While Drp1 does not harbor consensus SUMOylation sequences, our analysis identified2 clusters of lysine residues within the B domain that serve as noncanonical conjugation sites. Although initial analysis indicates that mitochondrial recruitment of ectopically expressed Drp1 in response to staurosporine is unaffected by loss of SUMOylation, we find that Drp1 SUMOylation is enhanced in the context of the K38A mutation. This dominant-negative mutant, which is deficient in GTP binding and hydrolysis, does not associate with mitochondria and prevents normal mitochondrial fission. This finding suggests that SUMOylation of Drp1 is linked to its activity cycle and is influenced by Drp1 localization.—Figueroa-Romero, C., Iñiguez-Lluhí, J. A., Stadler, J., Chang, C.-R., Arnoult, D., Keller, P. J., Hong, Y., Blackstone, C., Feldman, E. L. SUMOylation of the mitochondrial fission protein Drp1 occurs at multiple nonconsensus sites within the B domain and is linked to its activity cycle.
Keywords: Ubc9, small ubiquitin-like modifier, post-translational modification
Kolliker first described mitochondria as granules over 150 years ago in 1857, but it is only within the past several decades that the important role of mitochondrial malfunction in disease has been appreciated. Mitochondria are highly dynamic organelles, and their proper function relies on orchestrated fusion and fission events (1).
An evolutionary conserved set of molecules comprising members of the dynamin family of proteins regulates the balance between mitochondrial fusion and fission. In mammalian cells, the mitochondrial outer membrane proteins Mitofusin 1 and 2 (Mfn1, Mfn2) and inner mitochondrial membrane-associated protein optic atrophy protein (OPA) 1 regulate fusion (2, 3). Conversely, the Fission1 protein (Fis1) acts as a receptor at the outer mitochondrial membrane for the dynamin-related protein (Drp1) (4,5,6).
Mitochondrial dynamics have a significant effect in the development of mitochondrial diseases, particularly for muscle and neuronal cells (7,8,9,10,11). Mutations in OPA1 and Mfn2 lead to autosomal dominant optic atrophy type 1 and two autosomal dominant forms of Charcot-Marie-Tooth (CMT) neuropathy, respectively (12,13,14). Recently, a mutation in Drp1 was found to lead to phenotypic characteristics overlapping those of CMT and optic atrophy but with a fulminate prenatal onset and death shortly after birth (15).
Drp1 is predominantly a cytoplasmic protein composed of an N-terminal GTPase domain, which binds and hydrolyzes GTP, followed by a Middle domain important for multimerization, a variable B or coiled-coiled domain, and a GTPase-effector domain (GED) (16). Drp1 associates with mitochondrial fission sites upon oligomerization. Powered by its GTPase activity, it provides the driving force for mitochondrial membrane constriction in a manner similar to the role of dynamin in endocytosis (17,18,19). Drp1 also facilitates apoptotic mitochondrial fission, mainly by promoting mitochondrial outer membrane permeabilization (MOMP) induced by members of the Bcl-2 proapoptotic family of proteins (Bax/Bak) prior to physical fragmentation of mitochondria (20,21,22).
Although it is widely accepted that Drp1 plays a key role in driving mitochondrial fission, the molecular regulators of Drp1 that enable it to perform this task during different biological activities are only beginning to be revealed. Recent data indicate that Drp1 is the target for multiple post-translational modifications, such as phosphorylation, ubiquitination, S-nitrosylation, and the conjugation of small ubiquitin-like modifier proteins, or SUMOylation (23,24,25,26,27,28,29,30,31,32). These modifications are known to modulate protein-protein interactions, subcellular localization, protein degradation, and activation of signaling pathways. Such modifications may work in cooperation or antagonistically in response to environmental cues (33).
SUMOylation is a conserved post-translational modification process that uses a distinct but enzymologically parallel pathway to ubiquitination. SUMOylation, however, exerts unique functional roles. Four mammalian SUMO isoforms have been identified (SUMO1, 2, 3, and 4). SUMO2 and SUMO3 are closely related, whereas SUMO1 shares 48% identity to either SUMO2 or SUMO3 (34, 35). A fourth isoform closely related to SUMO2/3 (36) harbors a Pro residue at position 90 that prevents initial processing by known SUMO protease enzymes and subsequent conjugation (37). Whether this member functions exclusively through noncovalent interactions remains to be determined. SUMO conjugation involves specific E1-activating (SAE1/SAE2) and E2-conjugating (Ubc9) enzymes. Ubc9 catalyzes the formation of an isopeptide bond between the carboxyl terminus of SUMO and the ε-amino group of the target lysine (38). This step is facilitated by SUMO E3 ligases, such as RanBP2, and members of the protein inhibitor of activated STAT (PIAS) family (39,40,41). SUMOylation is reversible, and specific isopeptidases release the SUMO moiety (42). The functional consequences of SUMOylation are target specific, but in many cases the effects of SUMO depend on a distinct effector surface (43) that recognizes SUMO-interacting motifs in target proteins (44).
Although the effects of SUMO modification have been best characterized for nuclear proteins, such as sequence-specific transcription factors, recent evidence indicates that cytosolic as well as integral membrane proteins (45) are SUMOylated and that this modification exerts important regulatory roles. How this modification influences mitochondrial function, however, remains largely unknown. In this regard, despite the identification of Ubc9 and SUMO1 as Drp1-interacting proteins in a yeast 2-hybrid screen (28) and subsequent data implicating the SUMO protease SENP5 and the mitochondrial-anchored protein ligase (MAPL) in mitochondrial dynamics (31, 32), the exact targets of the SUMOylation machinery in this context have not been fully established. While multiple Drp1 species of various molecular masses have been proposed to be SUMO1-modified forms of Drp1 (28, 31), which of these unambiguously represent SUMO conjugated forms of Drp1 has not been resolved. In addition, whether other SUMO isoforms are involved in this process is unknown. Clearly, a specific assessment of the effect of SUMOylation on the function of Drp1 is hampered by the absence of knowledge of the sites of modification, especially since sequence analysis indicates that Drp1 does not contain canonical SUMO conjugation sites. To address these issues, we have undertaken a systematic examination of Drp1 SUMOylation. Our results provide important molecular tools to investigate directly the biological significance of Drp1 SUMOylation in mitochondrial function.
MATERIALS AND METHODS
Plasmids and antibodies
Human pcDNA3-Drp1 wild-type (WT) and dominant negative (DN) mutant pcDNA3-Drp1 K38A (19, 22) were obtained from Dr. Richard J. Youle (National Institutes of Health, Bethesda, MD, USA) and used as templates to generate pcDNA3 vectors driving the expression of N-terminal V5 and hexahistidine-tagged WT Drp1 and its derivatives as well as the yeast 2-hybrid constructs. Eukaryotic expression vectors (pGW1) for Myc-and HA-epitope-tagged Drp1 have been described previously (16). Expression vectors for N-terminal HA-tagged SUMO isoforms are pcDNA3 based and have been described previously (43). Similarly, human SUMO1 and ubiquitin cDNAs were cloned into the EcoRI site of pGW1 with an N-terminal Myc-epitope tag. pcDNA3-Ubc9 was a kind gift of Dr. Kim Orth (University of Texas Southwestern Medical Center, Dallas, TX, USA). pcDNA3-V5-Ubc9 was generated by amplification of the Ubc9 sequence by RT-PCR from the human neuroblastoma cell line SH-EP using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA) and subcloned into a pcDNA3 derivative bearing an N-terminal V5 tag. The CS2+ plasmid was a kind gift of Dr. David Turner (University of Michigan, Ann Arbor, MI, USA). Mutations were introduced into all vectors using the QuikChange method (Stratagene, La Jolla, CA, USA).
The pLex and VP16 yeast expression vectors were kindly provided by Dr. Anne B. Vojtek (University of Michigan) and were used to generate VP16-Ubc9, pLex-Drp1 full-length, pLex-Drp GTPase domain (aa 1–225), pLex-Drp1 Middle domain (aa 227–521), pLex-Drp B domain (aa 502–626), pLex-Drp GED domain (aa 627–736), and pLex-Drp No GTPase domain (aa 227–736) vectors. All constructs were confirmed by sequencing.
Sources of antibodies were as follows: anti-Drp1, mouse monoclonal H00010059 (Abnova, Walnut, CA, USA); anti-HA, mouse monoclonal 12CA5 (Abcam, Cambridge, MA, USA), rabbit polyclonal HA.11 (Covance, Princeton, NJ, USA); anti-GAPDH, mouse monoclonal (Chemicon, Temecula, CA, USA); anti-Myc, monoclonal 9E10 (Santa Cruz Biotechnology, Santa Cruz, CA, USA); anti-HSP60, mouse monoclonal (Sigma-Aldrich, St. Louis, MO, USA). Secondary antibodies were horseradish peroxidase-conjugated sheep anti-mouse (Pierce, Rockford, IL, USA) and goat anti-rabbit (Santa Cruz Biotechnology).
Cell culture and transient transfections
Human embryonic kidney (HEK)-293 cells were grown at 37°C in 10% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; Cellgro, Manassas, VA, USA), supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA), penicillin, and streptomycin. Transient transfections were performed using LipofectAmine 2000 reagent (Invitrogen, Carlsbad, CA, USA) with a total of 3 μg of DNA: 0.5 μg of Drp1, SUMO, and Ubc9 expression plasmids, 1.5 μg empty vector. Cos7 cell culture and transfections were as described previously (16).
In vivo SUMOylation
Procedures using the His-tagged Drp1 constructs were adapted from Benson et al. (45). Briefly, HEK-293 cells were plated at 3.5 × 105 cells/35-mm dish and transiently transfected 24 h later. Transfection medium was removed and replaced 6 h later with fresh medium. Cells were lysed ∼22 h after transfection in CHAPS (3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate) lysis buffer [0.5 mM NaCl, 25 mM imidazole, 45 mM Na2HPO4, 5 mM NaH2PO4, and 1% CHAPS, ± 20 mM N-ethylmaleimide (NEM), pH 8.0] supplemented with aprotinin, phenylmethanosulfonyl fluoride (PMSF), leupeptin, and Na2VO4. Cell lysates were precleared by centrifugation. Protein extracts (45 μl) were mixed with 2× sample buffer (100 mM Tris, pH 6.8; 4% SDS; 10% glycerol; and 0.015% bromphenol blue). The remainder of the extracts was incubated with nickel-agarose (Ni2+) beads (Qiagen) for 1 h at 4°C. Beads were washed 3× with CHAPS wash buffer 1 (0.4 mM NaCl, 25 mM imidazole, 17.6 mM Na2HPO4, 32.4 mM NaH2PO4, and 0.1% CHAPS, pH 6.75); 2× with CHAPS wash buffer 2 (0.150 mM NaCl, 17.6 mM Na2HPO4, 32.4 mM NaH2PO4, 0.1% CHAPS, and 8M urea, pH 6.75); and 3× with buffer 3 (50 mM NaCl, 45 mM Na2HPO4, 5 mM NaH2PO4, and 0.1% CHAPS, pH 8.0). Beads were then resuspended in 70 μl 3× EDTA sample buffer [150 mM Tris, pH 6.8; 10 mM EDTA; 6% SDS; 15% glycerol; 0.0225% bromphenol blue; and 20 mM β-mercaptoethanol (β-ME)]. The eluates were incubated at 50°C for 20 min, and the extracts were boiled for 3 min. Samples were centrifuged prior to loading onto 7.5 or 12.5% SDS-polyacrylamide gels. Samples were processed for Western blot analysis and probed with the indicated antibodies.
For assays using HA- and Myc-tagged Drp1 forms, SUMOylation was assessed in immunoprecipitates. Briefly, extracts from COS7 cells cotransfected with HA-Drp1 and Myc-SUMO1 (WT or mutant, as indicated), or else transfected with Myc-SUMO1 alone, were immunoprecipitated using anti-HA antibodies, as described previously (16). Myc-Drp1 was expressed alone where indicated. Protein samples were resolved by SDS-PAGE, then transferred to nitrocellulose membranes and immunoblotted with the indicated antibodies, as described previously (16).
Yeast 2-hybrid assays
Transformation of the Saccharomyces cerevisiae reporter strain L40 was as described previously (46). Briefly, L40 was cotransformed with vectors expressing fusion proteins of LexA DNA binding domain (LexA) to full-length Drp1 or Drp1 deletions and vectors expressing Ubc9 as a fusion protein to the VP16 acidic activation domain (VP16). Cotransformed colonies were grown in medium lacking tryptophan and leucine (−Trp/Leu), and serial dilutions were plated. Recovery of cells bearing both plasmids was confirmed by growth on −Trp/Leu plates. The interaction between proteins was assessed by growth on minimal medium plates also lacking histidine (−Trp/Leu/His), indicating the activation of the Lex operator driven by the HIS reporter gene. The L40 strain was obtained from Dr. Anne B. Vojtek (University of Michigan).
Isolation of mitochondria
HeLa cells were transfected with WT or mutant Myc-Drp1 were either left untreated or else treated with 1 μM staurosporine (Sigma-Aldrich) for 8 h in the presence of 100 μM zVAD-fmk (Calbiochem, San Diego, CA, USA). Mitochondria were isolated intact from HeLa cells by sucrose density gradient centrifugation and subjected to SDS-polyacrylamide gel electrophoresis and immunoblotting, as described previously (47, 48).
RESULTS
Ubc9 forms a bipartite interaction with two different domains of Drp1
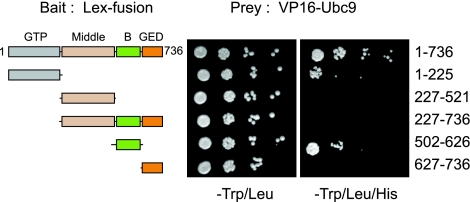
Recent reports indicate that post-translational modifications of Drp1, such as phosphorylation, ubiquitination, and SUMOylation, are important for proper mitochondrial morphology and function (49). The conjugation of SUMO to target proteins is carried out by the E2 enzyme Ubc9 and involves as a prerequisite the recruitment of Ubc9 to the modified protein (50, 51). We therefore examined the interaction between Ubc9 and Drp1. We expressed various domains of Drp1 singly or in combination as pLex fusion proteins and tested their interaction with VP16-Ubc9 in the yeast 2-hybrid system (Fig. 1). Our results indicate that Ubc9 binds to two distinct regions of Drp1 mapped to the GTPase and B domains. Interestingly, in the absence of the GTPase domain, the GED and Middle domains appear to interfere with the binding of Ubc9 to the B domain (Fig. 1). This indicates that the architecture of Drp1 is important for its interaction with Ubc9. Consistent with Drp1 serving as a target of SUMOylation, we also detect an interaction between Drp1 and conjugation competent forms of SUMO1, -2, and -3, although this interaction is substantially weaker (data not shown).
Figure 1.
Ubc9 interacts with the GTPase and B domains of Drp1 in the yeast 2-hybrid assay. L40 yeast cells were cotransformed with vectors expressing fusion proteins of LexA DNA binding domain (LexA) (bait) with full-length Drp1 (aa 1–736), GTPase (GTP) domain (aa 1–225), Middle domain (aa 227–521), no-GTPase domain (aa 227–736), B domain (aa 502–626), or GTPase-effector (GED) domain (aa 627–736) and vectors expressing Ubc9 as a fusion protein with the VP16 activation domain (VP16) (prey). Cotransformed colonies were grown on −Trp/Leu medium; serial yeast dilutions were plated. Expression of both plasmids is confirmed by growth in −Trp/Leu plates; interaction between proteins was assessed by growth of yeast on minimal medium plates (−Trp/Leu/His),indicating activation of the HIS reporter. None of the baits conferred histidine auxotrophy in the absence of the Ubc9 prey (data not shown).
Drp1 is a target of SUMOylation
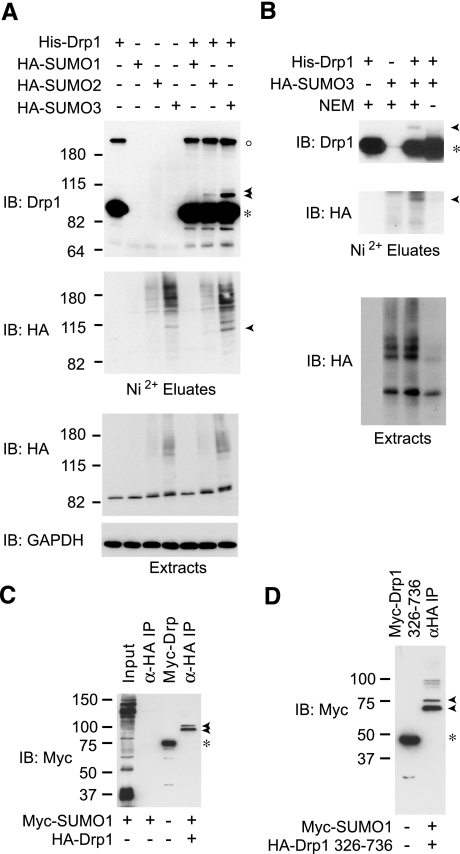
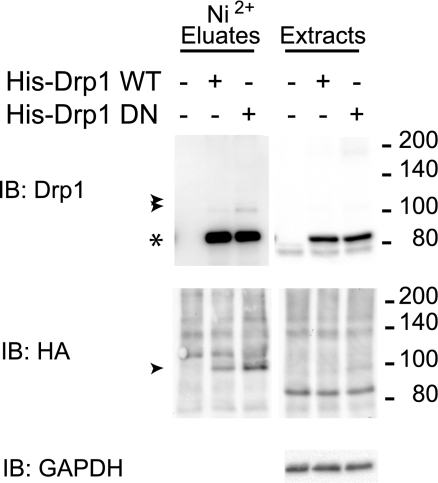
Recent data suggest that Drp1 is a target of SUMO1 modification (28), but the identity of SUMO conjugated forms as well as the SUMO isoform preference for Drp1 modification has not been examined. To this end, we have applied a validated cell culture expression approach that takes advantage of the covalent conjugation of SUMO and purification under denaturing conditions to examine under high-stringency conditions the SUMOylation of Drp1. As seen in Fig. 2A (Ni2+ eluate panels, arrowheads), analysis of hexahistidine-tagged Drp1 preparations isolated under denaturing conditions from HEK-293T cells coexpressing SUMO conjugating enzyme Ubc9 and each of the 3 major SUMO isoforms (SUMO1, -2, and -3) revealed that in addition to unmodified Drp1 (asterisk), a slower migrating Drp1 immunoreactive doublet is detected in cells expressing SUMO3 (arrowheads). On extended exposure, similar species are detected in SUMO2-expressing cells. Drp1-SUMO1 conjugates were difficult to identify under these conditions. The slower-migrating Drp1 species, with an apparent molecular mass of ∼100 kDa, are also immunoreactive toward the HA epitope present in the SUMO isoforms, consistent with their identity as SUMO-modified forms of Drp1.
Figure 2.
Drp1 is modified by SUMO1, SUMO2, and SUMO3. A) HEK-293 cells were transfected transiently with expression vectors for pcDNA3-Ubc9 and/or pcDNA3-HA-SUMO1, pcDNA3-HA-SUMO2, or pcDNA3-HA-SUMO3 and/or pcDNA3-V5-His-Drp1. Cells were lysed in the presence of 20 mM NEM, and His-Drp1 was isolated via Ni2+ chelate chromatography under denaturing conditions. Proteins from Ni2+ eluates and extracts were visualized by Western immunoblot (IB) analysis using antibodies against Drp1, HA-epitope, and GAPDH. Migrations of unmodified and SUMO-conjugated forms of Drp1 are indicated by asterisk and arrowheads, respectively. Open circle indicates larger apparent molecular mass Drp1 forms likely representing entangled SDS-resistant oligomers. B) SUMO modification of Drp1 is Ubc9 and SUMO3 dependent and NEM sensitive. HEK-293 cells transiently transfected with pcDNA3-Ubc9 and pcDNA3-HA-SUMO3 and/or pcDNA3-V5-His-Drp1 were lysed in the presence or absence of 20 mM NEM and analyzed as in A. C) Cells were cotransfected with HA-Drp1 and Myc-SUMO1, or else transfected with Myc-SUMO1 alone, and cell lysates were immunoprecipitated using anti-HA antibodies as indicated (α-HA IP), then immunoblotted using anti-Myc antibodies. Arrowheads indicate SUMO-modified Drp1 forms. Myc-Drp1 expression (asterisk) identifies the size of unmodified Drp1, as it is identical in size to HA-Drp1. D) Cells were cotransfected with HA-Drp1 (aa 326–736) and Myc-SUMO1, and extracts were immunoprecipitated with anti-HA antibodies and then immunoblotted for Myc as in C. Arrowheads indicate SUMO-modified Drp1 (aa 326–736) forms. Myc-Drp1 (aa 326–736) expression (asterisk) identifies the size of unmodified Drp1, which is identical in size to HA-Drp1 (aa 326–736). Migrations of molecular mass standards (kDa) are at left.
Interestingly, we do observe Drp1 immunoreactive species migrating at significantly higher positions in the gel (Fig. 2A, open circle). Such species, however, are not immunoreactive for the HA epitope (Fig. 2A, lanes 5–7) and are SUMO independent (Fig. 2A, lane 1). Thus, these high-molecular-mass species are unlikely to be SUMO modified. Whether these species correspond to the high-molecular-mass forms Harder et al. (28) interpreted as Drp1 modified by poly-SUMO1 chains remains to be determined. Given that Drp1 likely self-assembles into large multimeric complexes in vivo, as does dynamin, these apparent high-molecular-mass species may represent entangled SDS-resistant oligomers.
To further validate the identification of the slow-migrating ∼100-kDa doublets as Drp1-SUMO conjugates, we omitted the alkylating agent NEM during extract preparation. NEM is an agent that irreversibly inhibits SUMO proteases by alkylating a catalytic cysteine in their active site. As seen in the HA blot of cellular extracts (Fig. 2B, extracts), omission of this reagent leads to loss of most SUMO conjugates. Consistent with the assignment as SUMOylated Drp1, the slow migrating Drp1 and HA immunoreactive species are also lost in the absence of NEM despite the apparent higher recovery of Drp1 under these conditions (Fig. 2B, eluates).
Notably, although the above data indicate that SUMO3 conjugates are more readily detected, this is likely a simple reflection of the relative expression and overall conjugation of these tagged SUMO isoforms, as revealed by analysis of the cell extracts (Fig. 2A, extracts). Using an alternative approach in Cos7 cells, where Myc-epitope tagged SUMO1 conjugates are readily detected (Fig. 2C), we also recover the slower migrating doublet by immunoprecipitating HA-Drp1 from cells overexpressing Myc-SUMO1 (Fig. 2C). Notably, analysis of an N-terminally truncated form of Drp1 in this assay indicates that the slow-migrating SUMO-containing species correspond to SUMO1–Drp1 conjugates, since their apparent molecular mass is appropriately reduced in the case of the truncated Drp1 (326–736) form (Fig. 2D). Furthermore, the data indicate that the N-terminal GTPase region is dispensable for SUMO conjugation to Drp1. Taken together, our results indicate that Drp1 is a target of SUMO modification by all three conjugatable SUMO isoforms and that this modification preferentially occurs in the C-terminal half of the protein.
Drp1 is SUMO-modified at two lysine clusters located in the B domain
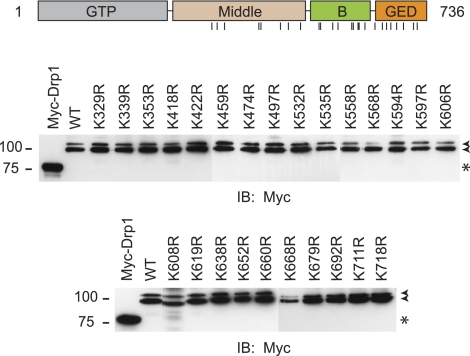
The above data indicate that an N-terminally truncated Drp1 form lacking the GTPase and part of the Middle domain is still SUMO-modified (Fig. 2D). Analysis of the Drp1 sequence and of this fragment indicates no clear instances of the SUMOylation consensus sequence, suggesting that the modification may occur at nonconsensus sites. In an effort to identify such modification sites, we performed a systematic mutational analysis. We replaced individually all 25 lysine residues present within the region comprising residues 326–736 and examined the SUMOylation of the resulting full-length protein. Remarkably, the slow-migrating doublet remains clearly present in all mutants (Fig. 3). This finding suggests that Drp1 SUMOylation likely occurs at multiple sites, and given their noncanonical nature, significant redundancy may be present.
Figure 3.
Mutational analysis of potential SUMO-acceptor lysines. Schematic diagram of Drp1 domain organization (top panel); vertical lines identify lysine residues within aa 326–736 mutated to arginines in the bottom panels. Cells were cotransfected with Myc-SUMO1 and WT HA-Drp1 (aa 1–736; splice variant 1) or the indicated Drp1 mutants (single amino acid letter code). Extracts were immunoprecipitated with anti-HA antibodies and then immunoblotted for Myc. Arrowheads indicate SUMO-modified Drp1. Myc-Drp1 expression (asterisk) identifies the size of unmodified HA-Drp1. Migrations of molecular mass standards (kDa) are at left.
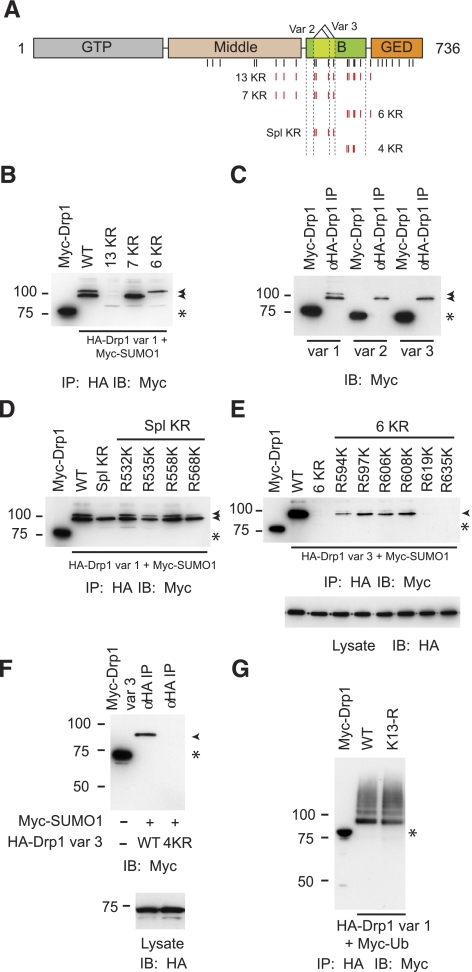
We next took a group-mutational approach to identify lysines required for SUMO modification (Fig. 4A, B). We found that joint substitution of 13 lysine residues to arginines within this region (13 KR) led to a complete elimination of the doublet of SUMO conjugated species (Fig. 4B). Mutation of the 7 N-terminal lysines (7 KR) resulted in loss of the upper SUMO-conjugated species, while replacement of the 6 C-terminal lysines (6 KR) led to the corresponding loss of the lower one (Fig. 4B). This suggests that the two modified forms likely represent Drp1 species modified at two different sets of lysines within this region, with the N-terminal ones responsible for the upper species and the C-terminal ones for the lower species.
Figure 4.
Noncanonical SUMOylation sites are present within the B domain of Drp1. A) Schematic diagram of Drp1; vertical lines identify lysine residues mutated to arginines in the other panels. In the 5 constructs at bottom, indicated Lys residues (red) were converted to Arg residues, and constructs were named as shown. B) Cells were cotransfected with Myc-SUMO1 and WT HA-Drp1 (aa 1–736; splice variant 1) or the indicated Drp1 mutants shown in A. Extracts were immunoprecipitated with anti-HA antibodies and then immunoblotted for Myc. Arrowheads indicate SUMO-modified Drp1, as in subsequent panels. Myc-Drp1 expression (asterisk) identifies size of unmodified HA-Drp1, as in subsequent panels. C) Cells were transfected and analyzed as in B, examining the 3 splice variants of WT Drp1. D) Cells were transfected and analyzed as in B, with indicated mutated residues in Drp1 Spl KR individually returned to the Lys residues found in the WT form. E) Cells were transfected and analyzed as in D, though HA-Drp1 was the variant 3 form, and the residues were reverted from the 6 KR mutant. Bottom panel: total cell lysates were immunoblotted for HA to demonstrate equal expression of all Drp1 forms. F) Cells were cotransfected with Myc-SUMO1 and either WT or 4 KR mutant HA-Drp1 (splice variant 3). Extracts were immunoprecipitated with anti-HA antibodies and then immunoblotted for Myc (top panel) or HA (bottom panel). G) Cells were cotransfected with Myc-ubiquitin and either WT or 13 KR mutant HA-Drp1 (splice variant 1). Extracts were immunoprecipitated with anti-HA antibodies and then immunoblotted for Myc.
Human Drp1 has three different splice variants (52) comprising the full-length 736-aa protein (variant 1) as well as forms lacking 27 (variant 2) or 37 (variant 3) amino acid residues in the B domain (Fig. 4A). Since four of the lysines within the 7 KR segment are within the differentially spliced region, we compared the SUMO modification of the three different variants. As shown in Fig. 4C, the doublet is present only in variant 1, which suggests that lysines necessary for the generation of the upper SUMO-conjugated species in the doublet are contained within the alternatively spliced region. Indeed, analysis of a mutant Drp1 (var. 1) in which the 4 lysines within the alternatively spliced region are replaced by arginines (Spl KR) indicated a loss of the upper modified species (Fig. 4D). To examine the contribution of each residue, we systematically reintroduced individual lysines in the context of the Spl KR mutant. As seen in Fig. 4D, reintroduction of any of the 4 lysines leads to a restoration of the upper modified species, indicating that lysines 532, 535, 558, and 568 can function as SUMO-acceptor residues within this region.
We followed a similar approach to identify the lysines within the 6 KR region that contribute to the generation of the lower modified species. We performed the analysis using Drp1 variant 3, which lacks the upper form. As expected, replacement of all six lysine residues in the context of variant 3 led to an essentially complete loss of SUMOylation. Reintroduction of any of the four N-terminal lysines (K594, K597, K606, K608) led to a partial restoration of SUMOylation, whereas reintroduction of either of the two C-terminal ones did not (Fig. 4E). Each of these variants was expressed at comparable levels, as revealed by the HA immunoblot of cellular extracts (Fig. 4E, bottom). Since reintroduction of single lysine residues did not fully restore SUMOylation to the level observed for variant 3, the data suggest that all four lysines likely contribute to the lower modified species. Consistent with this analysis, block substitution of these 4 lysines in variant 3 led to a complete loss of Drp1 SUMOylation (Fig. 4F). Since in some cases, lysine residues are sites for both SUMOylation and ubiquitination, we also examined the ubiquitination of the SUMOylation-deficient Drp1 (variant1) 13 KR mutant. As shown in Fig. 4G, it appears that the lysine residues within Drp1 involved in SUMO modification are dispensable for ubiquitination, since the 13 KR mutant is ubiquitinated to levels comparable to WT Drp1. Taken together, the mapping analysis clearly indicates that Drp1 SUMOylation occurs mainly at two lysine clusters within the B domain. Given the noncanonical nature of these sites, it is likely that the ability of Ubc9 to interact with this region (Fig. 1) allows the modification of redundant lysines without strict adherence to the consensus. These sites, however, are dispensable for ubiquitination of Drp1.
SUMOylation-deficient Drp1 is recruited to mitochondria in response to staurosporine
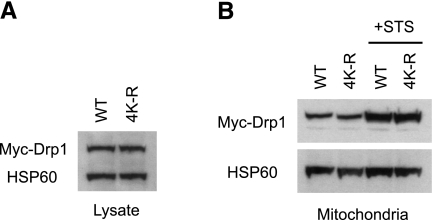
Our mapping of the major SUMOylation sites in Drp1 provides the opportunity to begin exploring the role of this modification in Drp1 function. Mitochondrial recruitment is central to the role of Drp1 in normal mitochondrial fission and apoptotic MOMP in response to proapoptotic agents such as staurosporine (22). Analysis of cells overexpressing WT or SUMOylation deficient (4 KR) Myc-Drp1 (var. 3) revealed that the basal and staurosporine-stimulated recruitment of ectopically expressed Drp1 to mitochondria is not affected by loss of SUMO modification (Fig. 5). The data, therefore, indicate that preventing SUMO modification of exogenous Drp1 does not lead to an overall dominant-negative effect on recruitment of Drp1 to the mitochondria.
Figure 5.
Preventing Drp1 SUMOylation does not alter its translocation to mitochondria. HeLa cells were transfected with Myc-tagged, WT Drp1 variant 3 or the 4 KR mutant form that cannot be SUMOylated. A) Total cell lysates were immunoblotted for Myc. HSP60 levels were monitored on the same immunoblot to ensure equal protein loading. B) Cells expressing WT or 4 KR mutant Drp1 were either left untreated or else treated with staurosporine (STS) in the presence of zVAD-fmk for 8 h. Mitochondria were isolated; equal protein aliquots were resolved by SDS-PAGE, then immunoblotted for Myc and HSP60.
Enhanced SUMOylation of a dominant-negative form of Drp1
The GTPase activity of Drp1 is essential for its ability to promote mitochondrial fission (19). The SUMO modification of Drp1 could therefore influence Drp1 function through an association with its GTPase cycle. Within the GTPase domain of Drp1, the conserved P loop lysine at position 38, which in dynamin forms hydrogen bonds with residues from switch II (53), plays a critical role in Drp1 function. An alanine substitution at this position severely hampers Drp1 function and confers to the protein a dominant-negative behavior. The resulting block in fission leads to unopposed mitochondrial fusion (19). We therefore examined the effect of the K38A substitution on Drp1 SUMOylation. Notably, as shown in Fig. 6, we consistently observed an increase in SUMO3 modification of the dominant-negative K38A mutant compared to WT Drp1, revealing that the SUMOylation of Drp1 is linked to its activity cycle.
Figure 6.
Enhancement of SUMO modification of dominant-negative Drp1. HEK-293 cells were transiently transfected with pcDNA3-V5-Ubc9, pcDNA3-HA-SUMO3, and/or pcDNA3-V5-His-Drp1 WT or pcDNA3-V5-His-Drp1 K38A, and CS2+empty vector. Protein extracts were obtained in the presence of 20 mM NEM, processed as in Fig. 2A, and analyzed by immunoblotting. Arrowheads indicate modified doublet; asterisk indicates unmodified Drp1.
DISCUSSION
The dynamin-related protein, Drp1, is a fission protein and a key molecular regulator of mitochondria and peroxisome function (54,55,56). Drp1 is post-translationally modified, and some of these modifications are thought to regulate the ability of Drp1 to promote fission (49). Although the SUMO family of proteins is widely distributed (57), the subcellular localizations are distinct. SUMO1 is localized at the nuclear membrane and nuclear bodies, SUMO2 is mainly cytoplasmic, and SUMO3 is mostly found in the nuclear bodies (58, 59). Although this finding suggests individual forms may perform specific biological roles, a significant overlap exists, since knockout of SUMO1 in mice is not lethal (60). The members of the SUMO protein family are also regulated distinctively by post-translational modifications (61). SUMO2 and SUMO3 form multimers in vitro and in vivo due to internal SUMOylation consensus sites. SUMO1 is incorporated into these chains, but the lack of SUMOylation sites is thought to prevent SUMO1-chain formation in vivo, although such chains are readily formed in vitro (59, 61, 62). Drp1 has been reported to be modified by SUMO1 in an “all-or-none” manner, and very high molecular mass forms were interpreted as Drp1-harboring covalent SUMO1 chains (28). However, we have clearly demonstrated that Drp1 is a target of SUMOylation by all three SUMO isoforms but without evidence of large chain formation, since these species migrate as a ∼100-kDa doublet. With regard to the different SUMO isoforms, it is important to note that oxidative stress generally leads to significantly enhanced SUMO2/3 modification. Given the central role of mitochondria in pathological oxidative stress, it will be interesting to examine whether SUMO2/3 modification of Drp1 or other mitochondrial components plays a role in the adaptive and maladaptive mitochondrial response to oxidative stress.
The dynamin family of proteins is characterized by distinct functional domains, including a GTPase domain, a Middle domain, and an effector (or GED) domain. The B domain in Drp1, which is absent in dynamin, is the most variable region through evolution, and no specific role has been assigned to this domain. Here we show that the E2 SUMO-conjugating enzyme Ubc9 binds to the B domain of Drp1 in a yeast 2-hybrid assay (Fig. 1). Recruitment of Ubc9 is a prerequisite for SUMO modification, and this occurs by direct recognition by Ubc9 of canonical SUMOylation sites. Ubc9 may also be recruited via alternative interactions not involving its active site. In this case, SUMO modification often occurs at multiple, noncanonical sites. Drp1 appears to follow the latter pattern, given the multivalent interaction between Drp1 and Ubc9 and the nonconsensus nature of the modification sites. For example, Ubc9 interaction with the GTPase domain and the B domain may orient the active site of Ubc9 in proximity to the lysine clusters within the B domain that are targeted for SUMOylation. This is particularly attractive given the extensive intra- and intermolecular interactions that Drp1 engages in (16). Given that lysine 597 in the second cluster resides within the sequence SKAEE, it is possible that the interaction of Ubc9 with the B domain involves in part the recognition of this sequence, which has some of the features of the canonical SUMOylation motif.
Recently, analyses of proteins that can interact with Ubc9 in a similar manner, such as DAXX (63) or Kap1 (64), indicate that they act as SUMO E3 ligases by serving as bridging partners to recruit Ubc9 to target proteins. This is analogous to the mechanism of action of ubiquitin E3 ligases of the Skp1-Cullin-F-Box complex (SCF) type. Notably, DAXX, and Kap1 are SUMOylated at multiple noncanonical sites, as is the case for Drp1. It is therefore plausible that Drp1 recruitment of Ubc9 may facilitate the SUMOylation of other proteins it interacts with during its normal functions. Thus, the interaction of Drp1 with Ubc9 may have functional consequences that extend beyond the SUMOylation of Drp1 itself.
Our mutational analysis indicates that 8 lysines within 2 clusters in the B domain function as SUMO-acceptor residues. Notably, four of these residues are located within, or directly abutting, the differentially spliced region of the B domain. Although no clear functional difference has been detected between the three Drp1 variants, the reduced SUMO modification of variants 2 and 3 argue that differential SUMOylation may contribute to potential functional heterogeneity among variants. During the review of this report, Zunino et al. (65) indicated that a SUMO-modified Drp1 species corresponding to the slow migrating band that we observe in our experiments is modified in a cell cycle-dependent manner. It will be interesting to determine whether the Drp1 variants play different roles in this process.
Our identification of the major SUMO modification sites in Drp1 is an essential step toward a direct assessment of the role of this modification in Drp1 function. On the basis of correlational data, it has been proposed that during apoptosis, “locking” of SUMO1-modified Drp1 to the mitochondrial membrane alters Drp1 cycling between the cytoplasm and mitochondria in a Bax/Bak-dependent manner prior to apoptotic fission as well as on onset of mitosis (31, 66). Our data show that in cells ectopically expressing a SUMOylation-deficient form of Drp1, the basal and staurosporine-stimulated Drp1 translocation to mitochondria is unaffected, indicating that loss of SUMO modification in the ectopic Drp1 pool does not lead to a dominant effect with respect to recruitment in an unsynchronized population of actively growing cells. SUMO modification, however, may influence other aspects of Drp1 function downstream of mitochondrial recruitment. Alternatively, a full assessment of the role of Drp1 SUMOylation may require the replacement of the endogenous complement of Drp1 with a non-SUMOylatable form. SUMO modification also alters the half-life of target proteins both in a positive or negative direction. Although it has been proposed that SUMOylation may stabilize Drp1 (28, 31, 32, 66), our results indicate that disruption of SUMO acceptor lysines in Drp1 affects neither protein stability nor ubiquitination of Drp1, arguing that Drp1 SUMOylation is not directly linked to its degradation.
The GTPase activity of members of the dynamin family of proteins is required for their ability to promote vacuolar endocytosis and mitochondrial fission (19, 67, 68). Mutations in the GTP-binding (GTPase) domain alter GTP binding and confer dominant-negative characteristics (67). A dominant form of Drp1 containing the K38A mutation, corresponding to the K44A mutation in dynamin, is defective in GTP binding and hydrolysis. Overexpression of this mutant alters mitochondrial morphology by increasing tubular mitochondria at the center of the cell (19, 67, 69). Cellular localization studies indicate that Drp1 K38A forms aggregates in the cytoplasm that also likely contain WT endogenous Drp1. Thus, an intact GTPase is critical for Drp1 function and localization (16, 19, 70,71,72,73,74). Our analysis of the SUMO modification of Drp1 K38A indicates that the mutation leads to significantly enhanced SUMO3 modification. This observation argues that the SUMO modification of Drp1 is linked to its activity cycle. Since sedimentation experiments indicate that the dominant-negative K38A Drp1 may be impaired in its ability to disassemble higher-order complexes (16), SUMO modification may, therefore, be part of a signal or mechanism for disassembly, a function reminiscent of the role of SUMOylation in the disassembly of septins during cell division in S. cerevisiae (75). The mechanism for enhanced SUMOylation of this mutant, however, is unclear. In this regard, Zunino et al. (65) have recently provided evidence that the SUMO protease SENP5 translocates from the nucleoli to the mitochondria at the onset of mitosis. In this context, the enhanced SUMO3 modification of Drp1 K38A that we observe in our experiments may be a consequence of its failure to translocate to mitochondria where SENP5-catalyzed de-SUMOylation may normally take place. Notably, recent data indicate that, similar to Drp1, dynamin-1 also associates with SUMO1 and Ubc9. Unlike Drp1, however, this interaction appears to be mediated by the GED domain (76, 77). Although the in vivo SUMO acceptor lysines in dynamin have not been identified, it will be interesting to examine whether SUMOylation of dynamin-1 is also enhanced in the case of the analogous K44A dominant-negative mutant.
Clearly, post-translational modifications such as phosphorylation and ubiquitination regulate Drp1, and the molecular interplay between such modifications may be important for Drp1 function (23, 24). It will therefore be interesting to test whether phosphorylation of Drp1 alters its SUMOylation, or vice versa. Our identification of the principal sites of SUMOylation in Drp1 provides critical tools to define the biological importance of this post-translational modification and its relation to the function of Drp1.
Recent advances in understanding the role of the fission protein Drp1 in mitochondria function indicate Drp1 may be a central regulator of devastating neurodegenerative disorders such as Parkinson’s disease, Alzheimer’s disease and other neuropathies (15, 25, 78,79,80,81). In light of recent data linking SUMOylation with the pathogenesis of neurodegenerative diseases (82, 83), identifying signaling events and molecular mechanisms regulating Drp1 post-translational modifications responsible for mitochondria homeostasis will facilitate the identification of potential therapeutic targets.
Acknowledgments
This work was supported by U.S. Public Health Service grant DK61656 (J.A.I.), the Program for Neurology Research and Discovery, the A. Alfred Taubman Medical Research Institute, U.S. National Institutes of Health (NIH) grant NIHU01 DK076160 (C.F.-R., E.L.F., Y.H., P.J.K.), and in part by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke (NINDS), NIH (J.S., C.-R.C., C.B.) as well as the Agence Nationale de La Recherche sur le SIDA, Fondation pour La Recherche Medicale, the Ligue contre le Cancer, and the Université Paris Sud (D.A.). This work utilized the sequencing core of the Michigan Diabetes Research and Training Center, funded by National Institute of Diabetes and Digestive and Kidney Diseases grant NIH5P60 DK20572. The authors also thank J. Nagle and D. Kauffman (NINDS DNA Sequencing Facility) for DNA sequencing.
References
- Cerveny K L, Tamura Y, Zhang Z, Jensen R E, Sesaki H. Regulation of mitochondrial fusion and division. Trends Cell Biol. 2007;17:563–569. doi: 10.1016/j.tcb.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Griparic L, van der Wel N N, Orozco I J, Peters P J, van der Bliek A M. Loss of the intermembrane space protein Mgm1/OPA1 induces swelling and localized constrictions along the lengths of mitochondria. J Biol Chem. 2004;279:18792–18798. doi: 10.1074/jbc.M400920200. [DOI] [PubMed] [Google Scholar]
- Santel A, Frank S, Gaume B, Herrler M, Youle R J, Fuller M T. Mitofusin-1 protein is a generally expressed mediator of mitochondrial fusion in mammalian cells. J Cell Sci. 2003;116:2763–2774. doi: 10.1242/jcs.00479. [DOI] [PubMed] [Google Scholar]
- Wells R C, Picton L K, Williams S C, Tan F J, Hill R B. Direct binding of the dynamin-like GTPase, Dnm1, to mitochondrial dynamics protein Fis1 is negatively regulated by the Fis1 N-terminal arm. J Biol Chem. 2007;282:33769–33775. doi: 10.1074/jbc.M700807200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, Krueger E W, Oswald B J, McNiven M A. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy A D, McCaffery J M, Shaw J M. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chan D C. Critical dependence of neurons on mitochondrial dynamics. Curr Opin Cell Biol. 2006;18:453–459. doi: 10.1016/j.ceb.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Baloh R H. Mitochondrial dynamics and peripheral neuropathy. Neuroscientist. 2008;14:12–18. doi: 10.1177/1073858407307354. [DOI] [PubMed] [Google Scholar]
- Chan D C. Mitochondrial dynamics in disease. N Engl J Med. 2007;356:1707–1709. doi: 10.1056/NEJMp078040. [DOI] [PubMed] [Google Scholar]
- Ghafourifar P, Mousavizadeh K, Parihar M S, Nazarewicz R R, Parihar A, Zenebe W J. Mitochondria in multiple sclerosis. Front Biosci. 2008;13:3116–3126. doi: 10.2741/2913. [DOI] [PubMed] [Google Scholar]
- Zeviani M, Carelli V. Mitochondrial disorders. Curr Opin Neurol. 2007;20:564–571. doi: 10.1097/WCO.0b013e3282ef58cd. [DOI] [PubMed] [Google Scholar]
- Alexander C, Votruba M, Pesch U E, Thiselton D L, Mayer S, Moore A, Rodriguez M, Kellner U, Leo-Kottler B, Auburger G, Bhattacharya S S, Wissinger B. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet. 2000;26:211–215. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- Delettre C, Lenaers G, Griffoin J M, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, Astarie-Dequeker C, Lasquellec L, Arnaud B, Ducommun B, Kaplan J, Hamel C P. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- Zuchner S, Mersiyanova I V, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali E L, Zappia M, Nelis E, Patitucci A, Senderek J, Parman Y, Evgrafov O, Jonghe P D, Takahashi Y, Tsuji S, Pericak-Vance M A, Quattrone A, Battaloglu E, Polyakov A V, Timmerman V, Schroder J M, Vance J M. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- Waterham H R, Koster J, van Roermund C W, Mooyer P A, Wanders R J, Leonard J V. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- Zhu P P, Patterson A, Stadler J, Seeburg D P, Sheng M, Blackstone C. Intra- and intermolecular domain interactions of the C-terminal GTPase effector domain of the multimeric dynamin-like GTPase Drp1. J Biol Chem. 2004;279:35967–35974. doi: 10.1074/jbc.M404105200. [DOI] [PubMed] [Google Scholar]
- Ingerman E, Perkins E M, Marino M, Mears J A, McCaffery J M, Hinshaw J E, Nunnari J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland D L, van der Bliek A M. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Shurland D L, Ryazantsev S N, van der Bliek A M. A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Stone A, Chipuk J E, Ingerman E, Song C, Yoo C, Kuwana T, Kurth M J, Shaw J T, Hinshaw J E, Green D R, Nunnari J. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etxebarria A, Terrones O, Yamaguchi H, Landajuela A, Landeta O, Antonsson B, Wang H G, Basanez G. Endophilin B1/Bif-1 stimulates BAX activation independently from its capacity to produce large-scale membrane morphological rearrangements. J Biol Chem. 2009;284:4200–4212. doi: 10.1074/jbc.M808050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S, Gaume B, Bergmann-Leitner E S, Leitner W W, Robert E G, Catez F, Smith C L, Youle R J. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- Cereghetti G M, Stangherlin A, Martins de Brito O, Chang C R, Blackstone C, Bernardi P, Scorrano L. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci U S A. 2008;105:15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C R, Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem. 2007;282:21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- Cho D H, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, Lipton S A. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribbs J T, Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 2007;8:939–944. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X J, Lu Y F, Li S A, Kaitsuka T, Sato Y, Tomizawa K, Nairn A C, Takei K, Matsui H, Matsushita M. CaM kinase I alpha-induced phosphorylation of Drp1 regulates mitochondrial morphology. J Cell Biol. 2008;182:573–585. doi: 10.1083/jcb.200802164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder Z, Zunino R, McBride H. Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr Biol. 2004;14:340–345. doi: 10.1016/j.cub.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- Wasiak S, Zunino R, McBride H M. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J Cell Biol. 2007;177:439–450. doi: 10.1083/jcb.200610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunino R, Schauss A, Rippstein P, Andrade-Navarro M, McBride H M. The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J Cell Sci. 2007;120:1178–1188. doi: 10.1242/jcs.03418. [DOI] [PubMed] [Google Scholar]
- Ulrich H D. Mutual interactions between the SUMO and ubiquitin systems: a plea of no contest. Trends Cell Biol. 2005;15:525–532. doi: 10.1016/j.tcb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- Su H L, Li S S. Molecular features of human ubiquitin-like SUMO genes and their encoded proteins. Gene. 2002;296:65–73. doi: 10.1016/s0378-1119(02)00843-0. [DOI] [PubMed] [Google Scholar]
- Bohren K M, Nadkarni V, Song J H, Gabbay K H, Owerbach D. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J Biol Chem. 2004;279:27233–27238. doi: 10.1074/jbc.M402273200. [DOI] [PubMed] [Google Scholar]
- Owerbach D, McKay E M, Yeh E T, Gabbay K H, Bohren K M. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem Biophys Res Commun. 2005;337:517–520. doi: 10.1016/j.bbrc.2005.09.090. [DOI] [PubMed] [Google Scholar]
- Sampson D A, Wang M, Matunis M J. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- Chun T H, Itoh H, Subramanian L, Iniguez-Lluhi J A, Nakao K. Modification of GATA-2 transcriptional activity in endothelial cells by the SUMO E3 ligase PIASy. Circ Res. 2003;92:1201–1208. doi: 10.1161/01.RES.0000076893.70898.36. [DOI] [PubMed] [Google Scholar]
- Pichler A, Gast A, Seeler J S, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- Sachdev S, Bruhn L, Sieber H, Pichler A, Melchior F, Grosschedl R. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 2001;15:3088–3103. doi: 10.1101/gad.944801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E T, Gong L, Kamitani T. Ubiquitin-like proteins: new wines in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- Chupreta S, Holmstrom S, Subramanian L, Iniguez-Lluhi J A. A small conserved surface in SUMO is the critical structural determinant of its transcriptional inhibitory properties. Mol Cell Biol. 2005;25:4272–4282. doi: 10.1128/MCB.25.10.4272-4282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Durrin L K, Wilkinson T A, Krontiris T G, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson M D, Li Q J, Kieckhafer K, Dudek D, Whorton M R, Sunahara R K, Iniguez-Lluhi J A, Martens J R. SUMO modification regulates inactivation of the voltage-gated potassium channel Kv1.5. Proc Natl Acad Sci U S A. 2007;104:1805–1810. doi: 10.1073/pnas.0606702104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- Arnoult D, Grodet A, Lee Y J, Estaquier J, Blackstone C. Release of OPA1 during apoptosis participates in the rapid and complete release of cytochrome c and subsequent mitochondrial fragmentation. J Biol Chem. 2005;280:35742–35750. doi: 10.1074/jbc.M505970200. [DOI] [PubMed] [Google Scholar]
- Arnoult D, Rismanchi N, Grodet A, Roberts R G, Seeburg D P, Estaquier J, Sheng M, Blackstone C. Bax/Bak-dependent release of DDP/TIMM8a promotes Drp1-mediated mitochondrial fission and mitoptosis during programmed cell death. Curr Biol. 2005;15:2112–2118. doi: 10.1016/j.cub.2005.10.041. [DOI] [PubMed] [Google Scholar]
- Santel A, Frank S. Shaping mitochondria: The complex posttranslational regulation of the mitochondrial fission protein DRP1. IUBMB Life. 2008;60:448–455. doi: 10.1002/iub.71. [DOI] [PubMed] [Google Scholar]
- Bernier-Villamor V, Sampson D A, Matunis M J, Lima C D. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–356. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- Rodriguez M S, Dargemont C, Hay R T. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- Kamimoto T, Nagai Y, Onogi H, Muro Y, Wakabayashi T, Hagiwara M. Dymple, a novel dynamin-like high molecular weight GTPase lacking a proline-rich carboxyl-terminal domain in mammalian cells. J Biol Chem. 1998;273:1044–1051. doi: 10.1074/jbc.273.2.1044. [DOI] [PubMed] [Google Scholar]
- Niemann H H, Knetsch M L, Scherer A, Manstein D J, Kull F J. Crystal structure of a dynamin GTPase domain in both nucleotide-free and GDP-bound forms. EMBO J. 2001;20:5813–5821. doi: 10.1093/emboj/20.21.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain M, Mathai J P, McBride H M, Shore G C. Endoplasmic reticulum BIK initiates DRP1-regulated remodelling of mitochondrial cristae during apoptosis. EMBO J. 2005;24:1546–1556. doi: 10.1038/sj.emboj.7600592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader M. Shared components of mitochondrial and peroxisomal division. Biochim Biophys Acta. 2006;1763:531–541. doi: 10.1016/j.bbamcr.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Shin H W, Shinotsuka C, Torii S, Murakami K, Nakayama K. Identification and subcellular localization of a novel mammalian dynamin-related protein homologous to yeast Vps1p and Dnm1p. J Biochem. 1997;122:525–530. doi: 10.1093/oxfordjournals.jbchem.a021784. [DOI] [PubMed] [Google Scholar]
- Li M, Guo D, Isales C M, Eizirik D L, Atkinson M, She J X, Wang C Y. SUMO wrestling with type 1 diabetes. J Mol Med. 2005;83:504–513. doi: 10.1007/s00109-005-0645-5. [DOI] [PubMed] [Google Scholar]
- Dorval V, Fraser P E. SUMO on the road to neurodegeneration. Biochim Biophys Acta. 2007;1773:694–706. doi: 10.1016/j.bbamcr.2007.03.017. [DOI] [PubMed] [Google Scholar]
- Matic I, van Hagen M, Schimmel J, Macek B, Ogg S C, Tatham M H, Hay R T, Lamond A I, Mann M, Vertegaal A C. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol Cell Proteomics. 2008;7:132–144. doi: 10.1074/mcp.M700173-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F P, Mikkonen L, Toppari J, Palvimo J J, Thesleff I, Janne O A. Sumo-1 function is dispensable in normal mouse development. Mol Cell Biol. 2008;28:5381–5390. doi: 10.1128/MCB.00651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich H D. The fast-growing business of SUMO chains. Mol Cell. 2008;32:301–305. doi: 10.1016/j.molcel.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Tatham M H, Jaffray E, Vaughan O A, Desterro J M, Botting C H, Naismith J H, Hay R T. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- Lin D Y, Huang Y S, Jeng J C, Kuo H Y, Chang C C, Chao T T, Ho C C, Chen Y C, Lin T P, Fang H I, Hung C C, Suen C S, Hwang M J, Chang K S, Maul G G, Shih H M. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol Cell. 2006;24:341–354. doi: 10.1016/j.molcel.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Ivanov A V, Peng H, Yurchenko V, Yap K L, Negorev D G, Schultz D C, Psulkowski E, Fredericks W J, White D E, Maul G G, Sadofsky M J, Zhou M M, Rauscher F J., 3rd PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell. 2007;28:823–837. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunino R, Braschi E, Xu L, McBride H M. Translocation of SenP5 from the nucleoli to the mitochondria modulates DRP1-dependent fission during mitosis. J Biol Chem. 2009;284:17783–17795. doi: 10.1074/jbc.M901902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braschi E, Zunino R, McBride H M. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 2009;10:748–754. doi: 10.1038/embor.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Bliek A M, Redelmeier T E, Damke H, Tisdale E J, Meyerowitz E M, Schmid S L. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vater C A, Raymond C K, Ekena K, Howald-Stevenson I, Stevens T H. The VPS1 protein, a homolog of dynamin required for vacuolar protein sorting in Saccharomyces cerevisiae, is a GTPase with two functionally separable domains. J Cell Biol. 1992;119:773–786. doi: 10.1083/jcb.119.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damke H, Baba T, Warnock D E, Schmid S L. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleazard W, McCaffery J M, King E J, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw J M. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrousse A M, Zappaterra M D, Rube D A, van der Bliek A M. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- Pitts K R, Yoon Y, Krueger E W, McNiven M A. The dynamin-like protein DLP1 is essential for normal distribution and morphology of the endoplasmic reticulum and mitochondria in mammalian cells. Mol Biol Cell. 1999;10:4403–4417. doi: 10.1091/mbc.10.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, Pitts K R, McNiven M A. Mammalian dynamin-like protein DLP1 tubulates membranes. Mol Biol Cell. 2001;12:2894–2905. doi: 10.1091/mbc.12.9.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Sheu S S, Robotham J L, Yoon Y. Mitochondrial fission mediates high glucose-induced cell death through elevated production of reactive oxygen species. Cardiovasc Res. 2008;79:341–351. doi: 10.1093/cvr/cvn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E S, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147:981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pountney D L, Raftery M J, Chegini F, Blumbergs P C, Gai W P. NSF, Unc-18-1, dynamin-1 and HSP90 are inclusion body components in neuronal intranuclear inclusion disease identified by anti-SUMO-1-immunocapture. Acta Neuropathol. 2008;116:603–614. doi: 10.1007/s00401-008-0437-4. [DOI] [PubMed] [Google Scholar]
- Mishra R K, Jatiani S S, Kumar A, Simhadri V R, Hosur R V, Mittal R. Dynamin interacts with members of the sumoylation machinery. J Biol Chem. 2004;279:31445–31454. doi: 10.1074/jbc.M402911200. [DOI] [PubMed] [Google Scholar]
- Leinninger G M, Backus C, Sastry A M, Yi Y B, Wang C W, Feldman E L. Mitochondria in DRG neurons undergo hyperglycemic mediated injury through Bim, Bax and the fission protein Drp1. Neurobiol Dis. 2006;23:11–22. doi: 10.1016/j.nbd.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Wang X, Su B, Fujioka H, Zhu X. Dynamin-like protein 1 reduction underlies mitochondrial morphology and distribution abnormalities in fibroblasts from sporadic Alzheimer’s disease patients. Am J Pathol. 2008;173:470–482. doi: 10.2353/ajpath.2008.071208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ouyang Y, Yang L, Beal M F, McQuibban A, Vogel H, Lu B. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci U S A. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men X, Wang H, Li M, Cai H, Xu S, Zhang W, Xu Y, Ye L, Yang W, Wollheim C B, Lou J. Dynamin-related protein 1 mediates high glucose induced pancreatic beta cell apoptosis. Int J Biochem Cell Biol. 2009;41:879–890. doi: 10.1016/j.biocel.2008.08.031. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Thomas M, Dadgar N, Lieberman A P, Iniguez-Lluhl J A. SUMO modification of the androgen receptor attenuates polyglutamine-mediated aggregation. J Biol Chem. 2009:M109.011494. doi: 10.1074/jbc.M109.011494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S, Sixt K M, Barrow R, Snyder S H. Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science. 2009;324:1327–1330. doi: 10.1126/science.1172871. [DOI] [PMC free article] [PubMed] [Google Scholar]