Summary
Kinesins in the mitotic spindle play major roles in determining spindle shape, size, and bipolarity, although specific regulation of these kinesins at distinct locations on the spindle is poorly understood. To balance the forces that are required for spindle bipolarity, microtubule-depolymerizing kinesins are tightly regulated. Aurora B kinase phosphorylates the neck regions of the kinesin-13 family microtubule depolymerases Kif2a and MCAK (Mitotic Centromere Associated Kinesin), and inhibits their depolymerase activities. How they are reactivated, and how this is controlled independently on different kinetochore fibers is unknown. We show that Inner Centromere Kin-I Stimulator (ICIS), which stimulates the related depolymerase MCAK, can reactivate Kif2a after Aurora B inhibition. When antibodies that block the ability of ICIS to activate Kif2a are injected into cells, monopolar spindles are generated. This phenotype is rescued by co-injection of anti-Nuf2 antibodies. We have performed a structure-function analysis of the ICIS protein, and find that the N-terminus of ICIS binds Aurora B and its regulators INCENP and TD60, while a central region binds MCAK, Kif2a and microtubules, suggesting a scaffold function for ICIS. These data argue that ICIS and the CPC (Chromosomal Passenger Complex) regulate Kif2a depolymerase activity.
Keywords: kinesin-13, microtubules, mitotic spindle, centromere
Results and Discussion
The C-terminus of ICIS Reverses Aurora B Inhibition of Kif2a in vitro
The microtubule depolymerase activities of Kif2a and MCAK are inhibited through phosphorylation by Aurora kinases [1–3]. On MCAK, this phosphorylation event has been mapped to the neck region, which lies proximal to the kinesin domain. Not only does Aurora regulate MCAK activity, but it also regulates its localization on the spindle [1,3,4]. Surprisingly, phosphorylated MCAK is often found at locations where MCAK activity is required [5,6]. Therefore, there must be tight spatial regulation of the activity of these proteins in their different subcellular locations as well as mechanisms to reactivate depolymerase activity after Aurora phosphorylation.
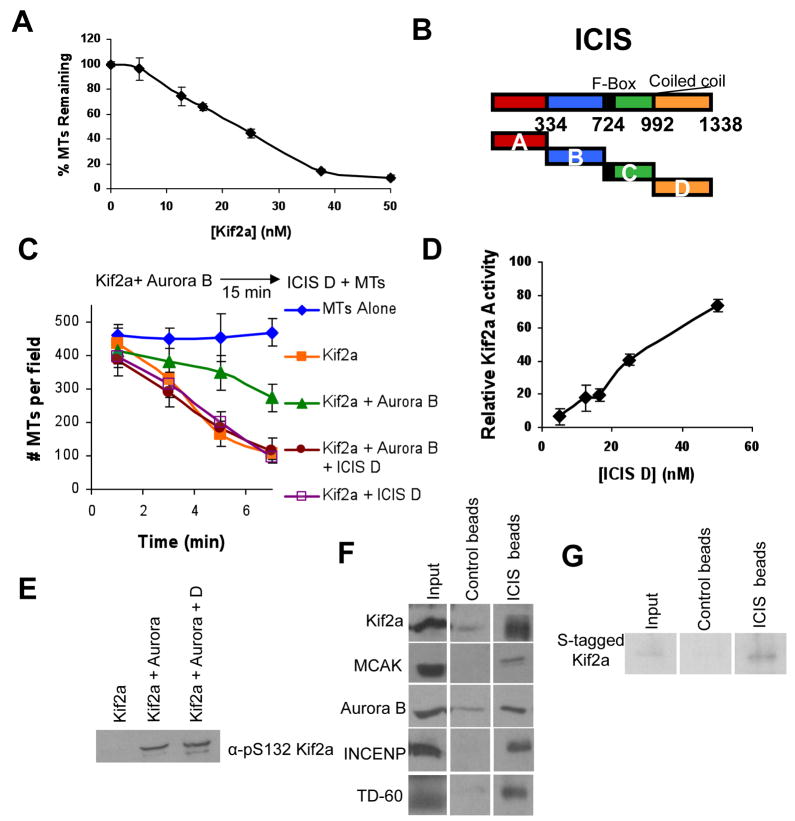
We adapted a system to measure Kif2a microtubule depolyerase activity in vitro[1]. Rhodamine-labeled taxol-stabilized microtubules were incubated with recombinant Kif2a aa118–530, which is the minimal domain required for depolymerase activity (Kif2a minimal domain). Although this protein construct was cloned from the human sequence, it is 93% identical to the Xenopus sequence (Supplemental Figure 1). Titration of Kif2a into the reaction shows concentration-dependent depolymerase activity with linear responses, and saturation of activity after 40nM (Figure 1A).
Figure 1. ICIS reverses Aurora inhibition of Kif2a in vitro.
A. Titration of Kif2a into a visual MT depolymerase assay, demonstrates depolymerase activity is concentration dependent at 3 minutes. Error bars represent standard deviation.
B. Map of Xenopus ICIS, showing the different truncation proteins that were cloned and bacterially expressed.
C. ICIS D reactivates Kif2a that has been inhibited by Aurora kinase. Schematic of the in vitro depolymerase assay and quantification of the mean number of microtubules per field in the presence of recombinant Kif2a, Aurora B, and the C-terminal fragment of ICIS (ICIS D). Error bars represent standard deviation.
D. Depolymerase assays titrating the amount of ICIS D added after prephosphorylation of Kif2a with Aurora B. Relative Kif2a activity was calculated using Kif2a alone as 100% activity, and Kif2a + Aurora B as 0% activity. Error bars represent standard deviation.
E. ICIS does not reactivate by dephosphorylating Kif2a. Immunoblot of α-pS132 Kif2a in an in vitro depolymerase reaction as in C.
F. Immunoblots showing that ICIS D interacts with Kif2a, MCAK, Aurora B, INCENP, and TD-60 in Xenopus mitotic extracts. ICIS D or 6His-GST was covalently attached to beads, incubated in mitotic extract and the washed beads probed for the indicated proteins.
G. ICIS D interacts directly with Kif2a in vitro. Recombinant S-tagged-Kif2a was incubated with the beads used in F, and bound Kif2a was detected with S-protein HRP. Input represents 20% of the reaction.
Since ICIS was shown to regulate MCAK microtubule depolymerase activity in vitro, we tested if it could similarly regulate Kif2a. Four truncation proteins were made from the full length Xenopus ICIS protein: ICIS A (ICIS1–334), B (ICIS334–724), C (ICIS724–992) and D (ICIS992–1338) (Figure 1B). The C fragment contains the protein’s F-Box, and the D fragment is the protein’s coiled-coil domain. All proteins were expressed in E. coli as 6-His fusion proteins and purified on Ni2+ columns. 50nM Kif2a depolymerized most microtubules by 7 minutes (Figure 1C, Supplemental Figure 2). Surprisingly, none of the ICIS fragments could stimulate this activity (Figure 1C, Supplemental Figure 2, data not shown).
Phosphorylation of Kif2a by Aurora B for 15 minutes before the addition of microtubules inhibited Kif2a depolymerase activity (Figure 1C, Supplemental Figure 2). To test its effects on phosphorylated Kif2a, equimolar amounts of ICIS fragments were then added to the reactions after Aurora B phosphorylation, and depolymerase activity was measured. The ICIS D fragment (the C-terminal, coiled-coil region) did not have an effect on Kif2a alone, but it could restore Kif2a activity after it was first inhibited by Aurora B phosphorylation (Figure 1C). The other fragments had no detectable effect on Kif2a activity (not shown). A titration of ICIS D into this reaction demonstrates that Kif2a activity increases with ICIS D concentration and activity approaches that of unphosphorylated Kif2a at stoichiometric amounts of ICIS D (Figure 1D). It appears that Kif2a cannot be stimulated above the unphosphorylated amount of activity because adding 5 or 10 times stoichiometric amounts of ICIS did not activate Kif2a above its uninhibited activity (Supplemental Figure 3). These data suggest that ICIS binding restores the activity of Kif2a that has been inhibited by Aurora B phosphorylation.
ICIS could reactivate Kif2a by removing the inhibitory phosphorylation or by direct stimulation. Aurora B inhibits MCAK activity by phosphorylation on serine 196, in the neck region of the protein. We found an equivalent site in the neck region of Kif2a, serine 132, and tested whether this site was phosphorylated by Aurora B (Supplemental Figure 4A). Unfortunately, recombinant Kif2a-S132A mutant protein has no depolymerase activity, making it impossible to conclusively show that this is the key inhibitory site on Kif2a. We made a polyclonal antibody against phospho-S132 of Kif2a. Antibody recognition of Kif2a in vitro required prior phosphorylation by Aurora kinase, demonstrating that this antibody is phospho-specific (Supplemental Figure 4B, Figure 1E). Moreover, the antibody recognizes a protein at ~85kD in Xenopus M-phase extracts containing phosphatase inhibitors, which is the expected size of Xenopus Kif2a. To determine if ICIS is dephosphorylating Kif2a to reactivate activity, we immunoblotted the recombinant microtubule depolymerase reaction with the pS132 antibody (Figure 1E). Phosphorylation of the presumptive inhibitory site did not decrease when ICIS stimulates activity. These data argue that ICIS D activates phosphorylated Kif2a not by dephosphorylation, but by direct binding.
To characterize the interactions of ICIS D in vivo, recombinant ICIS D was covalently linked to sepharose beads, these beads were incubated in mitotic Xenopus extracts, and associated proteins were probed by immunoblot. As expected from biochemical experiments, ICIS D binds MCAK and Kif2a (Figure 1F). In addition, ICIS D bound Aurora B and its activators INCENP and TD-60. ICIS D beads were also incubated with purified recombinant Kif2a, which showed that ICIS D and Kif2a interact directly in vitro (Figure 1G).
These data argue for dual regulation of kinesin-13 microtubule depolymerases. First, Aurora B kinase phosphorylates the neck domain of these proteins to inhibit depolymerase activity. Second, a coiled-coil domain on ICIS then reactivates inhibited Kif2a. It is likely that MCAK is regulated in the same way, even though ICIS has been demonstrated to stimulate intrinsic MCAK depolymerase activity [7]. We contend that the purified MCAK used in the original experiments contained both inhibited and uninhibited protein. MCAK purified from baculovirus (which was used in the original experiments) is phosphorylated on S196 and thus is partially inhibited (Supplemental Figure 5). MCAK from baculovirus is not fully phosphorylated, as we could more than double the pS196 staining after incubation with Aurora B kinase (data not shown). Because we used recombinant Kif2a from bacteria (which is not phosphorylated), it can only be activated by ICIS after Aurora B phosphorylation. However, it is important to test this idea in the future, since another important difference between the two experiments is that we used the catalytic portion of Kif2a, whereas previous experiments were performed with full-length MCAK.
Anti-ICIS injection results in a monopolar spindle phenotype
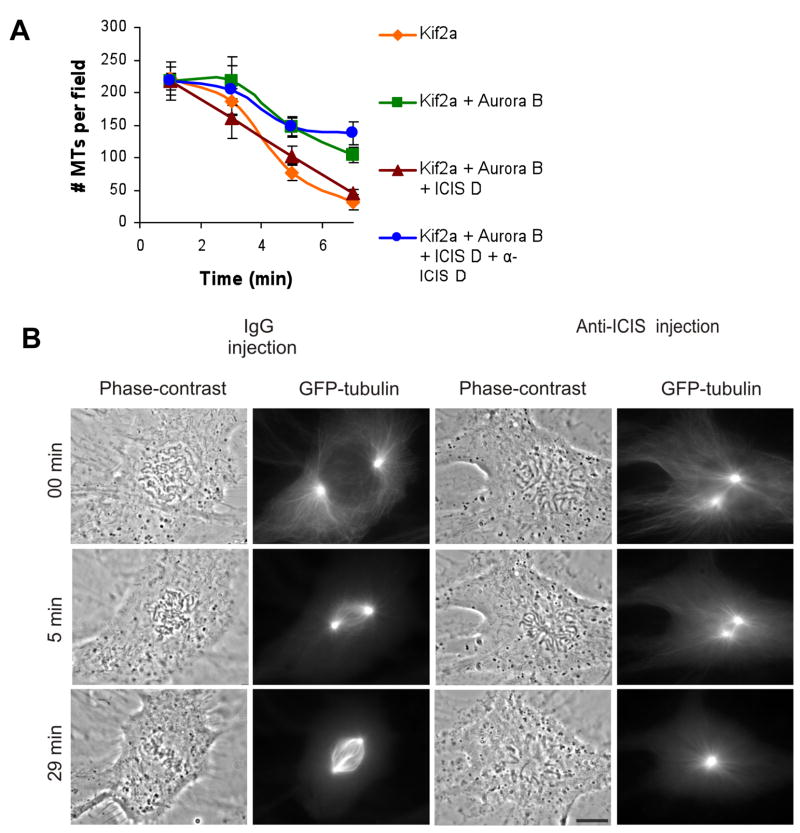
To determine if the C-terminus of ICIS regulates kinesin-13s in vivo we generated a polyclonal antibody against ICIS D (Supplemental Figure 6). After affinity purification, this antibody predominantly recognized a protein at the expected molecular weight for ICIS (160kD). Several smaller bands are also recognized by the antibody, but it only depletes the 160kD band, suggesting that ICIS is the only protein that is recognized in the native conformation. The antibody against the ICIS D region was added to a microtubule depolymerase reaction and the antibody blocked the reactivation of Kif2a by ICIS D (Figure 2A).
Figure 2. Injection of α-ICIS antibodies (AB) into cells gives a monopolar spindle phenotyope.
A. ICIS D antibodies inhibit Kif2a reactivation in vitro. Quantification of the mean number of microtubules per field in the presence of recombinant Kif2a, Aurora B, ICIS D, and α-ICIS. Error bars represent standard deviation.
B. IgG control-injected and α-ICIS injected in prophase Xenopus S3 cells expressing GFP-tubulin. Time lapse images from Supplemental movies 1 and 2. 0 minutes=time of injection. Scale bar represents 10μm.
To determine the importance of the C-terminus of ICIS in cultured cells, we injected this antibody into prophase Xenopus S3 cells stably expressing GFP-tubulin (Figure 2B, Supplemental movie 1). While control IgG-injected cells assemble bipolar spindles by 6–10 minutes (Figure 2B), and complete anaphase by 45 minutes, α-ICIS injected cells (n=8) form monopolar spindles and maintain a mitotic arrest at least 70 min (Supplemental movie 2). This result phenocopies siRNA knockdown of Kif2a in U2OS cells [8], although there is no evidence supporting Kif2a’s role in bipolarity in Xenopus cells. The injected antibodies primarily localize to spindle poles (Supplemental Figure 7). We note that the requirement for ICIS D may not be universal as this monopolar spindle phenotype could not be recapitulated in Xenopus extracts (not shown). Since an antibody that blocks the reactivation activity of ICIS also generates spindle defects, these data argue that ICIS reactivation of kinesin-13s is required for proper spindle function.
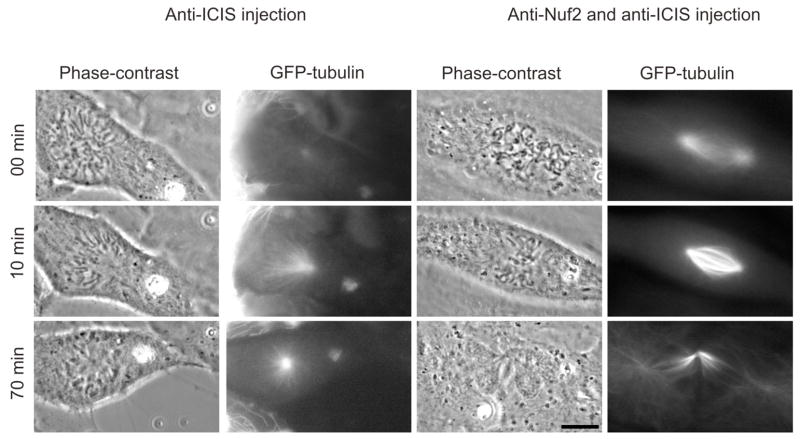
Monopolar spindles generated by α-ICIS injection are rescued by co-injecting α-Nuf2
Monopolar spindles can be generated by defects in centrosome separation, regulators of microtubules dynamics (Kif2a, Aurora A) or by inhibition of kinesins that cross-link pole to pole microtubules (Eg-5, KSP-1). In the α-ICIS injected cells we see separation of poles before nuclear envelope breakdown, arguing for proper centrosome segregation. Previous experiments have shown that inhibition of a class of proteins that regulate K-fiber dynamics can be rescued by co-inhibition of kinetochore-microtubule binding, while monopolarity generated by Eg-5 inhibition is not rescued by this treatment [8]. To classify the monopolarity generated by anti-ICIS injection, we co-injected α-Nuf2 antibodies along with α-ICIS D antibodies into GFP-tubulin expressing S3 cells (n=6, Figure 3, Supplemental movie 3). We found that co-injection did indeed rescue spindle bipolarity, and the cells proceeded through mitosis with a “cut” phenotype, which is expected in a cell with defective kinetochores. These data suggest that the knocking out the kinesin-13 reactivating function of ICIS in Xenopus cells may generate monopolarity through a similar pathway as siRNA knockdown of Kif2a in U2OS cells.
Figure 3. α-ICIS injection phenotype can be rescued by co-injection with α-Nuf2.
S3 cells expressing GFP-tubulin were co-injected with α-ICIS and α-Nuf2 antibodies, filmed and spindle morphology was observed. The co-injected cells had a restored bipolar spindle phenotype. Images are stills from supplemental movie 3. Scale bar represents 10μm.
Kif2a is localized to centromeres in Xenopus cells
ICIS and MCAK are localized to both centromeres and spindle poles in Xenopus [7,9,10]. In human U2OS cells, Kif2a localizes to spindle poles during mitosis and Kif2b localizes to centromeres [11]. Xenopus appears to only have a gene for Kif2a. We generated a polyclonal antibody against Kif2a to determine its localization in Xenopus cells. The Kif2a antibody recognized a single band around 85kD, the expected size in Xenopus (Supplemental Figure 8A). When the antibody was used for immunofluorescence in Xenopus cells, it labeled spindle poles throughout mitosis (Supplemental Figure 8B, arrows). Kif2a is also localized to most centromeres during prometaphase, and to a smaller number of centromeres at metaphase (Supplemental Figure 8B, inset). Thus Kif2a, MCAK and ICIS concentrate at the same cellular locations in Xenopus cells.
ICIS interacts with Aurora B kinase, its substrates and its activators
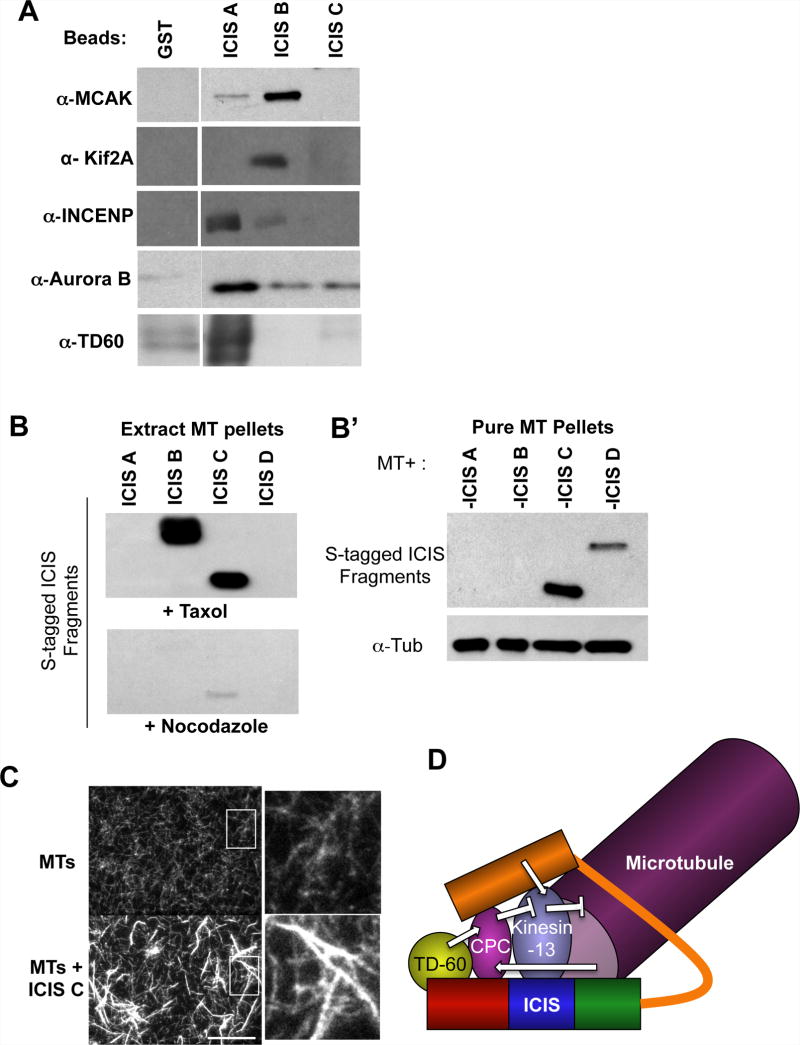
ICIS binds microtubules and coimmunoprecipitates with members of the CPC and MCAK [7]. We mapped these interactions by covalently attaching the four ICIS fragments to sepharose beads, incubating them in a Xenopus CSF-arrested extract and probing the interacting proteins by immunoblot (Figure 4A). We found that the N-terminus of ICIS (ICIS A) interacts with Aurora B, and with its activators INCENP and TD-60 [12]. A middle region (ICIS B fragment) interacts with both Kif2a and MCAK (Figure 4A). These data show that ICIS interacts with Aurora B kinase, its activators and two substrates, Kif2a and MCAK.
Figure 4. Mapping where proteins bind ICIS.
A. Mapping domains of ICIS interactions. The specified recombinant fragments of ICIS were purified from E. coli, covalently attached to beads, and incubated in a mitotic extract. Bound proteins were eluted and immunoblots performed on eluates.
B. Microtubule pelleting assays performed by adding ICIS fragments and taxol to mitotic extracts, pelleting microtubules and detecting ICIS by S-protein HRP (B), or measuring the affinity of ICIS fragments to purified taxol-stabilized microtubules (B′).
C. ICIS C bundles microtubules in vitro. Rhodamine-labeled microtubules were incubated with ICIS C and visualized by fluorescent microscopy. Fields taken with a 100× objective are shown, and magnified views are shown to the right. Scale Bar represents 10μm.
D. Model showing ICIS as a potential scaffold, that brings together Kinesin-13s with its activator (ICIS) and its inhibitor the Chromosomal Passenger Complex (CPC). ICIS also brings Aurora B into contact with its activators TD-60 and microtubules.
To determine which domain of ICIS is responsible for microtubule binding, we added taxol to Xenopus M-phase extracts and pelleted microtubules and their interacting proteins through sucrose cushions. Microtubules pelleted ICIS B and ICIS C (Figure 4B). ICIS C also bound purified taxol-stabilized microtubules; however, ICIS B did not and there was a weak interaction with ICIS D. (Figure 4B′).
We visualized complexes of ICIS C and purified rhodamine-labeled microtubules by fluorescence microscopy. ICIS C bundled microtubules (Figure 4C). The bundles appear to be aggregations of laterally associated microtubules, and the bundles are longer than the individual microtubules used in the assay. The other fragments of ICIS did not bundle microtubules.
Previous to this study, it was not understood why microtubule depolymerases would be phosphorylated on inhibitory sites in cellular locations where their activity is required [1,3,6]. This work reveals a novel form of regulation of these kinesins, whereby phosphorylated and inhibited depolymerases can be reactivated by ICIS without being dephosphorylated (Figure 4D). It remains to be seen whether phosphatase activity also plays a role in the reactivation of kinesin-13 family members. Our structure function analysis of ICIS implicates it as a potential scaffold, which could bring together the activator and the inhibitors of kinesin-13 depolymerases (Figure 4D), to facilitate tight spatial and temporal regulation of depolymerase activity in the mitotic spindle.
Experimental Procedures
Protein expression and antibody production
The full-length Xenopus laevis ICIS cDNA clone was obtained from Dr. Ryoma Ohi (Vanderbilt University, Nashville, TN). A series of bacterial expression constructs were generated by PCR and subcloned into pET30 using 5′ Nco I and 3′ Not I overhangs. The following primers were used to clone the different fragments of ICIS in to pET30: for ICIS A (ICIS1–334) CGCGGATCCATGGGATGAGTGTGCAAACAGCAACT and TCGAGTGCGGCCGCCTACACTTCTAGCATCACTTCACT, for ICIS B (ICIS334–724) CGCGGATCCATGGGGGTGGATGCTCGTAGGGTGATG and TCGAGTGCGGCCGCCTAATCTGCAGATACAGTGCCTTC, for ICIS C (ICIS724–992) CGCGGATCCATGGGGGATATGGATATCTTGGCCAAC and TCGAGTGCGGCCGCCTAGTTACTGCTGCTGAGGAGAT, and for ICIS D (ICIS992–1338) CGCGGATCCATGGGGAACCAAAGGTTCGAGGCACTG and TCGAGTGCGGCCGCTCATCTGGGAGACACCGTCGG. GST-Tagged ICIS fragments were cloned by digesting pET30 constructs with NcoI/NotI, and replacing the inserts into pGEX-4TS3
ICIS fragments were expressed in the E. coli strain BL21 (DE3 pLysS, Novagen). All 6His-tagged proteins were purified on Ni2+-NTA agarose as instructed by the manufacturer (Qiagen, Chatsworth, CA). Glutatione S-transferase (GST)-tagged proteins were purified on glutathione agarose beads. Purified proteins were covalently coupled to CNBr-activated Sepharose beads according to the manufacturer’s instructions (Amersham Biosciences, Piscataway, NJ).
Antibodies were produced from the 6His-ICIS D recombinant protein by Covance Research Products (Denver, PA). Antibodies raised against 6His-tagged proteins were purified over corresponding GST-tagged protein columns, and vice versa. Affinity purified antibodies were dialyzed into PBS.
The cDNA clone of human Kif2a (BC033842) was purchased from Open Biosystems (Huntsville, AL). Kif2a aa 530–679 was cloned into pET30a using primers with the following sequences-GCGGATCCCGGATGCCAACTGCTGCTGGTG and GATCGGAATTCGTTAAAGGGCACGGGGTCTCTT to generate the construct Pet30a-Kif2a (530–679). Recombinant protein was expressed in (pLysS) BL21 bacteria for 4 hours at room temperature. This 6His-tagged protein was purified using conventional nickel bead affinity (Qaigen) according to the manufacturer’s instructions, and the protein was used to make a polyclonal rabbit antibody (Covance, Denver, PA).
Kif2a minimal domain (aa 118–530) was cloned into pET30a as well using primers with the following sequences- GCGGATCCCGGATGGCAAAAAAGGAATTTGG and GATCGCGGCCGCTTATGGATCTACAGTCAATTCTTT, and was expressed and purified as above. The protein was dialyzed into supplemented kinase buffer (20mM tris pH 7.5, 1mM MgCl2, 25mM KCl, 50mM NaCl, 1mM DTT, 5% glycerol).
The pS132 antibody was raised against the peptide C-GPPSRRKpSN, (Synthesized in Yale’s W.M. Keck Protein Core facility, New Haven, CT). The peptide was attached to KLH using SMCC (Pierce), and the resulting conjugate was injected into rabbits to produce polyclonal serum (Covance). For affinity purification both phospho- and non-phospho columns were generated using Sulfo-beads (Pierce) according to the manufacturers instructions. Antibodies were affinity purified by first counter-selection on the non-phospho peptide before being affinity purified on the phosphopeptide column [13]. Immunoblots were performed as previously described [14] except for phospho-peptide antibodies were the PVDF membrane was blocked in 3% BSA.
Visual microtubule depolymerase assays
Microtubule depolymerase assays for Kif2a were performed as described for MCAK [1] using 50nM Kif2a minimal domain, 50nM ICIS fragments (unless otherwise noted), 1nM Aurora B/INCENP in the assays. Briefly, the proteins were incubated with microtubules in CSF-XB (10mM K-Hepes pH 7.7, 100mM KCl, 2mM MgCl2, 0.1 CaCl2, 50mM sucrose, 5mM EGTA), with 2mM ATP, 1mM DTT and 15mM taxol. At the time points indicated, 1ul samples were taken, diluted into 5ul fix solution, and spotted onto a slide. 4 fields were taken per time point using a 100X objective. The images were acquired as above, and analyzed using Metavue software.
Microtubule pelleting assays
For microtubule pelleting in CSF extracts, fresh extracts were treated with either 10 μM Taxol or 10 μg/ml Nocodazole (negative control) for 30 minutes at room temperature. Microtubules and associated proteins were spun through 1M sucrose cushion. For microtubule pelleting in vitro, pre-formed taxol-stabilized pure microtubules (Cytoskeleton, Denver, CO) were incubated with different ICIS mutants at room temperature for 30 min. The reaction mixtures were pelleted through 1M sucrose cushion, washed, and detected by S-protein-HRP or α-Tubulin antibody.
Cell injections
Xenopus laevis S3 cells stably transfected with α-tubulin-GFP were used for microinjections. Cells were kept at 23°C in 70% Leibovitz’s L-15 medium containing 15% FBS, L-Glutamine and penicillin-streptomycin. For microscopy, cells were grown on glass coverslips. Cells were injected with the following antibodies diluted in PBS: rabbit polyclonal anti-ICIS C-terminus at a needle concentration 9mg/ml and anti-xNuf2 at a needle concentration of 4mg/ml. Anti-xNuf2 antibodies were characterized previously [15]. Control cells were injected with non-specific rabbit IgG at a needle concentration of 4 mg/ml. All injections were done before nuclear envelope breakdown (NEB). The cells were analyzed using a Zeiss Axiovert 200M microscope equipped with Planapochromat 63× (N.A. 1.4) and 100× (N.A. 1.4) objectives and a Hamamatsu Orca ER CCD camera (Hamamatsu Photonics). Images were captured using Metamorph software (Molecular Devices).
Supplementary Material
Supplemental Figure 1. Alignment of Xenopus and human Kif2a neck and kinesin domains.
Green represents identical residues, and blue represents conservative substitutions.
Supplemental Figure 2: The C-terminus of ICIS reversed Aurora inhibition of Kif2a in vitro.
Visual microtubule depolymerase assay using rhodamine-labeled microtubules. Images were taken with a 100× objective. These are representative images from that data that were quantified and graphed in Figure 1C.
Supplemental Figure 3. ICIS D restimulation of Kif2a is saturated at stoichiometric levels.
Relative Kif2a activity after 15 minutes of Aurora B phosphorylation, and 5 minutes of depolymerization in the presence of ICIS D. Stoichiometric levels (50nM ICIS D) and above stoichiometric levels (250 nM and 500 nM ICIS D) all gave the same level of restimulation. Relative Kif2a activity was calculated using Kif2a alone as 100% activity, and Kif2a + Aurora B as 0% activity. Error bars represent standard deviation.
Supplemental Figure 4. Characterization of pS132 Kif2a antibody.
A. Alignment of MCAK S196 Aurora consensus site with S132 on Kif2a. Both phosphorylation sites are in the neck regions of the proteins. B. Immunoblot of Interphase extract (IE), mitotic extract (CSF), and mitotic extract + phosphatase inhibitors (MOE) showing specificity of the antibody for phospho-Kif2a.
Supplemental Figure 5. MCAK purified from baculovirus is partially phosphorylated on S196.
Immunoblot of baculovirus-purified MCAK showing that the protein is phosphorylated on S196 after purification.
Supplemental Figure 6. Characterization of α-ICIS D antibody.
Characterization of α-ICIS D antibody. CSF is Xenopus laevis M-phase egg extract. Control immunodepletion with non-specific IgG and immunodepletion with this antibody are also shown.
Supplemental Figure 7. Localization of injected ICIS antibodies
Prophase (left) and a prometaphase (right) cell stained for rabbit IgG after being injected with α-ICIS D. The antibody distributes widely in the cytoplasm, and concentrates at spindle poles (arrows). Scale bar represents 10μm.
Supplemental Figure 8. Kif2a is localized to centromeres in Xenopus cells.
A. α-Kif2a antibody recognizes a single 80 kD protein in Xenopus extracts. Immunoblot shows that the antibody recognizes a single band at the expected molecular weight of Kif2a.
B. Immnuofluorescence images of Xenopus S3 cells co-stained for Kif2a (green) and the kinetochore protein Ndc80 (red). Arrows point to spindle poles. Scale bar represents 10μm.
Supplemental Video 1. Control IgG injection into prophase Xenopus S3 cell expressing GFP-tubulin does not block normal progression through mitosis. Selected frames from this video are presented in the control IgG injection panels in Figure 2. Time is depicted in min:sec
Supplemental Video 2. Injection of anti-ICIS antibody during prophase or early prometaphase induces collapse of the bipolar spindle into a monaster. Selected frames from this video are presented in the anti-ICIS injection panels in Figure 2. Time is depicted in min:sec.
Supplemental Video 3. Co-injection of anti-Nuf2 antibody with anti-ICIS prevents spindle collapse. With time, cells undergo mitotic exit and cytokinesis without anaphase chromosome movement, an expected consequence of anti-Nuf2 antibody injection. Selected frames from this video are presented in the anti-ICIS and anti-Nuf2 injection panels in Figure 3.
Acknowledgments
We thank Claire Walczak for the generous gift of the MCAK antibody and the recombinant MCAK protein, and Ryoma Ohi for the ICIS cDNA clone. We also thank members of the Burke and Foltz labs and Stefan Bekiranov for helpful comments. This work was supported by NIH Grants 5R01GM50412 to GJG and RO1GM063045 to PTS. ALK was supported by Training Grant # T32 GM08136 from NIGMS for Cell and Molecular Biology at the University of Virginia.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Lan W, Zhang X, Kline-Smith SL, Rosasco SE, Barrett-Wilt GA, Shabanowitz J, et al. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr Biol. 2004;14:273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- 2.Ohi R, Burbank K, Liu C, Mitchison TJ. Nonredundant functions of kinesin-13s during meiotic spindle assembly. Current Biology. 2007;17:953–959. doi: 10.1016/j.cub.2007.04.057. [DOI] [PubMed] [Google Scholar]
- 3.Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L, et al. Aurora B regulates MCAK at the mitotic centromere. Dev Cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X, Lan WJ, Ems-McClung SC, Stukenberg PT, Walczak CE. Aurora B phosphorylates multiple sites on mitotic centromere-associated kinesin to spatially and temporally regulate its function. Molecular Biology of the Cell. 2007;18:3264–3276. doi: 10.1091/mbc.E07-01-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kline-Smith SL, Khodjakov A, Hergert P, Walczak CE. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol Biol Cell. 2004;15:1146–1159. doi: 10.1091/mbc.E03-08-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knowlton AL, Lan WJ, Stukenberg PT. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Current Biology. 2006;16:1705–1710. doi: 10.1016/j.cub.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 7.Ohi R, Coughlin ML, Lane WS, Mitchison TJ. An inner centromere protein that stimulates the microtubule depolymerizing activity of a KinI kinesin. Dev Cell. 2003;5:309–321. doi: 10.1016/s1534-5807(03)00229-6. [DOI] [PubMed] [Google Scholar]
- 8.Ganem NJ, Compton DA. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J Cell Biol. 2004;166:473–478. doi: 10.1083/jcb.200404012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wordeman L, Mitchison TJ. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J Cell Biol. 1995;128:95–104. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walczak CE, Mitchison TJ, Desai A. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- 11.Manning AL, Ganem NJ, Bakhoum SF, Wagenbach M, Wordeman L, Compton DA. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Molecular Biology of the Cell. 2007;18:2970–2979. doi: 10.1091/mbc.E07-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosasco-Nitcher SE, Lan WJ, Khorasanizadeh S, Stukenberg PT. Centromeric Aurora-B activation requires TD-60, microtubules, and substrate priming phosphorylation. Science. 2008;319:469–472. doi: 10.1126/science.1148980. [DOI] [PubMed] [Google Scholar]
- 13.Harlow EaLD. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 12-1, 1988. Ref Type: Generic. [Google Scholar]
- 14.Bolton MA, Lan W, Powers SE, McCleland ML, Kuang J, Stukenberg PT. Aurora B kinase exists in a complex with survivin and INCENP and its kinase activity is stimulated by survivin binding and phosphorylation. Mol Biol Cell. 2002;13:3064–3077. doi: 10.1091/mbc.E02-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCleland ML, Gardner RD, Kallio MJ, Daum JR, Gorbsky GJ, Burke DJ, et al. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 2003;17:101–114. doi: 10.1101/gad.1040903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Alignment of Xenopus and human Kif2a neck and kinesin domains.
Green represents identical residues, and blue represents conservative substitutions.
Supplemental Figure 2: The C-terminus of ICIS reversed Aurora inhibition of Kif2a in vitro.
Visual microtubule depolymerase assay using rhodamine-labeled microtubules. Images were taken with a 100× objective. These are representative images from that data that were quantified and graphed in Figure 1C.
Supplemental Figure 3. ICIS D restimulation of Kif2a is saturated at stoichiometric levels.
Relative Kif2a activity after 15 minutes of Aurora B phosphorylation, and 5 minutes of depolymerization in the presence of ICIS D. Stoichiometric levels (50nM ICIS D) and above stoichiometric levels (250 nM and 500 nM ICIS D) all gave the same level of restimulation. Relative Kif2a activity was calculated using Kif2a alone as 100% activity, and Kif2a + Aurora B as 0% activity. Error bars represent standard deviation.
Supplemental Figure 4. Characterization of pS132 Kif2a antibody.
A. Alignment of MCAK S196 Aurora consensus site with S132 on Kif2a. Both phosphorylation sites are in the neck regions of the proteins. B. Immunoblot of Interphase extract (IE), mitotic extract (CSF), and mitotic extract + phosphatase inhibitors (MOE) showing specificity of the antibody for phospho-Kif2a.
Supplemental Figure 5. MCAK purified from baculovirus is partially phosphorylated on S196.
Immunoblot of baculovirus-purified MCAK showing that the protein is phosphorylated on S196 after purification.
Supplemental Figure 6. Characterization of α-ICIS D antibody.
Characterization of α-ICIS D antibody. CSF is Xenopus laevis M-phase egg extract. Control immunodepletion with non-specific IgG and immunodepletion with this antibody are also shown.
Supplemental Figure 7. Localization of injected ICIS antibodies
Prophase (left) and a prometaphase (right) cell stained for rabbit IgG after being injected with α-ICIS D. The antibody distributes widely in the cytoplasm, and concentrates at spindle poles (arrows). Scale bar represents 10μm.
Supplemental Figure 8. Kif2a is localized to centromeres in Xenopus cells.
A. α-Kif2a antibody recognizes a single 80 kD protein in Xenopus extracts. Immunoblot shows that the antibody recognizes a single band at the expected molecular weight of Kif2a.
B. Immnuofluorescence images of Xenopus S3 cells co-stained for Kif2a (green) and the kinetochore protein Ndc80 (red). Arrows point to spindle poles. Scale bar represents 10μm.
Supplemental Video 1. Control IgG injection into prophase Xenopus S3 cell expressing GFP-tubulin does not block normal progression through mitosis. Selected frames from this video are presented in the control IgG injection panels in Figure 2. Time is depicted in min:sec
Supplemental Video 2. Injection of anti-ICIS antibody during prophase or early prometaphase induces collapse of the bipolar spindle into a monaster. Selected frames from this video are presented in the anti-ICIS injection panels in Figure 2. Time is depicted in min:sec.
Supplemental Video 3. Co-injection of anti-Nuf2 antibody with anti-ICIS prevents spindle collapse. With time, cells undergo mitotic exit and cytokinesis without anaphase chromosome movement, an expected consequence of anti-Nuf2 antibody injection. Selected frames from this video are presented in the anti-ICIS and anti-Nuf2 injection panels in Figure 3.