Abstract
Monoamine oxidase A (MAO A), encoded by the X chromosome, catalyzes the oxidative deamination of monoamine neurotransmitters, such as serotonin, and plays a critically important role in brain development and functions. Abnormal MAO A activity has been implicated in several neuropsychiatric disorders, such as depression, autism, and attention deficit hyperactivity disorder, which show sexual dimorphism. However, the molecular basis for these disease processes is unclear. Recently, we found that MAO A was a putative target gene directly regulated by a transcription factor encoded by the sex-determining region Y (SRY) gene located on the Y chromosome. We demonstrated that SRY activates both MAO A-promoter and catalytic activities in a human male neuroblastoma BE(2)C cell line. A functional SRY-binding site in the MAO A core promoter was identified and validated by electrophoretic mobility shift and chromatin immunoprecipitation (ChIP) analyses. Coimmunoprecipitation and ChIP assays showed that SRY and Sp1 form a transcriptional complex and synergistically activate MAO A transcription. This is the first study demonstrating that the Y-encoded transcription factor SRY is capable of regulating an X-located gene, suggesting a novel molecular mechanism for sexual dimorphism in neural development, brain functions, and initiation/progression of neural disorders associated with MAO A dysfunction.—Wu, J. B., Chen, K., Li, Y., Lau, Y.-F. C., Shih, J. C. Regulation of monoamine oxidase A by the SRY gene on the Y chromosome.
Keywords: sex-determining region Y gene, transcriptional activation, sexual dimorphism, X chromosome gene, promoter regulation, Sp1 coactivation
Monoamine oxidase (MAO) catalyzes the oxidative deamination of a number of biogenic and dietary amines, including serotonin, norepinephrine, dopamine, and phenylethylamine. The degradation of monoamines by MAO produces neurotoxic hydrogen peroxide as a byproduct. Located in the outer mitochondrial membrane, MAO exists in two isoforms, MAO A and MAO B, encoded by two independent genes on Xp11.23-11.4 (1,2,3,4). MAO A preferentially oxidizes serotonin, norepinephrine, and dopamine, and is irreversibly inhibited by low concentrations of clogyline (4).
In light of the vital role that MAO plays in the metabolism of neurotransmitters, MAO A dysfunctions have been implicated in a variety of neuropsychiatric disorders, such as depression (4, 5), social anxiety (6), autism (5), and attention deficit hyperactivity disorder (ADHD) (7, 8). MAO A deficiency caused by a spontaneous mutation in the MAO A gene resulted in impulsive aggressive behavior and mild mental retardation in affected males in a Dutch family (9). Similarly, Mao a-knockout mice also show aggressive behavior (4, 10, 11). MAO inhibitors such as moclobemide (a reversible inhibitor of MAO A) have been developed and clinically used to treat depression and social anxiety, in view of their high mood-enhancing efficacy (5).
Interestingly, several psychiatric disorders related to MAO A dysfunctions show significant sex differences. For example, the prevalence of autism (12) and ADHD (13) is about 4 and 2 times as frequent in males as in females, respectively. Males with obsessive-compulsive disorder tend to have an earlier onset and have more symmetry and religious obsessions and miscellaneous compulsions (14, 15). Females with major depressive disorder (MDD) demonstrate poorer emotion processing than nondepressed females, whereas males with MDD show equivalent performance in emotion processing compared to nondepressed males (16). Emerging evidence from recent studies has brought into focus the implication of sex differences in the pathogenesis of several psychiatric disorders. Sex differences have been found in brain structure, chemistry, and function, as demonstrated by neuroanatomical and imaging approaches (17,18,19). Sex hormones, such as androgen (20) and estrogen (21), have been shown to influence various serotonin functions in humans and animals, and have been suggested to be potentially important in the sex differences in the prevalence, course, and treatment response characteristics of autism (22, 23), schizophrenia, and mood disorders (24). Moreover, skewed X-chromosome inactivation tends to be another potential cause for some psychiatric disorders that differ in the prevalence and heritability between sexes such as MDD (25). However, the roles of the Y-chromosome genes in such sexual dimorphisms are not yet explored in detail.
Recently, we have identified MAO A as a novel neural target for the sex-determining SRY (sex-determining region on the Y chromosome) gene in a genome-wide chromatin immunoprecipitation (ChIP) and promoter tiling microarray analysis (unpublished results). The SRY gene, encoding a putative transcription factor, is the master switch regulator responsible for initiating the testis determination and differentiation during embryogenesis (26,27,28). Although many of the pathways regulating sexual differentiation have been elucidated, direct downstream targets of SRY are still unclear. Here, we demonstrate the molecular mechanisms of MAO A gene regulation by SRY and propose that this sexually dimorphic regulation may provide new insights into the initiation/progression of neurodevelopmental disorders associated with MAO A dysfunctions and suggest the role of the Y-chomosome genes in these processes.
MATERIALS AND METHODS
Cell line and reagents
The human male neuroblastoma BE(2)C cell line was purchased from the American Type Culture Collection (Manassas, VA, USA). BE(2)C cells were grown in a medium containing 1:1 mixture of Eagle’s minimum essential medium (MEM) with Earle’s balanced salt solution and Ham’s F12 medium supplemented with 10% FBS, 1 mM sodium pyruvate, 0.05 mM nonessential amino acids, 100 U/ml penicillin, and 100 μg/ml streptomycin. All culture materials were purchased from Mediatech (Manassas, VA, USA). Monoclonal anti-FLAG (F1804) and anti-HA (H3663) antibodies were purchased from Sigma-Aldrich (St. Louis, MO, USA). Monoclonal anti-SRY (sc-69842) and anti-β-actin (sc-47778) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Polyclonal anti-Sp1 antibody (A300-133A) was purchased from Bethyl Laboratories (Montgomery, TX, USA). Human SRY siRNA (sc-38443) was purchased from Santa Cruz Biotechnology.
Plasmids
Human MAO A 2-, 1.6-, 1.3-, 0.24-, and deleted 2-kb (without Sp1 sites) promoter-luciferase (firefly) reporter constructs were generated as described previously (29, 30). FLAG-SRY was constructed by inserting the ORF of the human SRY in frame to that of the p3XFLAG-CMV-7 vector (Sigma-Aldrich), as described previously (31).
Stable cell line establishment
FLAG-SRY expression construct or pCMV empty vector carrying the neomycin-resistant gene was transfected into BE(2)C cells. After 24 h, 750 μg/ml of geneticin (G418) was added to the transfected cells. Resistant clones were isolated after 14 d and cultured under G418 selection and maintained continuously with 200 μg/ml of the selective agent.
RNA isolation and quantitative real-time RT-PCR
Total DNA-free RNA was purified with the RNeasy Mini Kit (Qiagen, Valencia, CA, USA), following the manufacturer’s instructions. Two micrograms of total RNA was used for reverse transcription by M-MLV reverse transcriptase (Promega, Madison, WI, USA), following the manufacturer’s instructions. The RT products were used as the template for quantitative real-time PCR. Quantitation of the PCR products was determined by SYBR Green reagent (Maxima SYBR Green qPCR Master Mix 2X; Fermentas, Glen Burnie, MD, USA) using the iCycler optical system (Bio-Rad, Hercules, CA, USA). The primers for MAO A were forward 5′-CTGATCGACTTGCTAAGCTAC-3′ and reverse 5′-ATGCACTGGATGTAAAGCTTC-3′ (fragment length, 102 bp). The primers for GAPDH were forward 5′-GACAACAGCCTCAAGATCATCAG-3′ and reverse 5′-ATGGCATGGACTGTGGTCATGAG-3′ (fragment length, 122 bp) (32). PCR conditions included an initial denaturation step of 3 min at 95°C, followed by 40 cycles of PCR consisting of 30 s at 94°C, 30 s at 60°C, and 40 s at 72°C. The PCR data were analyzed by 2−ΔΔCT method (33). Regular PCR was conducted to determine SRY mRNA expression in BE(2)C cells overexpressing SRY. The primers for SRY were forward 5′-CAACAGCGATGATTACAGTCCAGC-3′ and reverse 5′-GACCACACGATGAATGCGTTC-3′ (fragment length, 177 bp). The primers for β-actin were forward 5′-CCCAGCCATGTACGTTGCTATCCA-3′ and reverse 5′-CACGGAGTACTTGCGCTCA-3′ (fragment length, 612 bp).
MAO A catalytic activity assay
MAO A catalytic activity was determined in BE(2)C cells overexpressing SRY or pCMV empty vector. One hundred micrograms of total protein (∼1×106 cells) were incubated with 1 mM 14C-5-HT in the assay buffer (50 mM sodium phosphate buffer, pH 7.4) at 37°C for 20 min, and the reaction was terminated by the addition of 100 μl of 6 N HCl. The reaction products were extracted with benzene/ethyl acetate and centrifuged at 4°C for 7 min. The organic phase containing the reaction products was extracted, and the radioactivity was determined by liquid scintillation spectroscopy (30).
siRNA interference
siRNA was transfected into BE(2)C cells with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. The sequence to silence the translation of Sp1 was 5′-GGUAGCUCUAAGUUUUGAUUU-3′ (sense) (34). A nonsilencing RNA with sense strand as 5′-UUCUCCGAACGUGUCACGUUU-3′ was used as control (35).
Transient transfection and luciferase reporter assay
Transfections were performed with Lipofectamine 2000 (Invitrogen) in 12-well plates. pRL-TK (expressing Renilla luciferase) was cotransfected as an internal control (Promega). pcDNA was added to maintain an equivalent amount of DNA in each transfection. After 24–48 h incubation, cells were harvested and assayed for luciferase activity using the Dual-Luciferase Reporter 1000 Assay System (Promega).
ChIP assay and PCR
Confluent BE(2)C cells in a 10-cm dish were treated with formaldehyde at a final concentration of 1% at room temperature with gentle shaking for 10 min to cross-link nuclear proteins with genomic DNA. Cross linking was quenched by incubating with glycine at the final concentration of 2.5 M at room temperature with gentle shaking for another 5 min. Cells were quickly washed by cold PBS twice, harvested by scraping, and centrifuged at 2000 rpm at 4°C for 5 min. Cell pellets were lysed in 350 μl of SDS lysis buffer (1% SDS; 10 mM EDTA; 50 mM Tris-HCl, pH 8.0; and 2× protease inhibitor) on ice for 10 min, followed by sonication using the Branson 450 Sonifier (Branson Ultrasonics, Danbury, CT, USA) to shear genomic DNA into 500- to 1000-bp fragments. One to 10% of the supernatant was saved as input. Supernatant was diluted (1:10) in dilution buffer (0.01% SDS; 1.1% Triton X-100; 1.2 mM EDTA; 16.7 mM Tris-HCl, pH 8.0; and 167 mM NaCl), and blocked with 60 μl of sheared salmon sperm DNA/protein A/G agarose at 4°C for 2–4 h. The supernatant was immunoprecipitated with 2–5 μg of specific antibody at 4°C overnight. IgG or no addition of antibody was used as a negative control for IP. After incubating 40 μl of salmon sperm DNA/Protein A/G agarose with IP samples at 4°C for another 2 h, beads were sequentially washed by low-salt buffer (0.1% SDS; 1% Triton X-100; 2 mM EDTA; 20 mM Tris-HCl, pH 8.0; and 150 mM NaCl), high-salt buffer (0.1% SDS; 1% Triton X-100; 2 mM EDTA; 20 mM Tris-HCl, pH 8.0; and 0.5 M NaCl), LiCl buffer (0.25 M LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl, pH 8.0), and TE buffer (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA). The DNA-protein complex was eluted by elution buffer (1% SDS and 0.1 M sodium bicarbonate) with gentle rotation at room temperature for 15 min twice, reverse-cross-linked by incubating at 65°C for 5–8 h, and purified using QIAquick PCR Purification Kit (Qiagen). Purified DNA was used as the template for the following PCR analysis (36).
For the ChIP/re-ChIP assay to determine the simultaneous presence of Sp1 and SRY in the MAO A promoter, BE(2)C cells transiently transfected with FLAG-SRY were subjected to ChIP assay using anti-FLAG antibody, as described above. The remaining chromatin bound within the beads in the anti-FLAG immunoprecipitates was recovered in 50 μl of DTT buffer (2% SDS, 10 mM DTT, and 2× protease inhibitor in 1× TE buffer) at 37°C for 30 min twice and subjected to the second ChIP assay (re-ChIP) using anti-Sp1 antibody.
The primers used for the MAO A core promoter (−360/−17) were forward 5′-GTGCCTGACACTCCGCGGGGTT-3′ and reverse 5′-TCCTGGGTCGTAGGCACAGGAG-3′ (fragment length, 344 bp). Distilled H2O was used as a negative control for PCR. PCR products were analyzed by agarose gel electrophoresis, and the intensity of DNA bands was quantified by Labworks analysis software (UVP, Upland, CA, USA).
Site-directed mutagenesis of the human MAO A 0.24-kb promoter
Site-directed mutagenesis was used to mutate the putative SRY-binding site as identified in MAO A 0.24-kb promoter. Wild-type MAO A 0.24-kb-luc was used as the template. Mutagenesis was carried out using QuickChange XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) following the manufacturer’s instructions. The primers used for mutagenesis was forward 5′-GGCTCCCCCCGGGTACCCTGAGAAGGATCGGCTCC-3′ (mutated nucleotides underscored). Mutated nucleotides were verified by DNA sequencing.
In vitro translation
In vitro translation was conducted with TNT Coupled Reticulocyte Lysate System (Promega), following the manufacturer’s instructions. Both pcDNA-SRY-v5 and pcDNA empty vector carry T7 promoter. All in vitro translated products were verified by Western blot.
Electrophoretic mobility shift analysis (EMSA)
MAO A 0.24-kb-promoter-derived oligonucleotide with WT SRY-binding site was used as a probe and radioactively labeled by Klenow fill-in reaction (37, 38). 32P-labeled probes were purified using Nucleotide Removal Kit (Qiagen). For determining the DNA-protein binding, 2 μl of in vitro translated SRY was diluted with 5× binding buffer [20% glycerol; 5 mM MgCl2; 2.5 mM EDTA; 2.5 mM DTT; 250 mM NaCl; 50 mM Tris-HCl, pH 7.5; and 0.25 mg/ml poly(dI-dC)] in a total volume of 20 μl. One-hundred-fold excess unlabeled probes (competitor) were added, and the mixture was incubated at room temperature for 20 min. 32P-labeled probe (∼600,000 cpm) was then added, and the mixture was incubated at room temperature for another 20 min. Samples were analyzed on 5% nondenaturing polyacrylamide gel in 1× Tris borate/EDTA buffer at 150 V at room temperature for 3 h. Gel was dried and visualized by autoradiography.
Coimmunoprecipitation assay
Confluent BE(2)C cells in a 10-cm dish were lysed with 500 μl of lysis buffer (20 mM Tris-HCl, pH 7.5; 150 mM NaCl; 1 mM EDTA; 1 mM EGTA; and 1% Triton X-100) containing various protease inhibitors. Two hundred microliters of lysates was then incubated with polyclonal anti-Sp1 antibody (2–4 μg) at 4°C overnight and incubated with 40 μl of protein G agarose beads (GE Healthcare, Piscataway, NJ, USA) at 4°C for 3 h. The samples were washed with lysis buffer 5 times and resuspended in 2× SDS sample buffer. Rabbit normal IgG was used as control for IP. The samples were analyzed by Western blot using a mouse anti-SRY antibody.
RESULTS
SRY activates MAO A-promoter and catalytic activities
We initially identified Mao a as a potential target gene for Sry in E11.5 embryonic mouse gonadal cells using a genome-wide ChIP assay on a 2.5-kb mouse-promoter tiling microarray (ChIP-chip) (Supplemental Fig. 1) (39, 40). Because human and mouse MAO A 2.5-kb promoters share ∼70% sequence identity, we speculate that MAO A could be regulated by SRY in humans as well, which is also consistent with the result predicted from the human genome that multiple putative SRY-binding sites are present in MAO A 2-kb promoter. Recent studies suggested the brain as an important sexual organ that begins to develop differentially between males and females prior to the formation of the respective sex organs and synthesis of sex hormones during embryogenesis (41,42,43). Currently, the exact molecular mechanisms for such sexually dimorphic differentiation is uncertain; genes on the sex chromosomes, including the Y-encoded SRY, could potentially play important roles in the processes. In light of the importance of MAO A and SRY implicated in brain, we used a human male neuroblastoma BE(2)C cell line, in which both genes are well expressed, to explore the molecular mechanisms by which SRY could regulate MAO A expression.
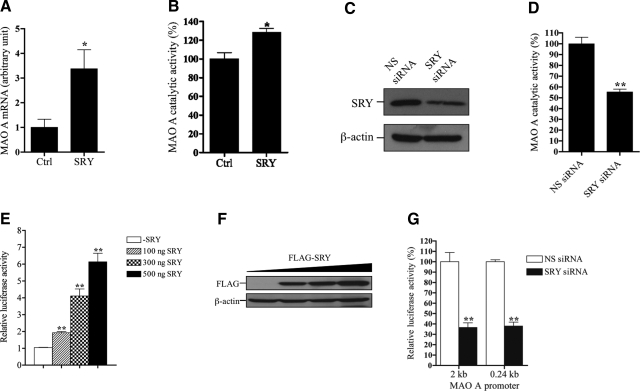
To study the SRY effect on MAO A expression, we established a stable BE(2)C cell line overexpressing SRY (Supplemental Fig. 2A, B). SRY overexpression resulted in significant increases of both MAO A mRNA (Fig. 1A) and catalytic activity (Fig. 1B) by 340 and 30%, respectively. We also knocked down the endogenous SRY synthesis in BE(2)C cells by siRNA interference technology. Successful SRY knockdown, as confirmed by Western blot analysis (Fig. 1C), dramatically inhibited MAO A catalytic activity by 45% (Fig. 1D). To examine the SRY effect on MAO A promoter, we transiently transfected a luciferase reporter gene directed by a 2-kb MAO A promoter (MAO A 2-kb-luc) together with various amounts of FLAG-tagged SRY expression construct into BE(2)C cells. After 24 h incubation, cells were harvested, and the luciferase activity was determined. As shown in Fig. 1E, SRY increased MAO A-promoter activity up to 6-fold in a concentration-dependent manner. The transfection efficiency of SRY used in the luciferase reporter assay was confirmed by Western blot (Fig. 1F). In addition, a knockdown of SRY expression substantially reduced both MAO A 2- and 0.24-kb core-promoter activities to <40% in the transfected cells (Fig. 1G). Previously, we demonstrated that the 0.24-kb MAO A core promoter (−303/−64) contains a 240-bp region immediately upstream from the transcription initiation site and exhibits the maximum MAO A-promoter activity (37). Taken together, these results demonstrate that SRY is capable of activating both MAO A-promoter and catalytic activities.
Figure 1.
SRY activates both MAO A-promoter and catalytic activities in BE(2)C cells. A) Effect of SRY overexpression on MAO A mRNA expression. MAO A mRNA levels were determined by quantitative real-time PCR in BE(2)C cells overexpressing SRY. In control (ctrl) cells, pCMV vector carrying neomycin-resistant gene was stably transfected. GAPDH was used as an internal control. Data were analyzed by 2−ΔΔCT method. MAO A mRNA expression level in control cells was arbitrarily set as 1. B) Effect of SRY overexpression on MAO A catalytic activity. MAO A catalytic activity was determined by MAO A catalytic activity assay in BE(2)C cells overexpressing SRY. MAO A catalytic activity in control cells was set as 100%. C) Knockdown of endogenous SRY in BE(2)C cells. SRY siRNA was transfected into BE(2)C cells; 48 h later, cells were harvested and analyzed by Western blot using anti-SRY antibody. β-Actin was used as loading control. Nonsense (NS) siRNA was transfected similarly in control cells. D) Effect of SRY knockdown on MAO A catalytic activity. BE(2)C cells were transfected with SRY siRNA; 48–72 h later, cells were harvested and analyzed by MAO A catalytic activity assay. MAO A catalytic activity in control cells was set as 100%. E) Effect of SRY on MAO A-promoter activity. MAO A 2-kb-luc was cotransfected with various amounts of FLAG-SRY expression construct into BE(2)C cells, 24–48 h later, cells were harvested, and the luciferase activity was determined. Activity of MAO A 2-kb-luc (expressing firefly luciferase) was normalized with cotransfected pRL-TK (expressing Renilla luciferase). pcDNA was added to keep the DNA amount of each transfection constant. Activity of MAO A 2-kb-luc without transfection of SRY was set as 1. F) Analysis of transfection efficiency of FLAG-SRY. Western blot was performed to document the FLAG-SRY expression in BE(2)C cells using anti-FLAG antibody. β-Actin was used as the loading control. G) MAO A 2- or 0.24-kb-luc was cotransfected with SRY siRNA into BE(2)C cells; 48 h later, cells were harvested, and the luciferase activity was determined. Activity of MAO A 2- or 0.24-kb-luc without transfection of SRY siRNA was set as 100%. All data are presented as the means ± sd from 3 independent experiments with triplicates for each experiment. Representative gels are shown. *P < 0.05, **P < 0.01.
SRY binds to a consensus SRY-binding site in the MAO A 0.24-kb core promoter in vitro and in vivo
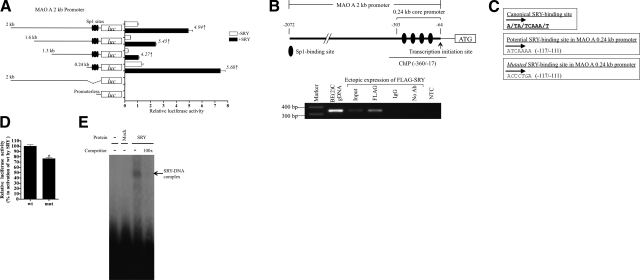
We postulate that SRY could potentially bind to promoter sequences of its target genes and mediate their expression (44,45,46). To locate putative SRY-binding sites in the 2-kb MAO A promoter, we transfected a SRY expression construct with a series of MAO A promoters of different lengths linked with a luciferase reporter gene into BE(2)C cells. Interestingly, MAO A-luc constructs consisting of 2-, 1.6-, 1.3-, or 0.24-kb promoter were activated by SRY to approximately the same extent (Fig. 2A), suggesting that SRY-binding sites are most likely located in the 0.24-kb core-promoter region. Significantly, MAO A 0.24-kb promoter contains 4 Sp1-binding sites (Table 1) (37), and deletion of these sites abolished basal MAO A 2-kb-promoter activity, as well as SRY-induced promoter activation (Fig. 2A).
Figure 2.
SRY binds to a functional SRY-binding site in MAO A core promoter both in vitro and in vivo. A) Serial deletion analysis of MAO A-promoter activation by SRY. Various deletions of MAO A-promoter-luc were cotransfected with SRY into BE(2)C cells; 24 to 48 h later, cells were harvested, and the luciferase activity was determined. Promoterless pGL2-basic vector was used as control. Fold of activation of MAO A-promoter-luc by SRY is indicated. Activity of MAO A 2-kb-luc without transfection of SRY was set as 1. B) Demonstration of SRY binding to MAO A 0.24-kb core promoter in vivo. Schematic representation of MAO A 2-kb (−2072/−64) and 0.24-kb (−303/−64) promoter structure is not proportional to the real promoter length. A in the start codon was set as +1. BE(2)C cells were transiently transfected with FLAG-SRY; 48 h later, cells were subjected to ChIP assay using anti-FLAG antibody and PCR with primers specific for MAO A core-promoter region (−360/−17). IgG was used as a negative control for IP. BE(2)C genomic DNA (gDNA) and ddH2O were used as positive and negative controls, respectively, for PCR. C) Sequence of canonical SRY-binding site (top panel), a potential SRY-binding site (−117/−111) in MAO A 0.24-kb promoter (middle panel), and the introduced point mutations (italic) used to inactivate the potential SRY-binding site (bottom panel). D) Effect of SRY on wild-type (wt) and mutated (mut) MAO A 0.24-kb-promoter-luc construct in BE(2)C cells. Fold of activation of wt MAO A 0.24-kb-luc by SRY was set as 100%. E) EMSA of in vitro translated SRY protein with MAO A 0.24-kb-promoter-derived, 32P-labeled oligonucleotide containing wt SRY-binding site. In vitro translation product using pcDNA vector as a template was used as mock. All data are presented as means ± sd from 3 independent experiments with triplicates for each experiment. Representative gels are shown. *P < 0.05.
TABLE 1.
Sp1- and SRY-binding sites in MAO A 0.24-kb promoter (−303/−64)
| Site | Description | Sequence | Location (bp) |
|---|---|---|---|
| 1 | Sp1-binding site | CTCCGCCC | −101/−94 |
| 2 | SRY-binding site | ATCAAAA | −117/−111 |
| 3 | Sp1-binding site | CCCGCCC | −148/−142 |
| 4 | Sp1-binding site | CTCCGCCC | −191/−184 |
| 5 | Sp1-binding site | TCCGCCC | −243/−237 |
Sequence and location of each site are indicated. Site numbers are presented from 3′ to 5′ in MAO A promoter. A in the start codon was set as +1.
To confirm an SRY binding to the MAO A promoter, we performed a ChIP assay with BE(2)C cells transiently transfected with FLAG-tagged SRY followed by PCR amplification of the precipitated chromatin DNA with primers specific for this 0.24-kb region (Fig. 2B). The binding of ectopically expressed SRY to this region was demonstrated only in chromatin DNA precipitated by an anti-FLAG antibody (Fig. 2B), which further supports the notation that SRY indeed binds to responsive elements in this region of the MAO A promoter in the transfected cells.
Sequence analysis of this 0.24-kb region identified a putative SRY-binding site (−117/−111) (Fig. 2C, middle panel) that is identical to the canonical motif (Fig. 2C, top panel) (38). Mutation of nucleotides at this site (Fig. 2C, bottom panel) reduced the SRY activation of MAO A 0.24-kb promoter by 30% (Fig. 2D), suggesting that this site is functional.
To determine whether SRY directly interacts with this element, we conducted an EMSA with in vitro translated SRY protein and radioactively labeled SRY-binding oligonucleotide as a probe. Our results showed a mobility-shifted band of the labeled SRY-binding oligonucleotide in the gel when SRY was incubated with the probe (Fig. 2E), indicating the presence of a SRY protein-DNA complex. This band was abolished in the presence of 100-fold excess of unlabeled probes as a competitor, suggesting it is specific for SRY (Fig. 2E). Furthermore, no such band was observed from the control, in which mock protein synthesis was conducted with vector DNA and the product was used in similar DNA-protein binding reaction (Fig. 2E). Taken together, these results indicate that SRY is indeed binding to this functional SRY-binding site both in vitro and in vivo.
Sp1 synergistically enhances the SRY activation of MAO A promoter
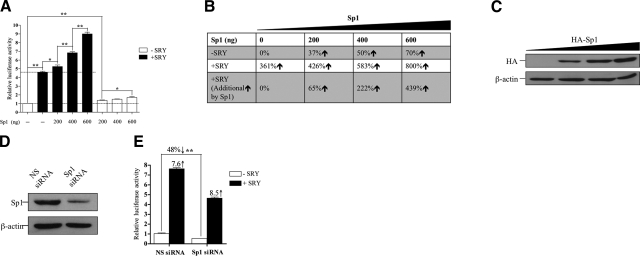
Both SRY- and Sp1-binding sites are located, but do not overlap, in the MAO A 0.24-kb core-promoter region (Table 1). SRY and Sp1 bind to AT-rich (38) and GC-rich sites (47, 48), respectively. Sp1 is a ubiquitous transcription factor in mammals (48) and is a key activator of MAO A promoter (37, 49, 50). Sp1 binds to GC boxes directly and transactivates synergistically with a large variety of transcription factors, such as Re1A (51) and Oct-1 (52). Moreover, Sp1 plays various roles in embryonic development, such as acting as a transcription regulator in spermatogenesis (53,54,55).
To study whether Sp1 is involved in the SRY activation of MAO A promoter, we cotransfected MAO A 0.24-kb-luc with various amounts of HA-tagged Sp1 in the absence or presence of SRY into BE(2)C cells and determined the luciferase reporter activity (Fig. 3A). The transfection efficiency of Sp1 was confirmed by Western blot (Fig. 3C). As summarized in Fig. 3B, in the absence of SRY, Sp1 increases MAO A-promoter activity by 37, 50, and 70% from 0 to 600 ng of DNA in a dosage-dependent manner. SRY itself activated the MAO A promoter by 361%, and this activation was synergistically enhanced to 426, 583, and 800% when 200, 400, and 600 ng of Sp1 was cotransfected. Thus, Sp1 increased the SRY-mediated transactivation of MAO A-luc reporter by 65, 222, and 439, respectively, in the same DNA dosages. To confirm such synergistic interactions between SRY and Sp1, we knocked down the endogenous Sp1 in BE(2)C cells, as shown by Western blot (Fig. 3D). Sp1 knockdown reduced its own transactivation, as well as its synergistic action on SRY transactivation of the MAO A promoter (Fig. 3E). However, such decreases in reporter activities seemed to be proportional in both siRNA knockdown of Sp1 and those with nonspecific siRNA control (Fig. 3E). Taken together, these data suggest that Sp1 synergistically enhances, but is not essentially required, for the SRY activation of MAO A promoter in BE(2)C cells.
Figure 3.
Sp1 synergistically enhances, but is not essential, for the SRY activation of MAO A promoter. A) Effect of Sp1 on MAO A-promoter activation by SRY. MAO A 0.24-kb-luc was cotransfected with various amounts of HA-Sp1 in the presence or absence of SRY into BE(2)C cells; 24 to 48 h later, cells were assayed for luciferase activity. Activity of MAO A 0.24-kb-luc transfected alone was set as 1. B) Summary of increases of MAO A-promoter activity obtained in A. C) Western blot analysis of transfected cells correlating the transfection efficiency of HA-Sp1 used in A. D) Knockdown of endogenous Sp1 in BE(2)C cells by siRNA interference technology. E) Effect of Sp1 knockdown on the SRY activation of MAO A promoter. MAO A 0.24-kb-luc was cotransfected with SRY together with or without Sp1 siRNA into BE(2)C cells; 48 h later, cells were assayed for luciferase activity. Activity of MAO A 0.24-luc transfected alone was set as 1. All data are presented as means ± sd from 3 independent experiments with triplicates for each experiment. *P < 0.05, **P < 0.01.
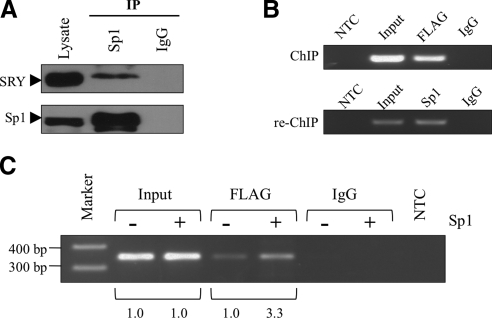
Sp1 enhances the SRY binding to MAO A promoter by interacting and forming a transcriptional regulatory complex with SRY at the MAO A core promoter in vivo
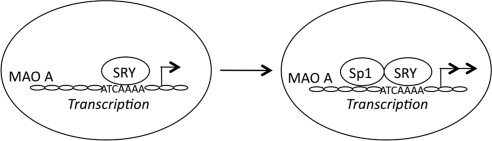
Previous studies suggest that SRY interacts with a variety of transcription factors in its transcriptional regulation of its target genes (29, 56, 57). Since SRY and Sp1 bind to the same region of the 0.24-kb promoter of MAO A, it is possible that they could physically interact and synergize in their transactivation of MAO A promoter. To explore this possibility, we performed a coimmunoprecipitation assay with anti-Sp1 antibody in BE(2)C cells. As demonstrated in Fig. 4A, Sp1 retained SRY in the anti-Sp1 immunoprecipitates, as revealed by Western blot using an anti-SRY antibody, suggesting that Sp1 indeed physically interacts with SRY. To further determine the simultaneous presence of SRY and Sp1 in the natural MAO A core promoter, we conducted a ChIP assay with BE(2)C cells transiently transfected with FLAG-tagged SRY using an anti-FLAG antibody, and subjected the resulting immunoprecipitates to re-ChIP assay using an anti-Sp1 antibody. As expected, SRY bound to MAO A core promoter in vivo (Fig. 4B, top panel), and Sp1 could be detected in the anti-FLAG immunoprecipitates (Fig. 4B, bottom panel), supporting the postulation that Sp1 does indeed form a complex with SRY at the natural MAO A core promoter. In addition, higher SRY occupancy in MAO A core promoter was observed in vivo when Sp1 was overexpressed (Fig. 4C), which is consistent with the synergistic transactivation of MAO A promoter by SRY and Sp1. Our results also suggest that the interaction between Sp1 and SRY has a physiological effect. Hence, Sp1 enhances SRY-induced MAO A transcriptional activation by interacting and forming a transcriptional regulatory complex with SRY in the MAO A core promoter (Fig. 5).
Figure 4.
Sp1 enhances SRY binding to MAO A promoter by interacting and forming a complex with SRY at the MAO A core promoter in vivo. A) Coimmunoprecipitation and Western blot analysis of interaction between SRY and Sp1 in BE(2)C cells. Rabbit polyclonal anti-Sp1 antibody was used for IP, and mouse monoclonal anti-SRY antibody was used for Western blot. IgG was used as a negative control for IP. 10% input was loaded as control. B) ChIP/re-ChIP analysis demonstrating the simultaneous presence of Sp1 and SRY in natural MAO A core promoter. BE(2)C cells were transiently transfected with FLAG-SRY; 48 h later, cells were subjected to ChIP assay using anti-FLAG antibody. Anti-FLAG immunoprecipitates were subjected to re-ChIP assay using anti-Sp1 antibody. IgG was used as a negative control for IP. PCR was performed using primers targeting MAO A core promoter region. ddH2O was used as a negative control (NTC) for PCR. C) Overexpression of Sp1 enhanced SRY binding to MAO A core promoter in vivo. BE(2)C cells were transiently transfected with FLAG-SRY together with or without Sp1; 48 h later, cells were subjected to ChIP assay using anti-FLAG antibody. PCR was performed using primers targeting MAO A core-promoter region. Intensity of DNA bands was quantified by Labworks analysis software (UVP). Representative gels are shown.
Figure 5.
Schematic diagram of MAO A transcriptional activation cooperatively regulated by SRY and Sp1.
DISCUSSION
The regulation of MAO A transcription has been studied extensively in recent years. Besides Sp1, two Sp-family proteins, Sp3 and Sp4, are also shown to regulate the MAO A promoter via the same Sp1-binding sites. Sp4 activates the MAO A core promoter as Sp1, whereas Sp3 represses this activation by competing with Sp1 and Sp4 (58). R1 (RAM2/CDCA7L/JPO2), as a novel transcription repressor, down-regulates MAO A gene expression by competing with Sp1 for binding to Sp1 sites as well (29).
For the first time, we have demonstrated the presence of a functional binding site for a Y-encoded SRY in the MAO A core promoter both in vitro and in vivo (Fig. 2). The SRY-binding site is located proximal to these well-characterized Sp1-binding sites (Table 1). The human SRY protein harbors a high mobility group (HMG) box DNA-binding domain, which can bind and bend target DNA sequences (38, 59). These DNA-binding and -bending activities are essential for SRY as a sex-determining factor, since mutations at the HMG box abolish such DNA binding properties and result in XY sex reversal (44, 60). Previous studies demonstrated that various cofactors interact with SRY to form transcription complexes that regulate SRY target genes (31, 57). The present study demonstrates that SRY is capable of interacting with Sp1 synergistically in transactivation of MAO A gene. Since other factors, such as Sp3, Sp4, and R1, also utilize the same Sp1 sites in their transcriptional regulation of MAO A, it will be interesting to determine whether these MAO A regulators could also interact with SRY, thereby exerting potentially complex transcriptional interplays and sexually dimorphic physiological effects.
In addition to SRY expression in neural tissues, cellular sublocalization of its protein is important for its actions on MAO A regulation. Interestingly, SRY acetylation by histone acetyltransferase p300 increases its nuclear localization, while specific deacetylation by HDAC3 induces a cytoplasmic localization of SRY (61). These observations suggest that SRY acetylation promotes its nuclear accumulation and may serve an important pathway for the SRY activation of MAO A gene. Previous studies showed that histone deacetylation (HDAC) inhibitors, such as sodium butyrate and trichostatin A (TSA), significantly increase MAO A-promoter activity in human glioblastoma cells. Hence, acetylation/deacetylation control of SRY subcellular localization could be an important modulatory mechanism for its sexually dimorphic regulation of MAO A transcription.
Genetic variants in certain X-located genes, such as MAO A, may have different effects on cognition and behavior in males and females, and may explain sex ratio differences in certain psychiatric disorders. Indeed, several studies have found various polymorphisms in MAO A (a 30-bp variable-number tandem repeat polymorphism in the promoter region, a GA repeat polymorphism in intron 2, and a G/T single-nucleotide polymorphism in exon 8) to be associated with certain psychiatric disorders, such as attention-deficit hyperactivity disorder (ADHD) (62,63,64,65). Further, epigenetic factors/mechanisms that regulate MAO A, such as methylation (66) and incomplete X inactivation (67), though not fully dissected, could contribute to sex differences of MAO A-related psychiatric disorders.
The identification of MAO A as a novel neural target for SRY has raised several significant issues regarding the potential contribution of this Y-chromosome-encoded transcription factor to neural development and mental disorders. MAO A is subjected to X inactivation, and hence, in principle, it is only active in one of the two X chromosomes in female (68). However, other studies have suggested that the levels of serotonin, the substrate for MAO A, could vary between males and females (69, 70), suggesting there might be sexual dimorphism in MAO A functions. Indeed, sexual dimorphisms in brain structures and manifestation of mental illnesses and neurodegenerative diseases have long been observed (71, 72). Hence, SRY influence on MAO A expression could be part of such a sexually dimorphic program in neural structure and physiology. Alternatively, the MAO A locus could be subjected to incomplete X inactivation, resulting in higher MAO A activities in females. The synergistic regulation of MAO A by SRY and Sp1 could be a normal physiological and compensational function in males, which raises the levels of MAO A to those in females. On the basis of this postulation, deficiency in X inactivation of MAO A locus could increase the levels of MAO A activities in females, which are prone to depressive disorders. Conversely, an inadequate up-regulation of MAO A by SRY-Sp1 could decrease MAO A activities in males. It is uncertain what effects, such as a decline in MAO A activity, might have on male physiology and cognitive functions. In addition to depressive disorders, MAO A, among other genes/genetic factors, has been implicated to play a role in the pathogenesis of some of other psychiatric disorders, such as autism, schizophrenia, and ADHD (4, 5, 71, 73,74,75,76). However, the exact mechanisms for these disorders have not yet been identified. Since SRY expression has been demonstrated in the brain during embryonic development and/or adulthood (41,42,43), it could be a male-specific genetic modifier in regulation of MAO A gene, thereby exerting sexual dimorphisms in neural development and/or development and progression of psychiatric/cognitive disorders associated with abnormal MAO A activities (5, 41, 77).
The fact that a Y-located transcription factor is capable of transactivating an X-located gene is intriguing. The molecular mechanisms leading to sexual dimorphisms in brain structures and cognitive disorders have been debated in numerous publications (19, 42, 78). One hypothesis suggests that dosage differences of X-located genes that escape X inactivation could have contributed to such physiological or disease phenomena (68). The current demonstration of the regulatory and exacerbating roles of SRY on MAO A transcription and enzymatic activities has clearly raised the possibility that Y-chromosome genes and epigenetic gene regulation could be significant players in sexual dimorphisms in both normal human physiology and diseases.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Mental Health grants R01MH67968 and R01MH39085 and the Boyd and Elsie Welin Professorship to J.C.S., and by a concept grant from the U.S. Department of Defense Autism Research Program to Y.-F.C.L. Y.-F.C.L. is a Research Career Scientist of the Department of Veterans Affairs. The authors thank Dr. Hans Rotheneder (Department of Medical Biochemistry and Molecular Biology, Medical University of Vienna, Vienna, Austria) for providing HA-Sp1 expression construct. The authors are also grateful to Dr. Marco Bortolato for critical reading of the manuscript.
References
- Bach A W, Lan N C, Johnson D L, Abell C W, Bembenek M E, Kwan S W, Seeburg P H, Shih J C. cDNA cloning of human liver monoamine oxidase A and B: molecular basis of differences in enzymatic properties. Proc Natl Acad Sci U S A. 1988;85:4934–4938. doi: 10.1073/pnas.85.13.4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsby J, Chen K, Wang L J, Lan N C, Shih J C. Human monoamine oxidase A and B genes exhibit identical exon-intron organization. Proc Natl Acad Sci U S A. 1991;88:3637–3641. doi: 10.1073/pnas.88.9.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan N C, Heinzmann C, Gal A, Klisak I, Orth U, Lai E, Grimsby J, Sparkes R S, Mohandas T, Shih J C. Human monoamine oxidase A and B genes map to Xp 11.23 and are deleted in a patient with Norrie disease. Genomics. 1989;4:552–559. doi: 10.1016/0888-7543(89)90279-6. [DOI] [PubMed] [Google Scholar]
- Shih J C, Chen K, Ridd M J. Monoamine oxidase: from genes to behavior. Annu Rev Neurosci. 1999;22:197–217. doi: 10.1146/annurev.neuro.22.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolato M, Chen K, Shih J C. Monoamine oxidase inactivation: from pathophysiology to therapeutics. Adv Drug Deliv Rev. 2008;60:1527–1533. doi: 10.1016/j.addr.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadic A, Rujescu D, Szegedi A, Giegling I, Singer P, Moller H J, Dahmen N. Association of a MAOA gene variant with generalized anxiety disorder, but not with panic disorder or major depression. Am J Med Genet B Neuropsychiatr Genet. 2003;117B:1–6. doi: 10.1002/ajmg.b.10013. [DOI] [PubMed] [Google Scholar]
- Jiang S, Xin R, Lin S, Qian Y, Tang G, Wang D, Wu X. Linkage studies between attention-deficity hyperactivity disorder and the monoamine oxidase genes. Am J Med Genet. 2001;105:783–788. doi: 10.1002/ajmg.10098. [DOI] [PubMed] [Google Scholar]
- Lawson D C, Turic D, Langley K, Pay H M, Govan C F, Norton N, Hamshere M L, Owen M J, O'Donovan M C, Thapar A. Association analysis of monoamine oxidase A and attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2003;116B:84–89. doi: 10.1002/ajmg.b.10002. [DOI] [PubMed] [Google Scholar]
- Brunner H G, Nelen M, Breakefield X O, Ropers H H, van Oost B A. Abnormal behavior associated with a point mutation in the structural gene for monoamine oxidase A. Science. 1993;262:578–580. doi: 10.1126/science.8211186. [DOI] [PubMed] [Google Scholar]
- Cases O, Seif I, Grimsby J, Gaspar P, Chen K, Pournin S, Muller U, Aguet M, Babinet C, Shih J C. Aggressive behaviors and altered amounts of brain serotonin and norepinephrine in mice lacking MAOA. Science. 1995;268:1763–1766. doi: 10.1126/science.7792602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A L, Bortolato M, Chen K, Shih J C. Novel monoamine oxidase A knock out mice with human-like spontaneous mutation. Neuroreport. 2008;19:739–743. doi: 10.1097/WNR.0b013e3282fd6e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J G, Allison C, Scott F J, Bolton P F, Baron-Cohen S, Matthews F E, Brayne C. The childhood autism spectrum test (CAST): sex differences. J Autism Dev Disord. 2008;38:1731–1739. doi: 10.1007/s10803-008-0558-6. [DOI] [PubMed] [Google Scholar]
- Dulcan M. Practice parameters for the assessment and treatment of children, adolescents, and adults with attention-deficit/hyperactivity disorder. American Academy of Child and Adolescent Psychiatry. J Am Acad Child Adolesc Psychiatry. 1997;36:85S–121S. doi: 10.1097/00004583-199710001-00007. [DOI] [PubMed] [Google Scholar]
- Jaisoorya T S, Reddy Y C, Srinath S, Thennarasu K. Sex differences in Indian patients with obsessive-compulsive disorder. Compr Psychiatry. 2009;50:70–75. doi: 10.1016/j.comppsych.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Pooley E C, Fineberg N, Harrison P J. The met(158) allele of catechol-O-methyltransferase (COMT) is associated with obsessive-compulsive disorder in men: case-control study and meta-analysis. Mol Psychiatry. 2007;12:556–561. doi: 10.1038/sj.mp.4001951. [DOI] [PubMed] [Google Scholar]
- Wright S L, Langenecker S A, Deldin P J, Rapport L J, Nielson K A, Kade A M, Own L S, Akil H, Young E A, Zubieta J K. Gender-specific disruptions in emotion processing in younger adults with depression. Depress Anxiety. 2009;26:182–189. doi: 10.1002/da.20502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Nanclares L, Gonzalez-Soriano J, Rodriguez J R, DeFelipe J. Gender differences in human cortical synaptic density. Proc Natl Acad Sci U S A. 2008;105:14615–14619. doi: 10.1073/pnas.0803652105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron-Cohen S, Knickmeyer R C, Belmonte M K. Sex differences in the brain: implications for explaining autism. Science. 2005;310:819–823. doi: 10.1126/science.1115455. [DOI] [PubMed] [Google Scholar]
- Cosgrove K P, Mazure C M, Staley J K. Evolving knowledge of sex differences in brain structure, function, and chemistry. Biol Psychiatry. 2007;62:847–855. doi: 10.1016/j.biopsych.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osran H, Reist C, Chen C C, Lifrak E T, Chicz-DeMet A, Parker L N. Adrenal androgens and cortisol in major depression. Am J Psychiatry. 1993;150:806–809. doi: 10.1176/ajp.150.5.806. [DOI] [PubMed] [Google Scholar]
- Sumner B E, Fink G. The density of 5-hydoxytryptamine2A receptors in forebrain is increased at pro-oestrus in intact female rats. Neurosci Lett. 1997;234:7–10. doi: 10.1016/s0304-3940(97)00651-4. [DOI] [PubMed] [Google Scholar]
- Knickmeyer R C, Baron-Cohen S. Fetal testosterone and sex differences. Early Hum Dev. 2006;82:755–760. doi: 10.1016/j.earlhumdev.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Knickmeyer R C, Baron-Cohen S. Fetal testosterone and sex differences in typical social development and in autism. J Child Neurol. 2006;21:825–845. doi: 10.1177/08830738060210101601. [DOI] [PubMed] [Google Scholar]
- Ozcan M E, Banoglu R. Gonadal hormones in schizophrenia and mood disorders. Eur Arch Psychiatry Clin Neurosci. 2003;253:193–196. doi: 10.1007/s00406-003-0424-7. [DOI] [PubMed] [Google Scholar]
- Mill J, Petronis A. Molecular studies of major depressive disorder: the epigenetic perspective. Mol Psychiatry. 2007;12:799–814. doi: 10.1038/sj.mp.4001992. [DOI] [PubMed] [Google Scholar]
- Chan W Y, Rennert O M. Molecular aspects of sex differentiation. Curr Mol Med. 2002;2:25–37. doi: 10.2174/1566524023363095. [DOI] [PubMed] [Google Scholar]
- Kim Y, Capel B. Balancing the bipotential gonad between alternative organ fates: a new perspective on an old problem. Dev Dyn. 2006;235:2292–2300. doi: 10.1002/dvdy.20894. [DOI] [PubMed] [Google Scholar]
- Wilhelm D, Palmer S, Koopman P. Sex determination and gonadal development in mammals. Physiol Rev. 2007;87:1–28. doi: 10.1152/physrev.00009.2006. [DOI] [PubMed] [Google Scholar]
- Chen K, Ou X M, Chen G, Choi S H, Shih J C. R1, a novel repressor of the human monoamine oxidase A. J Biol Chem. 2005;280:11552–11559. doi: 10.1074/jbc.M410033200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X M, Chen K, Shih J C. Glucocorticoid and androgen activation of monoamine oxidase A is regulated differently by R1 and Sp1. J Biol Chem. 2006;281:21512–21525. doi: 10.1074/jbc.M600250200. [DOI] [PubMed] [Google Scholar]
- Oh H J, Li Y, Lau Y F. Sry associates with the heterochromatin protein 1 complex by interacting with a KRAB domain protein. Biol Reprod. 2005;72:407–415. doi: 10.1095/biolreprod.104.034447. [DOI] [PubMed] [Google Scholar]
- Jiang H, Jiang Q, Liu W, Feng J. Parkin suppresses the expression of monoamine oxidases. J Biol Chem. 2006;281:8591–8599. doi: 10.1074/jbc.M510926200. [DOI] [PubMed] [Google Scholar]
- Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Jungert K, Buck A, von Wichert G, Adler G, Konig A, Buchholz M, Gress T M, Ellenrieder V. Sp1 is required for transforming growth factor-beta-induced mesenchymal transition and migration in pancreatic cancer cells. Cancer Res. 2007;67:1563–1570. doi: 10.1158/0008-5472.CAN-06-1670. [DOI] [PubMed] [Google Scholar]
- Salahpour A, Medvedev I O, Beaulier J M, Gainetdinov R R, Caron M G. Local knockdown of genes in the brain using small interfering RNA: a phenotypic comparison with knockout animals. Biol Psychiatry. 2007;61:65–69. doi: 10.1016/j.biopsych.2006.03.020. [DOI] [PubMed] [Google Scholar]
- Wu J B, Chen K, Ou X M, Shih J C. Retinoic acid activates monoamine oxidase B promoter in human neuronal cells. J Biol Chem. 2009;284:16723–16735. doi: 10.1074/jbc.M901779200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q S, Grimsby J, Chen K, Shih J C. Promoter organization and activity of human monoamine oxidase (MAO) A and B genes. J Neurosci. 1992;12:4437–4446. doi: 10.1523/JNEUROSCI.12-11-04437.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley V R, Lovell-Badge R, Goodfellow P N. Definition of a consensus DNA binding site for SRY. Nucleic Acids Res. 1994;22:1500–1501. doi: 10.1093/nar/22.8.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Dynlacht B D. Use of chromatin immunoprecipitation assays in genome-wide location analysis of mammalian transcription factors. Methods Enzymol. 2004;376:304–315. doi: 10.1016/S0076-6879(03)76020-0. [DOI] [PubMed] [Google Scholar]
- Taketo T, Lee C H, Zhang J, Li Y, Lee C Y, Lau Y F. Expression of SRY proteins in both normal and sex-reversed XY fetal mouse gonads. Dev Dyn. 2005;233:612–622. doi: 10.1002/dvdy.20352. [DOI] [PubMed] [Google Scholar]
- Dennis C. Brain development: the most important sexual organ. Nature. 2004;427:390–392. doi: 10.1038/427390a. [DOI] [PubMed] [Google Scholar]
- Dewing P, Chiang C W, Sinchak K, Sim H, Fernagut P O, Kelly S, Chesselet M F, Micevych P E, Albrecht K H, Harley V R, Vilain E. Direct regulation of adult brain function by the male-specific factor SRY. Curr Biol. 2006;16:415–420. doi: 10.1016/j.cub.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Mayer A, Lahr G, Swaab D F, Pilgrim C, Reisert I. The Y-chromosome genes SRY and ZNF are transcribed in adult human brain. Neurogenetics. 1998;1:281–288. doi: 10.1007/s100480050042. [DOI] [PubMed] [Google Scholar]
- Harley V R, Clarkson M J, Argentaro A. The molecular action and regulation of the testis-determining factors, SRY (sex-determining region on the Y chromosome) and SOX9 [SRY-related high-mobility group (HMG) box 9] Endocr Rev. 2003;24:466–487. doi: 10.1210/er.2002-0025. [DOI] [PubMed] [Google Scholar]
- Poulat F, Girard F, Chevron M P, Goze C, Rebillard X, Calas B, Lamb N, Berta P. Nuclear localization of the testis determining gene product SRY. J Cell Biol. 1995;128:737–748. doi: 10.1083/jcb.128.5.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner H M, Huth J R, Gronenborn A M, Clore G M. Molecular basis of human 46X, Y sex reversal revealed from the three-dimentional solution structure of the human SRY-DNA complex. Cell. 1995;81:705–714. doi: 10.1016/0092-8674(95)90532-4. [DOI] [PubMed] [Google Scholar]
- Briggs M R, Kadonaga J T, Bell S P, Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986;234:47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- Suske G. The Sp1-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- Shih J C, Zhu Q S, Chen K. Expression of human monoamine oxidase (MAO) A gene controlled by transcription factor Sp1. Prog Brain Res. 1995;106:49–56. doi: 10.1016/s0079-6123(08)61201-7. [DOI] [PubMed] [Google Scholar]
- Zhu Q S, Chen K, Shih J C. Bidirectional promoter of human monoamine oxidase A (MAO A) controlled by transcription factor Sp1. J Neurosci. 1994;14:7393–7403. doi: 10.1523/JNEUROSCI.14-12-07393.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins N D, Edwards N L, Duckett C S, Agranoff A B, Schmid R M, Nabel G J. A cooperative interaction between NF-κB and Sp1 is required for HIV-1 enhancer activation. EMBO J. 1993;12:3551–3558. doi: 10.1002/j.1460-2075.1993.tb06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom A C, Forsberg M, Lillhager P, Wester G. The transcription factors Sp1 and Oct-1 interact physically to regulate human U2 snRNA gene expression. Nucleic Acids Res. 1996;24:1981–1986. doi: 10.1093/nar/24.11.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T L, Alba D, Baxendale V, Rennert O M, Chan W Y. Application of transcriptional and biological network analyses in mouse germ-cell transcriptomes. Genomics. 2006;88:18–33. doi: 10.1016/j.ygeno.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Marin M, Karis A, Visser P, Grosveld F, Philipsen S. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell. 1997;89:619–628. doi: 10.1016/s0092-8674(00)80243-3. [DOI] [PubMed] [Google Scholar]
- Zhao C, Meng A. Sp1-like transcription factors are regulators of embryonic development in vertebrates. Dev Growth Differ. 2005;47:201–211. doi: 10.1111/j.1440-169X.2005.00797.x. [DOI] [PubMed] [Google Scholar]
- Lau Y F, Li Y, Kido T. Gonadoblastoma locus and the TSPY gene on the human Y chromosome. Birth Defects Res C Embryo Today. 2009;87:114–122. doi: 10.1002/bdrc.20144. [DOI] [PubMed] [Google Scholar]
- Li Y, Oh H J, Lau Y F. The poly(ADP-ribose) polymerase 1 interacts with Sry and modulates its biological function. Mol Cell Endocrinol. 2006;26:35–46. doi: 10.1016/j.mce.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Chen K. Organization of MAO A and MAO B promoters and regulation of gene expression. Neurotoxicology. 2004;25:31–36. doi: 10.1016/S0161-813X(03)00113-X. [DOI] [PubMed] [Google Scholar]
- Sinclair A H, Berta P, Palmer M S, Hawkins J R, Griffiths B L, Smith M J, Foster J W, Frischauf A M, Lovell-Badge R, Goodfellow P N. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:216–217. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- Pontiggia A, Rimini R, Harley V R, Goodfellow P N, Lovell-Badge R, Bianchi M E. Sex-reversing mutations affect the architecture of SRY-DNA complexes. EMBO J. 1994;13:6115–6124. doi: 10.1002/j.1460-2075.1994.tb06958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenet L, Mejean C, Moniot B, Bonneaud N, Galeotti N, Aldrian-Herrada G, Poulat F, Berta P, Benkirane M, Boizet-Bonhoure B. Regulation of human SRY subcellular distribution by its acetylation/deacetylation. EMBO J. 2004;23:3336–3345. doi: 10.1038/sj.emboj.7600352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M, Bhowmik A D, Sinha S, Chattopadhyay A, Chaudhuri K, Singh M, Mukhopadhyay K. MAOA promoter polymorphism and attention deficit hyperactivity disorder (ADHD) in indian children. Am J Genet B Neuropsychiatr Genet. 2006;141B:637–642. doi: 10.1002/ajmg.b.30385. [DOI] [PubMed] [Google Scholar]
- Domschke K, Sheehan K, Lowe N, Kirley A, Mullins C, O'sullivan R, Freitag C, Becker T, Conroy J, Fitzgerald M, Gill M, Hawi Z. Association analysis of the monoamine oxidase A and B genes with attention deficit hyperactivity disorder (ADHD) in an Irish sample: preferential transmission of the MAO-A 941G allele to affected children. Am J Genet B Neuropsychiatr Genet. 2005;134B:110–114. doi: 10.1002/ajmg.b.30158. [DOI] [PubMed] [Google Scholar]
- Guan L, Wang B, Chen Y, Yang L, Li J, Qian Q, Wang Z, Faraone S V, Wang Y. A high-density single-nucleotide polymorphism screen of 23 candidate genes in attention deficit hyperactivity disorder: suggesting multiple susceptibility genes among Chinese Han population. Mol Psychiatry. 2009;14:546–554. doi: 10.1038/sj.mp.4002139. [DOI] [PubMed] [Google Scholar]
- Rommelse N N, Altink M E, Arias-Vasguez A, Buschgens C J, Fliers E, Faraone S V, Buitelaar J K, Sergeant J A, Oosterlaan J, Franke B. Differential association between MAOA, ADHD and neuropsychological functioning in boys and girls. Am J Genet B Neuropsychiatr Genet. 2008;147B:1524–1530. doi: 10.1002/ajmg.b.30845. [DOI] [PubMed] [Google Scholar]
- Pinsonneault J K, Papp A C, Sadee W. Allelic mRNA expression of X-linked monoamine oxidase a (MAOA) in human brain: dissection of epigenetic and genetic factors. Hum Mol Genet. 2006;15:2636–2649. doi: 10.1093/hmg/ddl192. [DOI] [PubMed] [Google Scholar]
- Carrel L, Willard H F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Nordquist N, Oreland L. Monoallelic expression of MAOA in skin fibroblast. Biochem Biophys Res Commun. 2006;348:763–767. doi: 10.1016/j.bbrc.2006.07.131. [DOI] [PubMed] [Google Scholar]
- Chugani D C, Muzik O, Chakraborty P, Mangner T, Chugani H T. Human brain serotonin synthesis capacity measured in vivo with alpha-[C-11]methyl-L-tryptophan. Synapse. 1998;28:33–43. doi: 10.1002/(SICI)1098-2396(199801)28:1<33::AID-SYN5>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Nishizawa S, Benkelfat C, Young S N, Leyton M, Mzengeza S, de Montigny C, Blier P, Diksic M. Differences between males and females in rates of serotonin synthesis in human brain. Proc Natl Acad Sci U S A. 1997;94:5308–5313. doi: 10.1073/pnas.94.10.5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen I L, Liu X, Schutz C, White B N, Jenkins E C, Brown W T, Holden J J. Association of autism severity with a monoamine oxidase A functional polymorphism. Clin Genet. 2003;64:190–197. doi: 10.1034/j.1399-0004.2003.00115.x. [DOI] [PubMed] [Google Scholar]
- Spring S, Lerch J P, Henkelman R M. Sexual dimorphism revealed in the structure of the mouse brain using three-dimentional magnetic resonance imaging. Neuroimage. 2007;35:1424–1433. doi: 10.1016/j.neuroimage.2007.02.023. [DOI] [PubMed] [Google Scholar]
- Davis L K, Hazlett H C, Librant A L, Nopoulos P, Sheffield V C, Piven J, Wassink T H. Cortical enlargement in autism is associated with a functional VNTR in the monoamine oxiase A gene. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1145–1151. doi: 10.1002/ajmg.b.30738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hranilovic D, Novak R, Babic M, Novokmet M, Bujas-Petkovic Z, Jernej B. Hyperserotonemia in autism: the potential role of 5HT-related gene variants. Coll Antropol. 2008;32:75–80. [PubMed] [Google Scholar]
- Jonsson E G, Norton N, Forslund K, Mattila-Evenden M, Rylander G, Asberg M, Owen M J, Sedvall G C. Association between a promoter variant in the monoamine oxidase A gene and schizophrenia. Schizophr Res. 2003;61:31–37. doi: 10.1016/s0920-9964(02)00224-4. [DOI] [PubMed] [Google Scholar]
- Yoo H J, Lee S K, Park M, Cho I H, Hyun S H, Lee J C, Yang S Y, Kim S A. Family- and population-based association studies of monoamine oxidase A and autism spectrum disorders in Korean. Neurosci Res. 2009;63:172–176. doi: 10.1016/j.neures.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Wilson C A, Davies D C. The control of sexual differentiation of the reproductive system and brain. Reproduction. 2007;133:331–359. doi: 10.1530/REP-06-0078. [DOI] [PubMed] [Google Scholar]
- Arnold A P, Xu J, Grisham W, Chen X, Kim Y H, Itoh Y. Minireview: Sex chromosomes and brain sexual differentiation. Endocrinology. 2004;145:1057–1062. doi: 10.1210/en.2003-1491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.