Abstract
Cord blood (CB) hematopoietic stem cell transplantation can be successful even if donor and recipient are not fully matched for human leukocyte antigens (HLA). This may result from tolerance-inducing events during pregnancy but to date this concept has not been tested in CB transplantation. Hence we analyzed the impact of fetal exposure to noninherited maternal antigens (NIMA) of the HLA-A, -B antigens, or -DRB1 alleles on the outcome of CB transplants. The 1,121 patients studied were transplanted for hematological malignancy with a single CB unit: 1,059 received grafts mismatched for one or two HLA antigens. Of these patients, 79 patients had a mismatched antigen that was identical to a donor NIMA, 25 with one HLA mismatch (MM), and 54 with two. If there was a NIMA match, transplant-related mortality (TRM) was improved, especially in patients ≥10 years (P = 0.012) as were overall mortality and treatment failure (P = 0.022 and 0.020, respectively, in the older subset), perhaps related to improved neutrophil recovery, especially in patients who received a low total nucleated cell (TNC) dose (P = 0.031). Posttransplant relapse rate also tended to be reduced, especially in patients with myelogenous malignancies given units with a single HLA mismatch (P = 0.074). These findings represent unique evidence that donor exposure to NIMA can improve survival in unrelated CB transplantation and might reduce relapse, indicating that cord blood cells can mount an antileukemic effect. By matching for donor NIMAs in search algorithms of CB inventories, the probability of selecting a graft with an optimal outcome will increase significantly.
Keywords: cord blood transplantation, hematopoietic stem cell transplantation, relapse reducing mechanisms, tolerance
Despite initial scepticism, cord blood (CB) has become a widely accepted source of hematopoietic stem cells (HSC) for transplantation and, thus, accounted for 22% of the unrelated HSC transplants worldwide in 2007 (www.worldmarrow.org). This is not surprising because CB grafts offer many advantages such as almost immediate access, no risk to the donor, better representation of ethnic diversity, and, above all, less stringent requirements for HLA matching of donor and recipient, while the results can be similar (or even better in case of fully matched CB grafts) to those obtained with adult donors (1–5). Despite these advantages, CB transplantation has not yet been universally accepted, because of three important limitations: delayed engraftment, the small size of the current inventory of available CB units (just over 380,000) as compared to 13 million marrow donor volunteers (www.BMDW.org), and the inability to perform donor lymphocyte infusion (DLI) when a leukemic relapse occurs after transplantation.
Even with less stringent HLA matching requirements, selection of CB grafts remains a challenge because of our inability to predict which HLA mismatches (MM) will and which will not jeopardize patient survival. An obvious area to explore is whether in utero exposure to noninherited maternal antigens (NIMA), as a result of two-way traffic of cells and molecules such as soluble HLA antigens between mother and fetus during pregnancy and consequent development of immunity and tolerance, may affect graft responses to recipient HLA (6–12). We hypothesized that CB grafts with a NIMA match to the patient's mismatched antigen might have improved outcomes and, therefore, might guide us in selecting mismatched donors.
A large database is needed in retrospective studies to explore possible effects of NIMAs because informative donor–recipient pairs (i.e., ones in which the mismatched antigen in the recipient is identical to the respective NIMA antigen of the cord blood) were not selected a priori but occurred only by chance. The New York Blood Center (NYBC) National Cord Blood program has accumulated a sufficiently large database of transplanted CB units with the HLA typing of their respective donor mothers to provide data for initial answers to the above question. We, therefore, analyzed the outcomes of the 1,121 patients transplanted with a single NYBC CB for hematological malignancies, including myelodysplasia (MDS). Our primary study end point was transplant-related mortality (TRM) with secondary end points of neutrophil and platelet engraftment, acute and chronic graft vs. host disease (GVHD), relapse, overall mortality and treatment failure [relapse or death, the inverse of disease-free survival (DFS)]. Although the number of patients with a NIMA match analyzed is small, the results already suggest that, for patients who can find only mismatched CB units, choosing a CB unit with a NIMA identical to their own mismatched antigen(s) will improve outcome.
Results
Almost half of the patients in this study had a non-Caucasian background, 29% were 16 years of age or older, 22% suffered from advanced stage acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), or chronic myeloid leukemia (CML) (see Table S1). Most received myeloablative conditioning (78% of which was total body irradiation or busulfan based) and nearly all received a calcineurin inhibitor for GVHD prophylaxis. A total of 62 recipients received CB units that were HLA matched (6%). Among the 1,059 patients with mismatched grafts, 79 (7%) received a mismatched unit that had a NIMA that was identical to a mismatched antigen of the recipient. The latter are referred to as “NIMA matched transplants” (NMTs). The remaining 980 mismatched grafts had no NIMA match because either the NIMA was not identical to the patient's mismatched antigen (the majority of cases) or the patient or donor mother was homozygous at the mismatched locus (see Table S2). Together these are referred to as “no NIMA match transplants” (no-NMTs). Except for a higher proportion of high risk leukemia in NMTs with 2 HLA mismatches (P = 0.038), NMTs did not differ from the no-NMT group at the same HLA mismatch level in patient, treatment, or graft characteristics (Table S1). There were no significant differences between recipients of units with zero or one HLA mismatch, but recipients with two HLA mismatches were more likely to be older, have African ancestry, high risk leukemia, and to have received a lower TNC dose.
Relative risk comparisons for outcome end points are shown in Table 1, separately for pairs with zero or one HLA MM, with two HLA mismatches and pooling all cases together. The 3-year cumulative probability of TRM was 46% and was lower in NMT pairs than in the no-NMT pairs (P = 0.034), was especially apparent in patients 10 years of age or older (P = 0.012), and tended to have a lower relative risk among those that had one HLA mismatch (Table 1 and Fig. 1). Risks of overall mortality and treatment failure also were reduced, more prominently in the older patients (P = 0.022 and 0.020, respectively, Table 1).
Table 1.
Multivariate analyses of relative risk of study end points in patients with hematological malignancies with cord blood grafts having zero, one, or two HLA-A, -B, -DRB1 mismatch by match for a noninherited maternal antigen
| End point | Zero or one mismatch |
Two mismatches |
Zero, one, or two mismatches |
||||||
|---|---|---|---|---|---|---|---|---|---|
| N events/N patients | RR (95% CI) | P value | N events/N patients | RR (95% CI) | P value | N events/N patients | RR (95% CI) | P value | |
| Transplant-related mortality in 3 years | |||||||||
| Zero HLA mismatch | 11/62 | 0.4 (0.2–0.7) | 0.004 | NA | 11/62 | 0.4 (0.2–0.7) | 0.004 | ||
| HLA MM, NIMA match | 7/25 | 0.6 (0.3–1.3) | 0.170 | 25/54 | 0.7 (0.5–1.1) | 0.101 | 32/79 | 0.7 (0.5–0.97) | 0.034 |
| HLA MM, no NIMA match | 142/363 | Reference | 319/617 | Reference | 461/980 | Reference | |||
| Subset ≥10 years old | |||||||||
| Zero HLA mismatch | 4/22 | 0.3 (0.1–0.8) | 0.016 | NA | 4/22 | 0.3 (0.1–0.7) | 0.011 | ||
| HLA MM, NIMA match | 3/10 | 0.4 (0.1–1.3) | 0.131 | 17/31 | 0.6 (0.4–0.99) | 0.048 | 20/41 | 0.6 (0.3–0.9) | 0.012 |
| HLA MM, no NIMA match | 71/131 | Reference | 219/357 | Reference | 290/488 | Reference | |||
| Overall mortality in 3 years | |||||||||
| Zero HLA mismatch | 22/62 | 0.5 (0.3–0.8) | 0.002 | NA | 22/62 | 0.5 (0.3–0.8) | 0.002 | ||
| HLA MM, NIMA match | 10/25 | 0.5 (0.3–1.02) | 0.059 | 34/54 | 0.8 (0.5–1.1) | 0.180 | 44/79 | 0.7 (0.5–0.97) | 0.029 |
| HLA MM, no NIMA match | 218/363 | Reference | 406/617 | Reference | 624/980 | Reference | |||
| Subset ≥10 years old | |||||||||
| Zero HLA mismatch | 8/22 | 0.4 (0.2–0.9) | 0.021 | NA | 8/22 | 0.4 (0.2–0.8) | 0.011 | ||
| HLA MM, NIMA match | 5/10 | 0.5 (0.2–1.3) | 0.151 | 21/31 | 0.7 (0.4–1.04) | 0.073 | 26/41 | 0.6 (0.4–0.9) | 0.022 |
| HLA MM, no NIMA match | 93/131 | Reference | 260/357 | Reference | 353/488 | Reference | |||
| Treatment failure (relapse or death) in 3 years | |||||||||
| Zero HLA mismatch | 25/62 | 0.5 (0.3–0.8) | 0.003 | NA | 25/62 | 0.5 (0.3–0.8) | 0.002 | ||
| HLA MM, NIMA match | 11/25 | 0.5 (0.3–1.00) | 0.051 | 38/54 | 0.8 (0.6–1.2) | 0.289 | 49/79 | 0.7 (0.6–0.99) | 0.049 |
| HLA MM, no NIMA match | 231/363 | Reference | 429/617 | Reference | 660/980 | Reference | |||
| Subset ≥10 years old | |||||||||
| Zero HLA mismatch | 8/22 | 0.4 (0.2–0.9) | 0.021 | NA | 8/22 | 0.4 (0.2–0.8) | 0.009 | ||
| HLA MM, NIMA match | 5/10 | 0.5 (0.2–1.2) | 0.123 | 23/31 | 0.7 (0.4–1.04) | 0.072 | 28/41 | 0.6 (0.4–0.9) | 0.020 |
| HLA MM, no NIMA match | 97/131 | Reference | 274/357 | Reference | 371/488 | Reference | |||
| Absolute neutrophil count (ANC) 500 by day 77 (not reported on 61 patients) | |||||||||
| Zero HLA mismatch | 51/59 | 1.7 (1.2–2.3) | 0.001 | NA | 51/59 | 1.7 (1.2–2.3) | 0.001 | ||
| HLA MM, NIMA match | 18/21 | 1.3 (0.8–2.1) | 0.297 | 39/52 | 1.3 (0.95–1.9) | 0.094 | 57/73 | 1.3 (1.01–1.7) | 0.043 |
| HLA MM, no NIMA match | 254/345 | Reference | 421/583 | Reference | 675/928 | Reference | |||
| Subset TNC dose <2.5 × 107/kg (not reported on 6 patients) | |||||||||
| Zero HLA mismatch | 14/15 | 5.2 (2.5–11.1) | < 0.001 | NA | 14/15 | 3.8 (2.0–7.2) | < 0.001 | ||
| HLA MM, NIMA match | 3/4 | 2.1 (0.5–9.4) | 0.319 | 11/14 | 2.2 (1.1–4.3) | 0.024 | 14/18 | 1.9 (1.1–3.3) | 0.031 |
| HLA MM, no NIMA match | 48/72 | Reference | 90/147 | Reference | 138/219 | Reference | |||
| Relapse in 3 years | |||||||||
| Zero HLA mismatch | 14/62 | 0.7 (0.4–1.3) | 0.258 | NA | 14/62 | 0.7 (0.4–1.2) | 0.225 | ||
| HLA MM, NIMA match | 4/25 | 0.4 (0.2–1.2) | 0.100 | 12/54 | 1.1 (0.6–1.9) | 0.830 | 16/79 | 0.8 (0.5–1.3) | 0.336 |
| HLA MM, no NIMA match | 89/363 | Reference | 110/617 | Reference | 199/980 | Reference | |||
| Subset of myelogenous malignancies | |||||||||
| Zero HLA mismatch | 9/33 | 0.7 (0.3–1.4) | 0.299 | NA | 9/33 | 0.7 (0.4–1.5) | 0.383 | ||
| HLA MM, NIMA match | 1/13 | 0.2 (0.0–1.2) | 0.074 | 7/31 | 1.0 (0.4–2.1) | 0.917 | 8/44 | 0.6 (0.3–1.2) | 0.155 |
| HLA MM, no NIMA match | 54/193 | Reference | 67/343 | Reference | 121/536 | Reference | |||
| Subset of other hematological malignancies | |||||||||
| Zero HLA mismatch | 5/29 | 0.8 (0.3–2.0) | 0.616 | NA | 5/29 | 0.7 (0.3–1.8) | 0.480 | ||
| HLA MM, NIMA match | 3/12 | 1.0 (0.3–3.2) | 0.966 | 5/23 | 1.3 (0.5–3.3) | 0.598 | 8/35 | 1.1 (0.5–2.3) | 0.775 |
| HLA MM, no NIMA match | 35/170 | Reference | 43/274 | Reference | 78/444 | Reference | |||
RR, relative risk; MM, mismatch; NIMA, noninherited maternal antigen; CI, confidence interval; NA, not applicable; Reference, reference group.
Covariates included in multivariate models:
ANC 500: HLA mismatch level, TNC dose (log transformed continuous), GVHD prophylaxis (CSA + steroids vs. methotrexate vs. tacrolimus vs. other vs. unknown), prior transplant (none vs. allogeneic vs. autologous vs. unknown), center experience (U.S. with ≥50 NYBC CB transplants vs. U.S. with <50 NYBC CB transplants vs. non-U.S.) and year of transplantation (1993–2002 vs. 2003–2006).
Relapse: HLA mismatch level, diagnosis (ALL vs. AML vs. CML vs. other leukemia vs. MDS vs. other) and stage of disease at transplant (advance stage ALL, AML, or CML vs. other stage/other diseases vs. unknown ALL, AML, or CML).
TRM, overall mortality, and treatment failure: HLA mismatch level, TNC dose (log transformed continuous), patient ethnicity (Caucasian vs. non-Caucasian vs. unknown), prior transplant (none vs. allogeneic vs. autologous vs. unknown), patient's pretransplant antibody to cytomegalovirus (negative vs. positive vs. unknown), patient age at transplant (<10 vs. ≥10 years old), stage of disease at transplant (advance stage ALL, AML, or CML vs. other stage/other diseases vs. unknown ALL, AML, or CML), center experience (U.S. with ≥50 NYBC CB transplants vs. U.S. with <50 NYBC CB transplants vs. non-U.S.) and year of transplantation (1993–2002 vs. 2003–2006).
Fig. 1.
Probability of transplant-related mortality (TRM) for patients 10 years of age or older. Zero HLA mismatch (MM) = 22; one MM/NIMA match = 10; two MM/NIMA match = 31; one MM/no-NIMA match = 131; two MM/no-NIMA match = 357.
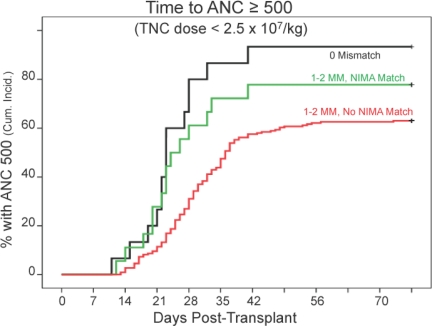
The cumulative probability of myeloid engraftment was 74%. NMTs engrafted faster than no-NMTs and almost as fast as the zero mismatched pairs, especially when unit TNC dose was low (<2.5 × 107/kg) (Table 1 and Fig. 2). The NIMA association with engraftment was seen in both patients whose grafts had one HLA mismatch and in those that had two. Platelet recovery was not associated with a NIMA match (see Table S3). Among patients who engrafted, the cumulative incidence of grade 2–4 acute GVHD was 46% (25% for grade 3–4) and, among engrafted patients who survived beyond day 100, the cumulative incidence of chronic GVHD was 34%. Acute and chronic GVHD incidence tended to be lower in NMTs but the difference was not significant (Table S3).
Fig. 2.
Time to absolute neutrophil count (ANC) ≥500/mm3 (at day 77) for the subset of patients transplanted with a CB unit with TNC per kilogram of body weight of <2.5 × 107. Zero HLA mismatch (MM) = 15; NIMA match = 18; no-NIMA match = 219.
The 3-year cumulative incidence of relapse was 22% and was associated with diagnosis and stage of disease at transplant (Table S3). Relapse rate tended to be lower in NMTs than in no-NMT among patients with myeloid malignancies, especially in those with one HLA mismatch (P = 0.074) and was lower than that of the zero mismatched donor recipient pairs, although these differences were not significant (Table 1 and Fig. 3).
Fig. 3.
Time to relapse for patients with myelogenous malignancies. Zero HLA mismatch (MM) = 33; one MM/NIMA match = 13; two MM/NIMA match = 31; one MM/no-NIMA match = 193; two MM/no-NIMA match = 343.
Discussion
This is a unique study that demonstrates in unrelated CB HSC transplantation that matching for the donor's NIMA can improve outcome and possibly lower the risk of leukemic relapse. The identification of the NMTs in this study was done after the transplants had been performed. Hence, NIMA matching was not used in donor selection and, consequently, our number of NIMA matches is low. However, availability of HLA typing of CB unit mothers would permit the preferential selection of NIMA matched grafts (as we plan to do) and should rapidly increase the number of NMT cases available to extend and confirm or refute the results of our study.
In general, NIMA match effects were more apparent in patients who were predicted to have a worse outcome and were seen in both one and two HLA mismatches, although tending to be stronger in those with one mismatch (Table 1). Thus, transplant-related mortality was reduced in NMT recipients, most significantly in older patients. While this study did not establish the mechanism, improved TRM in NMT may be related to improved engraftment seen in these recipients, especially in those whose CB unit had a TNC dose of <2.5 × 107/kg (91% of whom were also in the older age group). As a consequence of improved engraftment and TRM, overall mortality and treatment failure were reduced for NMT recipients.
The incidence of severe acute and chronic GVHD in our CB recipients was similar to rates reported in other series (3–5) but was not significantly associated with matching for NIMA. This observation is apparently discrepant with the findings of previous studies of NIMA matched haploidentical sibling HSC transplants that exhibited reduced acute and chronic GVHD (12, 13) and reports that reduced relapse (see next paragraph) is the fringe benefit of increased GVHD (5, 14). However, our CB cases are not comparable to the sibling transplants for the following reasons: Haploidentical sibling donors share a complete haplotype down to the allele level, facilitating MHC restriction, in contrast to unrelated CB grafts that are only phenotypically matched to the recipient (with only DRB1 matched to the allele level). Similarly, the NIMA is only phenotypically matched to the patient's antigen in CB transplants, but genotypically matched in the sibling-to-sibling transplants because donor and recipient share the same mother. Furthermore, the two mismatches in the sibling transplants were both NIMA matches, whereas in the 54 CB cases only 1 had two NIMA matches. Additionally, it has been shown recently that regulatory T cells are more numerous and more easily upregulated in CB (9)—hence the lower overall incidence of GVHD after CB transplantation—whereas such T cells are fewer in the adult donors. Further monitoring of anti-NIMA immunity and regulator T cell activity pre- and posttransplant is needed to evaluate these possibilities in CB transplantation. Chronic GVHD can be a serious problem in NIMA mismatched haploidentical-related transplantation (15), but was not increased in our NMT CB recipients.
One of the most intriguing findings in our study was the low rate of relapse in NMT pairs, a difference that approached significance compared with no-NMTs in patients with myeloid leukemia or MDS who had one HLA mismatch, suggesting that anti-NIMA immunity might exist or was upregulated after reexposure to the recipient mismatched antigen. This finding might be similar to the observation that after haploidentical sibling NIMA mismatched renal transplantation early acute rejection crises were significantly increased as compared to similar, but noninherited paternal antigen mismatched, renal grafts (16), and it is supported further by in vitro findings discussed below. While these data need to be confirmed, it might be appropriate to select HLA mismatched CB grafts with a NIMA match preferentially for their antileukemic effect. Whether infusion of NIMA matched CB units could also be used for treatment of posttransplant relapse to obtain an effect similar to that of DLI might also merit investigation (17).
In cadaveric renal transplantation of patients in whom the NIMA was known, donor HLA-A mismatches that were identical to the recipient's HLA-A NIMA had also improved graft survival (18). We did not see an association between engraftment or survival end points and a specific NIMA matched locus in our own study. However, the number of patients in each subset was small.
The authors recognize that the patients studied form a heterogeneous group, in diagnosis, stage of disease, treatment, and age. Nevertheless, these preliminary results indicate that matching for cord blood NIMA could improve disease-free survival and may reduce leukemic relapse in patients suffering from hematological malignancy. Obviously, the precise molecular and cellular mechanisms remain unclear from this clinical study. The reduced relapse risk could indicate that the fetus develops not only regulatory T cells but also modified CD4+ T cells that through the perforin pathway exert a “relapse reducing” effect, as suggested by preclinical studies (19). This concept gets further support from CB studies. Mommaes et al. observed that in man CD8+ anti-minor histocompatibility antigens (mHA) HA-1 cytotoxic T lymphocytes (CTL) identified in some cord blood samples can be isolated, boosted with phytohemaglutinin, and are capable of lysing mHA HA-1 positive target cells in vitro (20). Such CD8+ anti-mHA-1 CTLs have been shown to be boosted after DLI and might be essential in inducing a remission in patients with a leukemic relapse (21). Coexisting mHA-specific CD8-expressing regulatory T cells and CTL were described both in the nonphysiological setting of renal allograft tolerance (22) and in the physiological setting of pregnancy (23). Hence these, and recently described CD4+ regulatory T cells, which are induced during human pregnancy (9), are a logical area for further exploration and might be the explanation for the observation of reduced relapse without increased GVHD.
Many questions remain unanswered. Further studies are needed to assess more fully the benefit in grafts with two mismatches, those with either a single or two NIMA matches. The effect in double unit transplants would also be of interest as might be the impact of high resolution typing for HLA-A, -B, and matching for HLA-C, -DQ, and perhaps even -DP on the outcome of NMT. Likewise, the quantitative and qualitative aspects of microchimaerism (24), the effect of matching for NK ligands and NK alloimmunity should be assessed (25, 26) and the effect of homozygosity at one or more loci of the patient or CB unit and their effect in the NMT. In addition, this study did not include patients with diseases other than hematological malignancies.
Although much work lies ahead, our findings justify changing the match algorithm for CB transplants to identify NIMA matches as part of the CB unit selection process so that we can provide patients with “permissive” HLA mismatched but NIMA matched grafts predicted from this study to have an improved outcome. Such mismatched units might be chosen preferentially over other similar units, especially because there are no apparent adverse effects associated with NIMA matching. To illustrate the potential impact, Table 2 gives an example of the phenotypes (what we will call “virtual phenotypes”) for a CB unit based on substitutions of donor antigens for NIMAs at the HLA-A, -B, and -DRB1 loci. For a unit and mother with six identified antigens at these three loci, a single substitution produces six new virtual phenotypes, increasing the number of optimal CB units sixfold. Allowing for two NIMA substitutions for grafts with two HLA mismatches would increase these units by up to 18-fold. In practice, the actual increase will be lower because of duplicates or cases when mother and donor share the same antigens at a given locus or the patient or donor mother is homozygous. Even in the case of one HLA mismatch, however, the proposed strategy for CB unit selection might well resolve the problem of the small size of the presently available CB inventory. A 6-fold increase in the probability of finding an optimal unit (i.e., HLA-matched or one HLA mismatch with a NIMA match), for example, would be equivalent to expanding the effective current world inventory to over 2 million units.
Table 2.
HLA-A, -B, and -DRB1 virtual phenotypes
| CB unit: | A2 | A24 | B7 | B65 | DR0102 | DR1501 |
|---|---|---|---|---|---|---|
| A-NIMA | A-IMA | B-NIMA | B-IMA | DR-NIMA | DR-IMA | |
| Mother: | A1 | A24 | B57 | B65 | DR1305 | DR0102 |
| Virtual phenotypes with one substitution | ||||||
| VP1: | A1 | A2 | B7 | B65 | DR0102 | DR1501 |
| VP2: | A1 | A24 | B7 | B65 | DR0102 | DR1501 |
| VP3: | A2 | A24 | B7 | B57 | DR0102 | DR1501 |
| VP4: | A2 | A24 | B57 | B65 | DR0102 | DR1501 |
| VP5: | A2 | A24 | B7 | B65 | DR0102 | DR1305 |
| VP6: | A2 | A24 | B7 | B65 | DR1305 | DR1501 |
| Virtual phenotypes with two substitutions | ||||||
| VP7: | A1 | A2 | B7 | B57 | DR0102 | DR1501 |
| VP8: | A1 | A2 | B57 | B65 | DR0102 | DR1501 |
| VP9: | A1 | A24 | B7 | B57 | DR0102 | DR1501 |
| VP10: | A1 | A24 | B57 | B65 | DR0102 | DR1501 |
| VP11: | A1 | A2 | B7 | B65 | DR0102 | DR1305 |
| VP12: | A1 | A2 | B7 | B65 | DR1305 | DR1501 |
| VP13: | A1 | A24 | B7 | B65 | DR0102 | DR1305 |
| VP14: | A1 | A24 | B7 | B65 | DR1305 | DR1501 |
| VP15: | A2 | A24 | B7 | B57 | DR0102 | DR1305 |
| VP16: | A2 | A24 | B57 | B65 | DR0102 | DR1305 |
| VP17: | A2 | A24 | B7 | B57 | DR1305 | DR1501 |
| VP18: | A2 | A24 | B57 | B65 | DR1305 | DR1501 |
The virtual phenotypes (VP) are those that are created by substitution of one or more maternal NIMAs for antigens at each respective locus of the cord blood unit's HLA haplotype. Thus, a patient whose HLA-A, -B, and -DRB1 type corresponds to VP1, for example, would have one HLA mismatch to this cord blood (CB) unit and a match to the CB donor's NIMA at the A locus. Maternal NIMAs are identified in bold.
Materials and Methods
Patients.
Consecutive patients with hematological malignancy who were transplanted with a single CB unit from the NYBC National Cord Blood program between 1993 and 2006 were eligible for this study (n = 1,407). Recipients of grafts with three or four HLA antigen mismatches were excluded because of small numbers (85 and 5, respectively) and the fact that such units are now rarely selected as CB inventories have grown. Additionally, 7 patients were excluded because of inadequate HLA typing and 49 had no HLA typing of the donor mother. Patients signed an informed consent for CB transplantation at the respective transplant centers; the NYBC program operates under an investigational new drug exemption from the U.S. Food and Drug Administration.
Transplant centers provided data on patient demographics, diagnosis, and stage of disease at the time of transplant, conditioning regimen, prophylaxis for GVHD, and posttransplant events. Transplant and follow-up reports were reviewed for completeness and consistency by one of the coauthors (A.S.). Outcome data included information on myeloid and platelet engraftment, tests for donor chimerism, occurrence and severity of acute and chronic GVHD, relapse, survival, and causes of death. Of 1,261 eligible patients, 1,121 (89%) have had outcome data reported. Among survivors, median follow-up was 27 months. Patients with ALL, AML, or CML were classified as low, intermediate, or high risk using International Bone Marrow Transplant Registry criteria.
Cord Blood Units.
Methods for collecting, testing, processing, freezing, and storage of cord blood units have been described in detail previously (1, 2). Mothers signed institutional review board-approved informed consent to donate their babies' cord blood to the NYBC program.
Assignment of Patient–Donor Match Level and NIMA Match.
CB units and donor mothers were typed for HLA-A, -B, and -DRB1 using serological and DNA methods. CB units were selected on the basis of TNC dose and HLA match. Match grades for HLA-A and -B were assigned at low–intermediate resolution level (antigen level) and for DRB1 at high resolution (allele level) and were expressed as 0, 1, or 2 antigen mismatched. Among patient/donor pairs that were HLA mismatched, we evaluated whether the patient's mismatched antigen(s) matched that of the donor's NIMA at the mismatched loci. Table 2 shows the HLA typing of a CB unit and the donor mother for one of our study cases (see Table S2 for additional examples). When the NIMA at one or more loci matched to the patient's mismatched antigen, the case was assigned to the NMT group. There were a total of 79 cases with a NIMA match: 25 among patient–donor pairs with a single HLA mismatch and 54 with 2 mismatches (53 with a single NIMA match and 1 with 2 NIMA matches). In the case shown in Table 2, the patient's HLA type was the same as the second virtual phenotype (VP2). When the patient's mismatched locus was homozygous (88 of 1,597 mismatched loci), the pair was considered not to have a NIMA match because there was no patient antigen for the NIMA to match to. These cases were included in the no-NMT group. Similarly, when the mother was homozygous at the mismatched locus (170 of 1,597), the pair was categorized as no-NMT.
Transplant Outcome End Points.
Definitions of study end points followed standard conventions (2–5). Time to myeloid engraftment was defined as the first of 3 consecutive days of absolute neutrophil cell count of 500/μL (ANC 500) of donor-derived cells; analyses were limited to the first 77 days posttransplant. Analyses of acute GVHD were based on those reported as grade 2–4 (transplant center assessment) and of chronic GVHD combining limited or extensive disease. If no date for onset of acute GVHD was provided, the average reported time (28 days) was assigned (n = 94). Onset of chronic GVHD was assigned to day 100 posttransplant if no date was provided. The probability and risk of acute GVHD was assessed only in patients who engrafted and of chronic GVHD only in those who engrafted and survived to day 100 posttransplant. TRM was defined as any death after transplant while the patient was in remission. Overall mortality was based on deaths from all causes. Treatment failure was defined as death or relapse, whichever came first, and was the inverse of disease-free survival.
Data Analysis and Statistical Methods.
Differences between categorical variables were estimated by χ2 or Fisher's exact test (two-tailed) and between means by Student's t test. The probabilities of death and treatment failure were calculated by the Kaplan-Meier method. For other end points that had competing outcomes, event-specific hazard functions and free-from-event survival rates were calculated to obtain event-specific cumulative incidence rates (27). For myeloid and platelet engraftment, death was the competing event. For GVHD and relapse, the competing outcomes were graft failure (documented autologous reconstitution or subsequent transplant following CB graft failure) or death, whichever came first. Relapse was the competing outcome for TRM and death was the competing outcome for relapse. The Cox proportional hazard model was used to estimate relative risks of outcome end points in uni- and multivariate analyses (28). Initial multivariate models included variables already reported as associated with outcome of cord blood transplantation (patient age, sex, ethnicity, diagnosis, stage of ALL, AML, and CML, prior transplantation, conditioning regimen, GVHD prophylaxis, year of transplantation, and transplant center experience, as well as CB unit HLA match and total nucleated cell dose per kilogram of patient weight). To account for possible effects of transplant center experience with CB transplantation, U.S. centers that had given ≥50 patients cord blood units from our program were grouped together for comparison with patients transplanted in other U.S. and non-U.S. centers. We performed subset analyses on the basis of myelogenous and nonmyelogenous diseases because of reported stronger graft vs. leukemia effects in myelogenous malignancies. We also performed analyses on subsets predicted to have a worse outcome (i.e., donor/recipient pairs with low TNC dose and older patients), reasoning that potential benefits, if any, should be most apparent in these patients. High risk TNC (<2.5 × 107/kg) and age (≥10 years) categories were established by dividing the study population into 10 equal groups and identifying the cut point for engraftment and survival end points, respectively. A stepwise backward regression model was applied on the whole data set, where only variables of interest (HLA and NIMA match) were forced to remain in the model.
Supplementary Material
Acknowledgments.
We are grateful to the transplant centers that reported on outcome data and to NYBC National Cord Blood program staff who performed all of the tasks needed to ensure the quality of the CB units. We are also indebted to the obstetricians in collaborating hospitals who supported the program and to the mothers who generously donated their infant's cord blood to any patient who might need it. We thank Drs Mary Horowitz, Tatsuo Ichinohe, Astrid van Halteren, Frans Claas, Machteld Oudshoorn, and Anneke Brand for critical reading of and constructive comments on the manuscript. Finally, we thank Dr. Pablo Rubinstein for his conceptual contributions. The program was supported by grants from the U.S. Public Health Service, National Heart Lung and Blood Institute, and the Starr Foundation, and this study was supported by the Macropa Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910310106/DCSupplemental.
References
- 1.Rubinstein P, et al. Processing and cryopreservation of placental/umbilical cord blood for unrelated bone marrow reconstitution. Proc Natl Acad Sci USA. 1995;92:10119–10122. doi: 10.1073/pnas.92.22.10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rubinstein P, et al. Outcomes among 562 recipients of placental-blood transplants from unrelated donors. N Engl J Med. 1998;339:1565–1577. doi: 10.1056/NEJM199811263392201. [DOI] [PubMed] [Google Scholar]
- 3.Laughlin MJ, et al. Outcomes after transplantation of cord blood or bone marrow from unrelated donors in adults with leukemia. N Engl J Med. 2004;351:2265–2275. doi: 10.1056/NEJMoa041276. [DOI] [PubMed] [Google Scholar]
- 4.(for the Acute Leukemia Working Party of European Blood and Marrow Transplant Group; Eurocord-Netcord Registry) Roche V, et al. Transplants of umbilical-cord blood or bone marrow from unrelated donors in adults with acute leukemia. N Engl J Med. 2004;351:2276–2285. doi: 10.1056/NEJMoa041469. [DOI] [PubMed] [Google Scholar]
- 5.Eapen M, et al. Outcomes of transplantation of unrelated donor umbilical cord blood and bone marrow in children with acute leukaemia: A comparison study. Lancet. 2007;369:1947–1954. doi: 10.1016/S0140-6736(07)60915-5. [DOI] [PubMed] [Google Scholar]
- 6.Rubinstein P. Placental blood-derived hematopoietic stem cells for unrelated bone marrow reconstitution. J Hemother. 1993;2:207–210. doi: 10.1089/scd.1.1993.2.207. [DOI] [PubMed] [Google Scholar]
- 7.van Rood JJ, van Leeuwen A, van Santen MCT. Anti-HL-A2 inhibitor in normal human serum. Nature. 1970;226:366–367. doi: 10.1038/226366a0. [DOI] [PubMed] [Google Scholar]
- 8.Andrassy J, et al. Tolerance to noninherited maternal MHC antigens in mice. J Immun. 2003;171:5554–5561. doi: 10.4049/jimmunol.171.10.5554. [DOI] [PubMed] [Google Scholar]
- 9.Mold JE, et al. Maternal alloantigens promote the development of tolerogenic fetal regulatory T cells in utero. Science. 2008;322:1562–1565. doi: 10.1126/science.1164511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burlingham WJ. A lesson in tolerance—maternal instruction to fetal cells. N Engl J Med. 2009;360:1355–1357. doi: 10.1056/NEJMcibr0810752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Rood JJ, Roelen DL, Claas FH. The effect of noninherited maternal antigens in allogeneic transplantation. Semin Hematol. 2005;42:104–111. doi: 10.1053/j.seminhematol.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 12.van Rood JJ, et al. Effect of tolerance to noninherited maternal antigens on the occurrence of graft-versus-host disease after bone marrow transplantation from a parent or an HLA-haploidentical sibling. Blood. 2002;99:1572–1577. doi: 10.1182/blood.v99.5.1572. [DOI] [PubMed] [Google Scholar]
- 13.(for the Japanese Collaborative Study Group for NIMA-Complementary Haploidentical Stem Cell Transplantation) Ichinohe T, et al. Feasibility of HLA-haploidentical hematopoietic stem cell transplantation between noninherited maternal antigen (NIMA)-mismatched family members linked with long-term fetomaternal microchimerism. Blood. 2004;104:3821–3828. doi: 10.1182/blood-2004-03-1212. [DOI] [PubMed] [Google Scholar]
- 14.Horowitz MM, et al. Graft-versus-leukemia reactions after bone marrow transplantation. Blood. 1990;75:555–562. [PubMed] [Google Scholar]
- 15.Kanda J, et al. Long-term survival after HLA-haploidentical SCT from noninherited maternal antigen-mismatched family donors: Impact of chronic GVHD. Bone Marrow Transplant. 2009;44:327–329. doi: 10.1038/bmt.2009.18. [DOI] [PubMed] [Google Scholar]
- 16.Burlingham WJ, et al. The effect of tolerance to noninherited maternal HLA antigens on the survival of renal transplants from sibling donors. N Engl J Med. 1998;339:1657–1664. doi: 10.1056/NEJM199812033392302. [DOI] [PubMed] [Google Scholar]
- 17.Tokita K, Terasaki P, Maruya E, Saji H. Tumour regression following stem cell infusion from daughter to microchimeric mother. Lancet. 2001;358:2047–2048. doi: 10.1016/S0140-6736(01)07140-9. [DOI] [PubMed] [Google Scholar]
- 18.Smits JM, Claas FH, van Houwelingen HC, Persijn GG. Do noninherited maternal antigens (NIMA) enhance renal graft survival? Transplant Int. 1998;11:82–88. doi: 10.1007/s001470050109. [DOI] [PubMed] [Google Scholar]
- 19.Edinger M, et al. CD4+ CD25+ regulatory T cells preserve graft-versus-tumor activity while inhibiting graft-versus- host disease after bone marrow transplantation. Nat Med. 2003;9:1144–1150. doi: 10.1038/nm915. [DOI] [PubMed] [Google Scholar]
- 20.Mommaes B, et al. Cord blood comprises antigen-experienced T cells specific for maternal minor histocompatibility antigen HA-1. Blood. 2005;105:1823–1827. doi: 10.1182/blood-2004-07-2832. [DOI] [PubMed] [Google Scholar]
- 21.Marijt WAE, et al. Hematopoietic-restricted minor histocompatibility antigens HA-1 or HA-2-specific T cells can induce complete remissions of relapsed leukemia. Proc Natl Acad Sci USA. 2003;100:2742–2747. doi: 10.1073/pnas.0530192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai J, et al. Minor H antigen HA-1-specific regulator and effector CD8+ T cells, and HA-1 microchimerism, in allograft tolerance. J Exp Med. 2004;199:1017–1023. doi: 10.1084/jem.20031012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Halteren AGS, et al. Naturally acquired tolerance and sensitisation to minor histocompatibility antigens in healthy family members. Blood. 2009;114:2263–2272. doi: 10.1182/blood-2009-01-200410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ichinohe T, Maruya E, Saji H. Long-term feto-maternal microchimerism: Nature's hidden clue for alternative donor hematopoietic cell transplantation? Int J Hematol. 2002;76:229–237. doi: 10.1007/BF02982792. [DOI] [PubMed] [Google Scholar]
- 25.(Eurocord-Netcord and Acute Leukaemia Working Party of the EBMT) Willemze R, et al. KIR-ligand incompatibility in the graft-versus-host direction improves outcomes after umbilical cord blood transplantation for acute leukemia. Leukemia. 2009;23:492–500. doi: 10.1038/leu.2008.365. and erratum (2009) 23: 630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunstein CG, Wagner JE, Weisdorf DJ, Cooley S, Noreen H. Negative effect of KIR alloreactivity in recipients of umbilical cord blood transplant depends on transplantation conditioning intensity. Blood. 2009;113:5628–5634. doi: 10.1182/blood-2008-12-197467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smits JMA, van Houwelingen HC, De Meester J, Persijn GG, Claas FHJ. Analysis of the renal transplant waiting list. Application of a parametric competing risk method. Transplantation. 1998;66:1146–1153. doi: 10.1097/00007890-199811150-00006. [DOI] [PubMed] [Google Scholar]
- 28.Marubini E, Valsecchi M. Analysing Survival Data from Clinical Trials and Observational Studies. New York: Wiley; 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.