Abstract
Genes required for ribosome biogenesis in yeast, referred to collectively as the Ribi regulon, are tightly regulated in coordination with nutrient availability and cellular growth rate. The promoters of a significant fraction of Ribi genes contain one or more copies of the RNA polymerases A and C (PAC) and/or ribosomal RNA-processing element (RRPE) motifs. Prompted by recent studies showing that the yeast protein Dot6 and its homolog Tod6 can bind to a PAC motif sequence in vitro and are required for efficient Ribi gene repression in response to heat shock, we have examined the role of Dot6 and Tod6 in nutrient control of Ribi gene expression in vivo. Our results indicate that PAC sites function as Dot6/Tod6-dependent repressor elements in vivo. Moreover, Dot6 and Tod6 mediate different nutrient signals, with Tod6 responsible for efficient repression of Ribi genes after inhibition of the nitrogen-sensitive TORC1 pathway and Dot6 responsible for repression after inhibition of the carbon-sensitive protein kinase A signaling pathway. Consistently, Dot6 and Tod6 are required for efficient repression of Ribi gene repression immediately after nutrient deprivation and for successful adaptation to nutrient limitation. Thus, these results establish Dot6/Tod6 as a direct link between nutrient availability, Ribi gene regulation, and growth control.
Keywords: nutrient growth control, rapamycin, Saccharomyces cerevisiae, transcriptional control
Yeast cells can alter their growth rate in proportion to nutrient availability over a wide range of limiting nutrient concentrations (1–3). This nutrient control of cell growth essentially reduces to nutrient control of ribosome biogenesis, because the cohort of ribosomes in the cell represents the majority of the cell's mass and because ribosomes provide the biosynthetic capacity by which cells can increase their mass. Moreover, recent studies have suggested that the mechanism by which nutrients impinge on the control of cell size and the coupling of cell size to cell cycle progression proceeds predominantly through ribosome biogenesis (4, 5). Thus, understanding fundamental questions of cell growth and cell division rests on understanding ribosome biogenesis.
The budding yeast ribosome comprises 79 ribosomal proteins encoded by 138 genes (the RP regulon), and four rRNAs (5S, 5.8S, 18S, and 25S) encoded by ≈150 rDNA tandem repeats. Moreover, another ≈300 genes encoding nonribosomal proteins are involved in various aspects of ribosome assembly and translational capacity (the ribosome biogenesis, or Ribi, regulon): RNA polymerases I and III subunits, tRNA synthetases, rRNA processing and modifying enzymes, translation factors, etc. (6, 7). Synthesis of the translation machinery is very energetically expensive to the cell. For instance, in an exponentially growing cell, ribosome synthesis uses ≈90% of the total cellular energy (8). Not surprisingly, the cell carefully adjusts its ribosome biogenesis in response to changes in nutrient availability.
Genes in the Ribi regulon are rapidly and robustly induced after glucose addition to glucose-depleted cells or addition of glutamine to cells growing on a poor nitrogen source. This affect is mimicked by activation of protein kinase A (PKA) or by activation of Sch9, the yeast homolog of mammalian Akt/PKB (9). Conversely, expression of these genes is repressed by rapamycin addition or by inactivation of Sch9 or PKA (9, 10). PKA comprises a heterotetramer consisting of two cAMP-responsive regulatory subunits encoded by BCY1, and two kinase subunits encoded by the essentially redundant genes TPK1, TPK2, and TPK3. The small G protein Ras2 regulates PKA through activation of adenylyl cyclase in response to the presence of glucose. Sch9 is activated in yeast by direct phosphorylation by the nitrogen source-responsive Target of Rapamycin Complex 1 (TORC1), whose activity itself is quite sensitive to the macrolide antibiotic rapamycin (10). The pathways through which the PKA and Sch9 activities impinge on ribosomal RNA and ribosomal protein synthesis have been extensively explored (11). However, the direct link between the Ribi gene regulon and the PKA and Sch9 pathways has not previously been identified.
Ribi gene promoters are enriched for RNA polymerases A (I) and C (III) (PAC), and ribosomal RNA-processing element (RRPE) motifs in specific orientation to each other and to the transcriptional start site (7, 12, 13). The split finger transcription factor Sfp1 has been implicated in Ribi gene regulation: loss of Sfp1 results in reduced expression of Ribi genes, whereas induction of Sfp1 results in a rapid activation of the Ribi regulon (13). Moreover, Sfp1 partitions between the nucleus and the cytoplasm, and the relative fraction of Sfp1 in the nucleus increases in nutrient-rich medium and in strains expressing high PKA or Sch9 activity, conditions under which Ribi gene expression is enhanced. Despite the correlation of Sfp1 nuclear localization with Ribi gene expression, Sfp1 does not bind in vitro or in vivo to RRPE/PAC-containing promoters (6, 13–15). Thus, the mechanism of regulation of Ribi gene expression has remained elusive.
Several recent studies have identified proteins that bind to the PAC and RRPE sites. Liko et al. (16) identified Stb3 as a protein capable of binding to RRPE both in vitro and in vivo and provided evidence that Stb3 is required for efficient glucose activation of Ribi genes. More recently, Hughes and colleagues (17) and Bulyk and colleagues (18) used protein binding to DNA microarrays to demonstrate that the yeast protein Dot6 and its homolog encoded by the gene YBL056W, recently renamed TOD6, could bind to the PAC motif in vitro. Consistently, Freckleton et al. (19) used a novel phage display method to demonstrate that Dot6 binds to a canonical PAC site in vitro. Moreover, Zhu et al. (18) provided data demonstrating that Dot6 and Tod6 redundantly function to repress Ribi genes after heat shock. We have examined their role in nutrient regulation of Ribi gene expression. In results described below, we show that Dot6 and Tod6 are overlapping repressors of Ribi gene transcription, whose repressive activity is alleviated by the presence of rich nutrient sources mediated by the PKA and TORC1 nutrient signaling pathways.
Results
PAC Sites Mediate Dot6/Tod6 Transcriptional Repression.
Because Dot6 and Tod6 were identified as proteins that bind PAC sites in vitro and because heat shock-stimulated repression of Ribi genes, many of which have PAC sites in their promoters, is attenuated in a dot6 tod6 mutant strain, we tested directly whether PAC sites can elicit Dot6- and Tod6-dependent repression. We constructed a reporter plasmid in which the expression of lacZ, encoding Escherichia coli β-galactosidase, is driven either by the constitutively active promoter from the yeast HOP1 gene or the identical HOP1 promoter into which two copies of a 10-bp PAC motif were inserted. The two reporter plasmids were individually transformed into WT, dot6, tod6, and dot6 tod6 strains, and the level of β-galactosidase activity was measured for each transformant. As evident in Table 1, PAC sites substantially repress HOP1 promoter activity: β-galactosidase activity in the WT strain from the PAC site-containing reporter is one fiftieth of that from the promoter lacking PAC sites. Moreover, this repression requires either Dot6 or Tod6: although repression is slightly mitigated by deletion of either DOT6 or TOD6, it is substantially alleviated by deletion of both genes. From these results, we conclude that both Dot6 and Tod6 can independently act through PAC motifs to elicit transcriptional repression.
Table 1.
PAC sites are Dot6/Tod6-dependent repressor elements
| Strain | β-Galactosidase activity |
|
|---|---|---|
| PHOP1-lacZ | PHOP1-PAC2-lacZ | |
| WT | 143 ± 3 | 3.1 ± 0.4 |
| dot6 | 140 ± 2 | 5.1 ± 0.1 |
| tod6 | 144 ± 6 | 10.6 ± 0.2 |
| dot6 tod6 | 110 ± 2 | 42.2 ± 1.0 |
Strains Y2092 (WT), Y3705 (dot6), Y3706 (tod6), and Y3707 (dot6 tod6) were transformed with either plasmid B2558, a URA3 2-μ vector carrying lacZ under control of the constitutively active HOP1 promoter, or plasmid B2901, identical to B2558 except with two copies of PAC motifs inserted in the HOP1 promoter. Strains carrying each plasmid were grown in synthetic complete medium lacking uracil to mid-log and then harvested and assayed for β- galactosidase activity. Values are displayed in Miller units and are mean ± SD of three independent transformants for each strain.
Repression of Ribi Genes in Response to TORC1 Inactivation Requires Tod6.
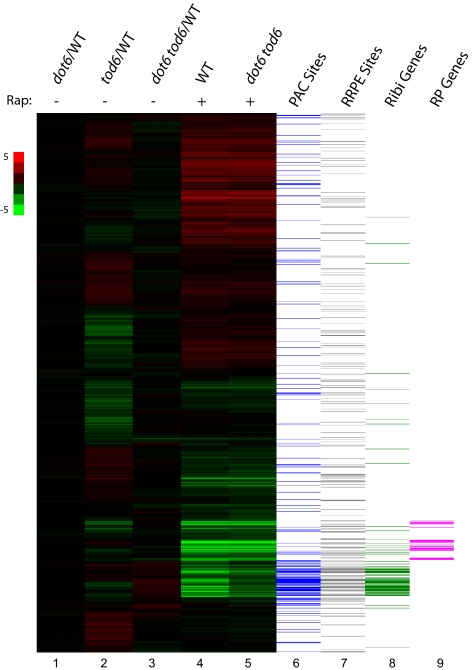
The promoters of Ribi genes are highly enriched in PAC motifs (7, 12, 13). The Saccharomyces Genome Database currently annotates 303 nonribosomal protein genes in the functional category of ribosome biogenesis, and 199 of these overlap with the 647 genes whose promoters contain one or more PAC sites (P < 10−190). Because Dot6 and Tod6 bind PAC sites in vitro and elicit repression through PAC sites in vivo, we investigated the role of Dot6 and Tod6 in nutrient control of Ribi gene expression. We examined the transcriptional profiles of WT cells and cells lacking Dot6, Tod6, or both during the early exponential phase of growth in rich medium and under conditions of impaired growth. Although deletion of Tod6 alone had a modest effect on expression of a number of genes during growth in rich medium, loss of Dot6 alone or in combination with Tod6 did not result in any attenuation in the expression of Ribi genes under these conditions (Fig. 1). In fact, the mRNA levels for most Ribi genes were slightly elevated in dot6 tod6 cells in comparison with WT cells, consistent with a repressive activity of these regulators. In contrast, dot6 tod6 cells showed a specific defect in Ribi gene repression by rapamycin treatment. As evident in Fig. 1, inhibition of TORC1 by rapamycin results in significant reprogramming of gene expression in yeast cells: Ribi and ribosomal protein genes are rapidly and extensively repressed while stress-responsive genes and those involved in use of alternative nitrogen sources are rapidly and substantially induced (20). Although most of those changes in gene expression also occur in dot6 tod6 cells, repression of most Ribi genes was attenuated in these cells. Thus, Ribi gene repression, but not other transcriptional changes in response to rapamycin treatment, depends on Dot6 and Tod6.
Fig. 1.
Dot6 and Tod6 specifically affect repression of ribosome biogenesis genes. Lanes 1–3: relative steady-state expression of yeast genes in mutant versus WT cells grown in rich medium. RNA isolated from Y3705 (dot6), Y3706 (tod6), or Y3707 (dot6 tod6) grown to early log phase in SC plus 2% glucose medium was labeled with Cy3 and mixed with Cy5-labeled RNA from Y2092 (WT) similarly grown and hybridized to Agilent microarray chips. The heat map organized by unsupervised hierarchical clustering shows the relative expression of each of 5,600 yeast genes (horizontal lines) in mutant versus WT (scale shown on the left). Lanes 4 and 5: strains Y2092 (WT) and Y3707 (dot6 tod6) were grown in SC plus 2% glucose medium to a density of OD600 = 0.25, at which point rapamycin was added to a final concentration of 100 nM. Heat map represents expression levels of yeast genes 80 min after rapamycin addition relative to cells just before addition for each of the two indicated strains. Lanes 6 and7 denote the presence of PAC (GCGATGAGNT) or RRPE (TGAAAATTT) motifs within 500 bp upstream of the transcriptional start site of the corresponding yeast gene, and lanes 8 and 9 indicate whether the corresponding genes are annotated as ribosome biogenesis (Ribi) or ribosomal protein (RP) genes.
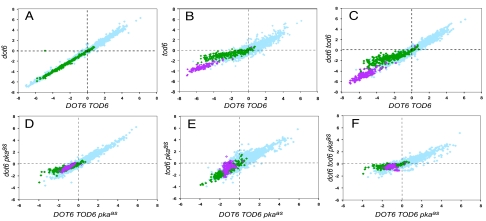
To examine in more detail the role of these repressor proteins on the global transcriptional response of cells to TORC1 inhibition, cells growing in rich medium were treated with 100 nM rapamycin for 80 min, and the expression changes in the strains lacking Dot6, Tod6, or both were compared to those in WT cells. As seen in Fig. 2A, elimination of Dot6 had no effect on rapamycin-induced transcriptional changes (R2 = 0.99 between the responses of DOT6 and dot6 strains). In contrast, loss of Tod6 significantly attenuated the repression of Ribi genes by rapamycin (Fig. 2B). A total of 80% of all Ribi genes are repressed by more than 2-fold by rapamycin addition to TOD6 cells, and the repression of all of these is attenuated in tod6 cells. Of those Ribi genes whose repression was attenuated by at least 4-fold, 90% contain at least one PAC motif in their promoter (Table S1). Attenuation in the expression response of tod6 cells was only observed in genes that were repressed upon rapamycin and was limited predominantly to Ribi genes containing PAC motifs. By contrast, ribosomal protein genes, which are dramatically repressed by rapamycin in TOD6 cells and generally lack PAC or RRPE motifs, still undergo substantial repression in tod6 cells. The responses to rapamycin of most genes lacking PAC sites are unaffected in tod6 cells, as are those of genes containing PAC sites that are not normally repressed by rapamycin. Finally, the response of dot6 tod6 strains to rapamycin treatment is essentially identical to that of tod6 cells (Fig. 2C). From these results, we conclude that Tod6 but not Dot6 is required for efficient repression of Ribi genes by rapamycin, and this effect is restricted predominantly to the PAC-containing Ribi genes.
Fig. 2.
Dot6 and Tod6 are differentially required for repression of Ribi genes after inactivation of TORC1 or PKA. Shown are scatter plots of microarray data for ≈5,600 yeast genes, obtained from two different strains after the same change in condition. Each point represents the log2 change in levels of the mRNA for a single gene for one strain in the horizontal dimension and the log2 change in mRNA levels for that gene in a different strain in the vertical dimension. (A–C) Expression changes of all genes in a dot6 (Y3705), tod6 (Y3706), or dot6 tod6 (Y3707) strain pregrown on SC plus 2% glucose, 80 min after addition of rapamycin to 100 nM relative to that before addition (vertical axis), versus an isogenic DOT6 TOD6 strain (Y2092) treated the same way (horizontal axis). (D–F) Expression changes of all genes in a dot6 pkaas (Y3708), tod6 pkaas (Y3709), or dot6 tod6 pkaas (Y3710) strain (pkaas designates the triple-mutant tpk1as tpk2as tpk3as) pregrown on SC plus 2% glucose, 80 min after addition of 1NM-PP1 to 100 nM relative to that before addition (vertical axis), versus an isogenic DOT6 TOD6 pkaas strain (Y3561) treated the same way (horizontal axis). Points corresponding to genes annotated as Ribi genes are shown in green, and those corresponding to RP genes are shown in pink.
Deletion of TOD6 has broader effects on repression immediately after rapamycin addition than at the later time point described above. As noted in Fig. S1, the level of repression of ribosomal proteins genes, which comprise most of the non-Ribi genes subject to rapamycin repression, is also attenuated in a tod6 strain 20 min after rapamycin addition relative to that in a WT strain. This enhanced effect of TOD6 deletion at early times is not due to exclusion of rapamycin from the cell or resistance of TORC1 to rapamycin inhibition, because genes subject to rapamycin induction are induced equally well at 20 min in both strains. Rather, this may reflect a coupling of ribosomal protein gene expression to Ribi gene expression, at least at early stages after rapamycin treatment. However, the long-term effect of Tod6 elimination is restricted predominantly to Ribi genes enriched in the PAC site.
Repression of Ribi Genes in Response to PKA Inactivation Requires Dot6.
As with rapamycin addition, PKA inactivation results in a massive transcriptional reprogramming of the cell, including repression of Ribi and RP genes (9, 21). Accordingly, we examined whether Dot6 or Tod6 mediates repression of Ribi genes in response to loss of PKA activity. To inactivate PKA specifically, we used a strain carrying a single mutation in each of TPK1, TPK2, and TPK3 that rendered each of the encoded kinases sensitive to inhibition by a modified kinase inhibitor, 1NM-PP1, which carries a bulky side group that precludes it from inhibiting WT PKA or any other kinase in the cell (22). We refer to the combination of tpk1as, tpk2as, and tpk3as mutations as pkaas. In the absence of the inhibitor, the mutant kinases function essentially normally in the cell, and pkaas strains exhibit normal growth and nutrient responses. We assessed the global transcriptional response of pkaas cells lacking Dot6, Tod6, or both after addition of 100 nM 1NM-PP1. Although repression of gene expression 80 min after inactivation of PKA in DOT6 TOD6 cells was not as extreme as that seen after rapamycin treatment, deletion of DOT6, whether alone or in combination with tod6, substantially attenuated that repression (Fig. 2). The degree of repression in DOT6 TOD6 cells was greater at 20 min than at 80 min (Fig. S1). At this time point as well, deletion of DOT6 attenuated Ribi gene repression, although we observed more substantial attenuation in the dot6 tod6 double mutant than in the dot6 single mutant. Deletion of TOD6 alone had no effect on repression at either time point. In all conditions, the genes whose repression was most affected were highly enriched for those involved in ribosome biogenesis and for those carrying PAC sites (Fig. 2 and Table S1). These results add weight to the hypothesis that Dot6 and Tod6 function as repressors of ribosome biogenesis by binding to PAC sites. The results also identify a distinction in the mechanism of Ribi gene repression resulting from TORC1 inactivation, which is mediated predominantly by Tod6, and Ribi gene repression resulting from PKA inactivation, which is mediated predominantly by Dot6.
We performed a similar series of experiments examining gene expression after inactivation of Sch9, a kinase regulated by TORC1 that functions in parallel with PKA in nutrient signaling (9). We replaced the WT allele of SCH9 with an analog-sensitive allele in strains deleted for DOT6, TOD6, or both and then measured gene expression globally after addition of 1NM-PP1. As with inactivation of PKA, gene expression changes were more pronounced 20 min after inactivation of Sch9 than at 80 min and, as previously reported, inactivation of Sch9 resulted in repression of Ribi genes (Fig. S2 and Table S1). This repression was essentially unaffected in either single-mutant background but was substantially attenuated in the dot6 tod6 double mutant. Thus, the role of Dot6 and Tod6 in Ribi gene repression after inactivation of Sch9 resembles neither that in PKA inactivation nor that in rapamycin treatment, despite the fact that Sch9 activity responds to rapamycin addition. These results are addressed in the Discussion.
dot6 tod6 Mutants Are Defective in Ribosomal Gene Repression After Nutrient Starvation.
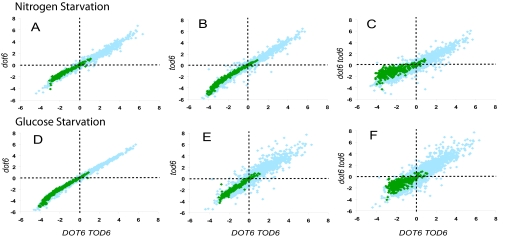
Because ribosome biogenesis is tightly coupled to nutrient availability and because two key nutrient-responsive signaling pathways affect expression of ribosome biogenesis genes through Dot6 and Tod6, we asked whether Dot6 and Tod6 also mediate the regulation of Ribi gene expression in response to changes in nutrient availability. To test this idea, we constructed prototroph strains lacking Dot6 and Tod6, both individually and in combination. We grew these prototroph mutants and their isogenic WT strain in synthetic media containing 2% glucose as sole carbon source and 0.1% glutamine as sole nitrogen source to early exponential phase and then transferred cells to media lacking either glucose or glutamine. As shown in Fig. 3, both of these transitions resulted in increased mRNA levels for a large number of genes and a decrease in mRNA levels for Ribi as well as other genes within 20 min, a pattern that persists for at least 80 min. Eliminating DOT6 alone or TOD6 alone did not have any significant effect on the change in Ribi gene repression or that of any other genes after either starvation condition (Table S1). In contrast, a significant fraction of the Ribi genes in the dot6 tod6 strain showed an attenuated reduction in mRNA levels relative to that seen in WT under both starvation conditions at both time points. Thus, although deletion of these two genes did not completely eliminate Ribi gene repression upon nutrient starvation, these results indicate that efficient repression after glucose or nitrogen starvation is compromised by deletion of both genes.
Fig. 3.
Dot6 and Tod6 are redundantly required for Ribi gene repression after nutrient deprivation. The indicated prototrophic strains were grown in synthetic medium containing 2% glucose and 0.1% glutamine as sole carbon and nitrogen sources, respectively, to an OD600 of 0.25, harvested by filtration nitrocellulose membranes via rapid vacuum filtration, washed with corresponding nutrient starvation medium, then transferred to identical prewarmed synthetic medium lacking glutamine (A–C; Nitrogen Starvation) or lacking glucose (D–F; Glucose Starvation). Samples were collected immediately before transfer (time point 0) and at 20 and 80 min after transfer and were analyzed by expression microarrays as described in Materials and Methods and the legend to Fig. 2. The resulting data are displayed as scatter plots with the log2 expression level of each gene 80 min after transfer relative to that before transfer in a mutant strain versus the corresponding value for that gene in the WT strain (Y3628). (A and D) dot6 strain (Y3714). (B and E) tod6 strain (Y3715). (C and F) dot6 tod6 strain (Y3716).
DOT6 and TOD6 Are Required for Efficient Adaptation to Nutrient Limitation.
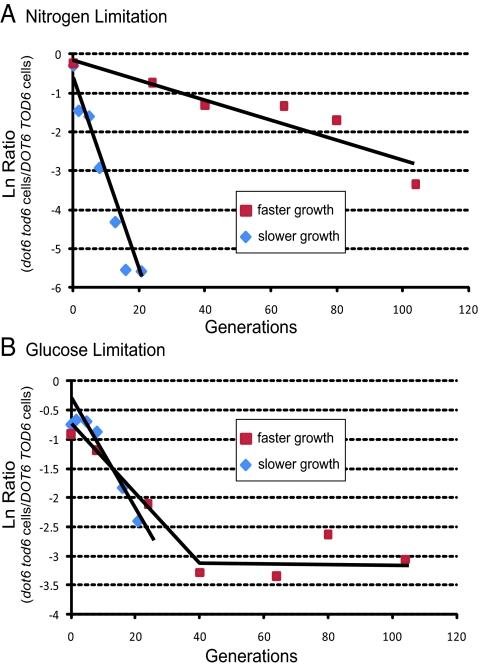
Because our results demonstrate that mutations in DOT6 and/or TOD6 attenuate repression of Ribi gene expression under a variety of growth-inhibiting conditions, we addressed whether elimination of these repressors had any obvious physiological consequence for cells confronted with nutrient limitation. We found that neither the double mutant nor either single mutant alone exhibited any apparent change in sensitivity to various levels of, or time of exposure to, rapamycin. Similarly, none of the mutants in a pkaas background exhibited any change in sensitivity to 1NM-PP1. However, in chemostat-based competition experiments under limiting nutrient conditions, DOT6 TOD6 rapidly outcompeted an isogenic dot6 tod6 strain, most notably in nitrogen-limiting conditions but also in glucose-limiting conditions (Fig. 4). In both cases, the effect was more pronounced under slower growth conditions than faster growth. Thus, Dot6 and Tod6 are necessary for successful adaptation to limiting nutrient conditions. We also determined that the dot6 tod6 strain showed modest but reproducible decrease in viability upon starvation for carbon or nitrogen, with single mutants showing viability intermediate between WT and the double mutant (Fig. S3). Moreover, we observed that nitrogen starvation, but not carbon starvation, induced petite formation, and this effect was enhanced in the dot6 tod6 strain (Fig. S4). From this, we conclude that Dot6 or Tod6 enhances fitness of yeast cells subject to nutrient deprivation.
Fig. 4.
Fitness competition between WT and dot6 tod6 strains under nutrient limitation. Competition under limiting nitrogen (A) or glucose (B) chemostat conditions between prototrophic WT and dot6 tod6 strains at either a dilution rate of 0.2 h−1 (red squares) or 0.05 h−1 (blue diamonds) was performed as described in Materials and Methods. Data are presented as the natural log of the ratio of dot6 tod6 colony-forming units to WT colony-forming units as a function of generations after initiation of competition in the chemostat. Linear regression trend lines are presented. For the glucose 0.2 h−1 culture, the trend line was calculated for only the first 40 generations.
Discussion
The promoters of genes encoding proteins required for biosynthesis of ribosomes are strongly enriched in two motifs, RRPE and PAC, a correlation that has prompted speculation that these motifs mediate binding of factors that regulate transcription of these genes. The recent identification of two proteins, Dot6 and Tod6, capable of binding the PAC motif sequence in vitro allowed us to test that hypothesis. The data presented in this report not only confirm that these proteins regulate ribosomal biogenesis gene expression but also demonstrate that these proteins function as nutrient-responsive transcriptional repressors.
Our data are consistent with a model in which Dot6 and Tod6 bind to and repress expression from PAC-containing Ribi promoters, and this repression activity or binding is eliminated by the action of two nutrient-responsive signaling pathways, mediated respectively by PKA and TORC1 (Fig. 5). First, we have directly demonstrated that Dot6 and Tod6 act specifically through PAC sites in vivo to effect transcriptional repression. Second, we recently analyzed several Ribi genes by chromatin immunoprecipitation and showed that Dot6 is bound to their promoters in vivo during growth under conditions of nutrient limitation but not during growth in rich medium (19). Moreover, Deminoff et al. (23) have shown that PKA phosphorylates Dot6 in vitro, and we have recently found that Dot6 and Tod6 exhibit increased phosphorylation on several PKA sites after addition of glucose to glycerol-grown cells (Table S2). Loewith and colleagues (24) have determined that phosphorylation of Dot6 and Tod6 is altered by rapamycin in a Sch9-dependent manner. Finally, both Dot6 and Tod6 have been identified as components of the Rpd3L complex, which likely possesses histone deacetylase activity, an association that may account for their repressive activity (25).
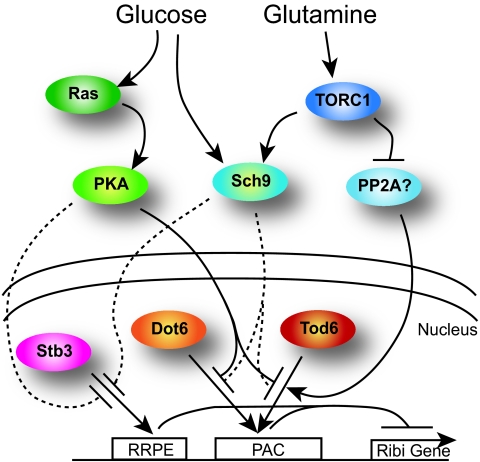
Fig. 5.
Model for nutrient regulation of Ribi gene expression. Dot6 and Tod6 bind to PAC sites to repress expression of Ribi genes in the absence of nutrients. Their binding is abrogated by nutrient availability, mediated in part through PKA and TORC1. Additional regulators must also participate in Ribi gene repression in the absence of nutrients, and we speculate that Stb3, acting through a second motif, RRPE, is at least one of those additional regulators. We also speculate that TORC1-dependent signaling components in addition to Sch9 modulate Tod6/Dot6 activity and propose that PP2A (or Sit4) may be one of those components.
Our results suggest that Dot6 and Tod6 function in response to distinct nutritional inputs but that they are not the sole components mediating nutrient regulation of Ribi genes. Although loss of Dot6 and Tod6 almost completely eliminates repression attendant on inactivation of PKA or TORC1 activity, such deletion strains only partially attenuate repression resulting from nutrient starvation. This would suggest that nutrient starvation engages Tod6 and Dot6 along with one or more additional transcriptional regulators. One likely candidate, indicated in Fig. 5, is Stb3, which has been recently shown to bind to the RRPE motif also present in promoters of Ribi genes (16). Recent data from our group and the Loewith group support a role for Stb3 in Ribi gene repression. Second, dot6 mutants are partially defective in repression after PKA inactivation but are unimpaired in repression after rapamycin addition. Tod6 mutants have the opposite properties. Further, neither mutant exhibits a defect after Sch9 inactivation, even though Sch9 lies downstream of TORC1 (10). These results indicate that Tod6 and Dot6 are not completely interchangeable. Moreover, their roles in regulation are not readily accommodated in a simple Boolean representation of a regulatory circuitry. Rather, our results are more readily interpreted assuming that PKA and Sch9 have quantitatively different effects on Tod6 and Dot6 activity and that Sch9 is not the only route through which TORC1 impinges on Dot6/Tod6 function. Clearly, further studies are needed to precisely define the connections from nutrient signaling to PKA and TORC1 activity, to Tod6 and Dot6 activity, and to Ribi gene repression.
Although PAC sites reside in promoters of genes other than those encoding ribosome biogenesis components, only Ribi genes among the PAC-containing promoters respond in a Dot6/Tod6-dependent manner to inactivation of PKA, rapamycin treatment, or nutrient limitation. This restriction suggests that Dot6/Tod6-mediated repression is limited primarily to those PAC sites resident in Ribi promoters. Several factors could contribute to this restricted spectrum of activity. Beer and Tavazoie (12) noted previously that the group of genes coregulated with the Ribi regulon contains not only PAC sites but also RRPE sites in a stereotypic organization relative to each other. Thus, an RRPE site positioned in a particular configuration relative to PAC sites could be required to render the associated PAC site responsive to Dot6/Tod6 regulation. In addition, we have recently shown that those genes with PAC sites residing in a nucleosome-free domain in the promoter are substantially more likely to respond to induction after nutrient upshift than those promoters in which the PAC site is occluded by nucleosomes (26). Thus, PAC site accessibility determined by nucleosome position could dictate whether a promoter carrying a PAC site could be regulated by Dot6/Tod6. These two factors—nucleosome-dictated accessibility and spatial coincidence with an RRPE site—could be functionally intertwined, because the RRPE sequence tends to exclude nucleosomes. Further studies will be needed to shed light on the ability of Dot6 and Tod6 to act on a specific subset of PAC sites.
Consistent with a role for Dot6 and Tod6 regulating ribosomal biogenesis in response to nutritional deprivation, we observed that dot6 tod6 mutants are less fit than WT cells under nutrient-limited conditions. Moreover, the degree of fitness diminishes with further limitation of available nutrients. We surmise that the reduced fitness of the mutants under nutrient-limiting conditions results from premature depletion of nutrient reserves, which are funneled into unproductive accumulation of unusable biosynthetic capacity. These observations suggest that cells must tightly regulate ribosome biogenesis to match nutritional availability and that a failure to achieve a precise correspondence diminishes the fitness of the cell. In addition, these observations add weight to our previous suggestion that cells control growth and biosynthetic capacity in response to their perception of their nutritional environment; namely, through the well-conserved, nutrient-sensing PKA and TORC1 pathways (11). Any disconnect between the cell's perception of nutrient availability and actual nutrient availability, as suggested by the results presented here, has deleterious consequences.
Materials and Methods
Strains and Media.
All strains used in this study are described in Table S3 and were derived from strain W303. Gene deletions were created by using one-step PCR-mediated gene disruption and standard yeast genetic techniques. Analogue-sensitive alleles of SCH9 and TPK1/2/3 were described previously (9, 13). All experiments were conducted in synthetic medium (S; 1.7 g/L yeast nitrogen base without amino acids or ammonium sulfate) supplemented with 2% glucose (D), 0.5% ammonium sulfate (N), and/or amino acids and bases (C), as indicated.
RNA Preparation and Microarray.
RNA preparation, amplification, microarray hybridization, and analysis were performed as described previously (9). cRNA was hybridized to either Agilent Yeast Gene Expression Microarray (V1, 4x44K, G2519F) or (V2, 8x15K G4813A) slides and scanned at 5-μm resolution. Data were extracted by using Agilent Feature Extraction Software version 9.5 with Linear Lowess dye normalization and no background subtraction and submitted to the Princeton University Microarray database for storage and analysis. Normalized data were filtered to exclude features flagged with low intensity and to include only features with both red and green intensities well above background. Finally, of the genes that passed the spot-filter criteria, only genes with greater than 80% good data across all experiments were included in subsequent analyses. All microarray data described in this report are available at http://puma.princeton.edu/cgi-bin/publication/viewPublication.pl?pub_no=530.
Chemostat Competition Assays.
Prototrophic WT (Y3628) and prototrophic dot6Δ tod6Δ (Y3716) strains were grown individually in chemostat at a dilution rate of 0.2 h−1 at 30 °C, 400 rpm, and 300-mL culture volume for 3 days at which steady-state growth was attained, either in carbon-limited media [1.7 g/L YNB lacking amino acids and (NH4)2SO4, 0.08 g/L glucose, 5 g/L (NH4)2SO4] or nitrogen-limited media [1.7 g/L YNB lacking amino acids and (NH4)2SO4, 20 g/L glucose, 0.3 g/L (NH4)2SO4] as described previously (1). A total of 150 mL each of the WT and dot6 tod6 cultures under carbon limitation were then mixed, as were 150 mL each of the nitrogen-limited cultures, and growth continued in chemostats at 0.2 h−1 (≈3-h doubling time) or 0.05 h−1 (≈14-h doubling time). Population measurements of the strains were conducted immediately after mixing and at every 1–2 days for 13 days. The populations of WT and dot6 tod6 strains in the mixed cultures were determined by plating appropriate dilutions of the chemostat culture on YEPD and YEPD plus G418 plates in triplicates, growing 2 days at 30 °C, and counting colony-forming units.
Supplementary Material
Acknowledgments.
We thank Olivier Elemento for assistance with bioinformatic analysis, Donna Storton for assistance with microarray technology, John Matese for assistance with storage and analysis of microarray datasets, Amy Caudy for assistance with petite formation analysis, and Sandy Silverman for assistance with chemostat procedures. This research was supported by National Institutes of Health Grant GM076562 (to J.R.B.) and Center for Quantitative Biology/National Institutes of Health Grant P50 GM071508.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All microarray data described in this report are available at http://puma.princeton.edu/cgi-bin/publication/viewPublication.pl?pub_no=530.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907027106/DCSupplemental.
References
- 1.Brauer MJ, et al. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castrillo JI, et al. Growth control of the eukaryote cell: A systems biology study in yeast. J Biol. 2007;6:4. doi: 10.1186/jbiol54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Regenberg B, et al. Growth-rate regulated genes have profound impact on interpretation of transcriptome profiling in Saccharomyces cerevisiae. Genome Biol. 2006;7:R107. doi: 10.1186/gb-2006-7-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jorgensen P, Tyers M. How cells coordinate growth and division. Curr Biol. 2004;14:R1014–R1027. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 5.Rudra D, Warner JR. What better measure than ribosome synthesis? Genes Dev. 2004;18:2431–2436. doi: 10.1101/gad.1256704. [DOI] [PubMed] [Google Scholar]
- 6.Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- 7.Wade CH, Umbarger MA, McAlear MA. The budding yeast rRNA and ribosome biosynthesis (RRB) regulon contains over 200 genes. Yeast. 2006;23:293–306. doi: 10.1002/yea.1353. [DOI] [PubMed] [Google Scholar]
- 8.Warner JR, Vilardell J, Sohn JH. Economics of ribosome biosynthesis. Cold Spring Harbor Symp Quant Biol. 2001;66:567–574. doi: 10.1101/sqb.2001.66.567. [DOI] [PubMed] [Google Scholar]
- 9.Zaman S, Lippman SI, Schneper L, Slonim N, Broach JR. Glucose regulates transcription in yeast through a network of signaling pathways. Mol Syst Biol. 2009;5:245. doi: 10.1038/msb.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urban J, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 11.Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
- 12.Beer MA, Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- 13.Jorgensen P, et al. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee TI, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 15.Marion RM, et al. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci USA. 2004;101:14315–14322. doi: 10.1073/pnas.0405353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liko D, Slattery MG, Heideman W. Stb3 binds to ribosomal RNA processing element motifs that control transcriptional responses to growth in Saccharomyces cerevisiae. J Biol Chem. 2007;282:26623–26628. doi: 10.1074/jbc.M704762200. [DOI] [PubMed] [Google Scholar]
- 17.Badis G, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu C, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freckleton G, Lippman SI, Broach JR, Tavazoie S. Microarray profiling of phage-display selections for rapid mapping of transcription factor-DNA interactions. PLoS Genet. 2009;5:e1000449. doi: 10.1371/journal.pgen.1000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasch AP, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, et al. Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol. 2004;2:E128. doi: 10.1371/journal.pbio.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bishop AC, Buzko O, Shokat KM. Magic bullets for protein kinases. Trends Cell Biol. 2001;11:167–172. doi: 10.1016/s0962-8924(01)01928-6. [DOI] [PubMed] [Google Scholar]
- 23.Deminoff SJ, Howard SC, Hester A, Warner S, Herman PK. Using substrate-binding variants of the cAMP-dependent protein kinase to identify novel targets and a kinase domain important for substrate interactions in Saccharomyces cerevisiae. Genetics. 2006;173:1909–1917. doi: 10.1534/genetics.106.059238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huber A, et al. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009;23:1929–1943. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shevchenko A, et al. Chromatin Central: Towards the comparative proteome by accurate mapping of the yeast proteomic environment. Genome Biol. 2008;9:R167. doi: 10.1186/gb-2008-9-11-r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zawadzki KA, Morozov AV, Broach JR. Chromatin-dependent transcription factor accessibility rather than nucleosome remodeling predominates during global transcriptional restructuring in Saccharomyces cerevisiae. Mol Biol Cell. 2009;20:3503–3513. doi: 10.1091/mbc.E09-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.