Summary
Proteasome-mediated protein turnover in all domains of life is an energy-dependent process that requires ATPase activity. Mycobacterium tuberculosis (Mtb) was recently shown to possess a ubiquitin-like proteasome pathway that plays an essential role in Mtb resistance to killing by products of host macrophages. Here we report our structural and biochemical investigation of Mpa, the presumptive Mtb proteasomal ATPase. We demonstrate that Mpa binds to the Mtb proteasome in the presence of ATPγS, providing the physical evidence that Mpa is the proteasomal ATPase. X-ray crystallographic determination of the conserved inter-domain showed a five-stranded double β-barrel structure containing a Greek key motif. The structure and mutagenesis indicate a major role of the inter-domain for Mpa hexamerization. Our mutational and functional studies further suggest that the central channel in the Mpa hexamer is involved in protein substrate translocation and degradation. These studies provide insights into how a bacterial proteasomal ATPase interacts with and facilitates protein degradation by the proteasome.
Keywords: proteasome, proteasomal ATPase, protein structure, X-ray crystallography, electron microscopy
INTRODUCTION
The proteasome carries out the majority of cytosolic protein degradation inside a eukaryotic cell. In doing so, the proteasome regulates a multitude of cellular functions such as protein homeostasis, signaling, cell cycle progression, and the production of antigenic peptides (Goldberg, 2007; Hershko and Ciechanover, 1998). The proteasome has emerged as an effective drug target, and the proteasome inhibitor bortezomib (Velcade®), a dipeptide boronate, has been approved for the treatment of multiple myeloma (Borissenko and Groll, 2007). The proteasome is found ubiquitously in eukaryotes and archaea and some bacteria (Voges et al., 1999). The structure of the 20S core particle and its proteolytic mechanism are highly conserved across all domains of life: the enzyme is a threonine protease composed of α- and β-subunits arranged in α7β7β7α7 fashion into a cylindrical structure (Groll et al., 1997).
The core particle alone degrades only small peptides or unstructured proteins. Degradation of normally folded protein substrates is usually an energy dependent process that requires ATPase activity provided by additional structures (Smith et al., 2006; Voges et al., 1999). The eukaryotic proteasome contains a 19S complex at one or both ends of the core particle. The yeast 19S cap contains at least 19 protein subunits, six of which, Rpt1-6, are ATPases that contact the core particle (Schmidt et al., 2005). Due to its large size and highly dynamic nature, crystallographic solution of the 19S cap or the entire 26S proteasome has not been achieved, leaving critical questions as to how proteasomal ATPases unfold and translocate protein substrates for degradation by the core particle.
Archaeal proteasomes have a simpler ATPase system in which six identical proteins form a homo-hexamer, loosely capping the seven-fold symmetrical core particle (Smith et al., 2007). These hexameric ATPases hydrolyze ATP to actively unfold and translocate substrates through a gate into the proteolytic chamber (Benaroudj et al., 2003; Medalia et al., 2009). Unlike archaea, most bacteria do not contain proteasomes, and instead, synthesize other protease complexes, such as ClpAP, ClpXP, HslUV, Lon, and FtsH (Baker and Sauer, 2006; Butler et al., 2006). However, proteasome genes, together with proteasomal ATPase genes, have been identified in bacteria of the order Actinomycetales (Nagy et al., 1998; Wolf et al., 1998; Zhang et al., 2004). The functions of these bacterial proteasomes are largely unknown. Recently, the proteasome and its associated ATPase in Mtb, a species of Actinomycetales, were found to play a critical role in bacterial resistance to killing by reactive nitrogen intermediates, which are products of host macrophages (Darwin et al., 2003). Moreover, suppression of expression of the Mtb proteasome led to death of the pathogen in mice (Gandotra et al., 2007). Thus, the Mtb proteasome is currently being targeted for the development of anti-tuberculosis agents (Nathan et al., 2008).
The Mtb proteasomal genes prcA and prcB encode bona fide proteasome subunits (Lin et al., 2006), and the Mtb 20S proteasome structure is essentially identical to eukaryotic and archaeal proteasomes (Hu et al., 2006). The small post-translational modifier Pup in Mtb covalently tags proteasome substrates for degradation in a manner that is functionally analogous to ubiquitylation in eukaryotes (Pearce et al., 2008). Thus, the Mtb 20S-Mpa pathway may be a simpler model system for the structural investigation of how certain tagged proteins are recognized, unfolded, and degraded by proteasomes.
Mpa was shown to have ATPase activity (Darwin et al., 2005), and degradation of proteasome substrates in Mtb required the presence of Mpa, together with an additional proteasome accessory factor (PafA), a Pup ligase (Festa et al., 2007; Pearce et al., 2006; Pearce et al., 2008; Striebel et al., 2009). However, there were no data that showed any bacterial ATPase interacted with its cognate proteasome. In this report, we demonstrate for the first time that Mpa could indeed directly interact with the Mtb 20S core particle. Mpa is a multi-domain structure, with an amino terminal predicted coiled-coil domain, a 150-amino acid inter-domain (Mpa-ID) that is unique to the proteasome-associated ATPases, a canonical AAA (ATPase associated with various activities) domain, and a small carboxyl terminal domain (Fig. 1A). We determined the crystal structure of the Mpa-ID, which formed a tightly packed ring-shaped hexamer in the crystal structure as well as in solution. Finally, we demonstrate that specific point mutations in Mpa selected based on our structural analysis could either abolish hexamerization in vitro, or proteasome substrate degradation in Mtb.
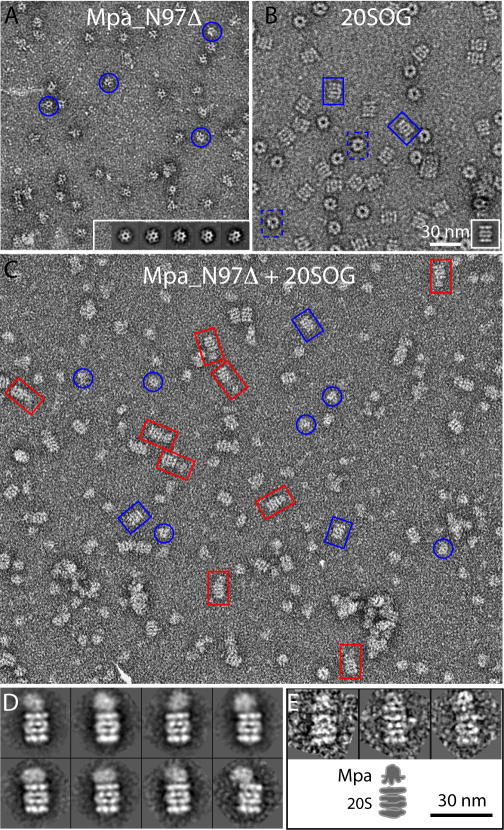
Fig. 1.
Mpa directly interacts with the open gate form (20SOG) of the Mtb proteasome. (A) A small region of a raw electron micrograph of negatively stained Mpa_N97Δ. Inset: Five class averages of the deletion mutant Mpa particles. The blue circles mark several particles. (B) A raw EM image of the Mtb open gate 20S proteasome. Inset: 2D class average of the proteasome particles in the side view. The blue rectangles mark individual particles in side (solid line) or top (dashed line) views. (C) A raw EM micrograph of 20SOG incubated with Mpa_N97Δ in the presence of ATPγS. The red rectangles mark several 20CP-Mpa complexes. Panels (A) and (B) are in the same scale. (D) Eight selected 2D class averages of the Mpa-20SOG complex particles showing the flexible nature of the interaction between Mpa and proteasome. (E) Upper panel: three raw particle images of the Mpa-20SOG complex in which the Mpa binds approximately axially on the proteasome. Lower panel: a sketch showing that the Mpa C-terminal portion with weaker density interfaces with the 20SOG. Panels (D) and (E) are in the same scale. Scale bar: 30 nm.
RESULTS
1. Mpa directly interacts with the Mtb 20S proteasome core particle
The requirement for Mpa in Mtb 20S proteolysis has been strongly implicated (Darwin et al., 2003; Pearce et al., 2006; Pearce et al., 2008). However, a direct interaction between the 20S core particle (CP) and Mpa has not been demonstrated. We investigated by EM the binding between the full-length Mpa or an N-terminal 97-residue deletion mutant Mpa (Mpa_N97Δ) with the wild type or the open gate (OG) mutant 20S (20SOG) CP in the presence of various nucleotides (See SI for sample preparation) (Fig. 1). Mpa_N97Δ has the N-terminal predicted coiled coil domain removed. The 20SOG CP is an N-terminal octapeptide deletion mutant at the α-subunit of the 20S CP. The proteolytic activity of this mutant increased by an order of magnitude, and the mutant was thus designated open gate (Lin et al., 2006). No significant binding was observed between Mpa or Mpa_N97Δ and the wild type 20S CP (data not shown). However, we found that both forms of Mpa were able to bind the 20SOG CP in the presence of 1 mM ATPγS (Fig. 1 and Fig. S1). Under these conditions, Mpa_N97Δ had higher affinity for the 20SOG than the full-length Mpa, based on the frequency with which the 20SOG was found to be in complex, which was ~15% and ~2% for Mpa_N97 and Mpa, respectively. The orientation of the bound Mpa on 20SOG appears to be highly flexible (Fig. 1D, Fig. S1C). From particles in which Mpa binds approximately along the axial position of the 20SOG CP, it appears that the C-terminal end of Mpa contacts the α-ring of the CP (Fig. 1E).
2. The conserved Mpa-ID forms an oligomer in solution
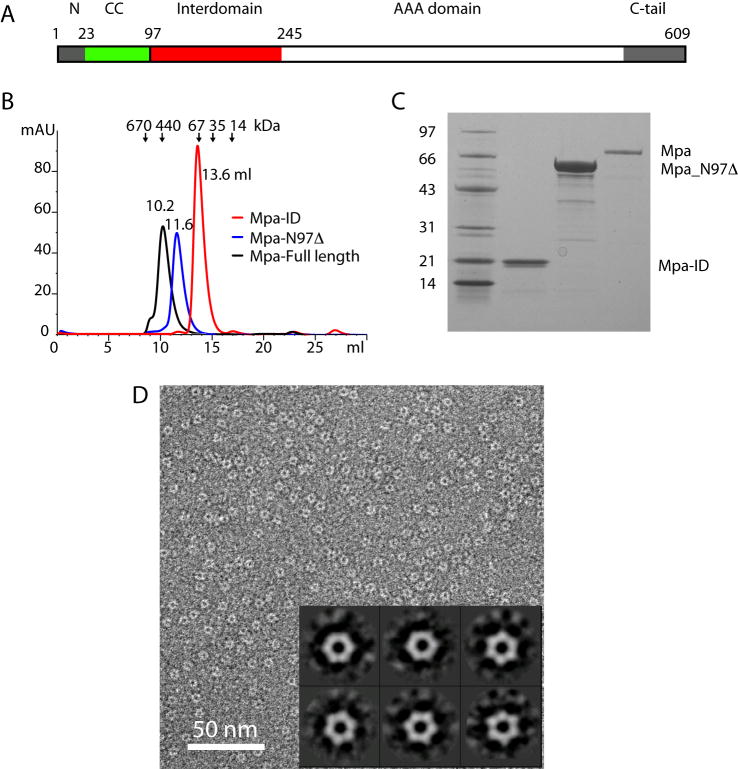
Bioinformatic analysis of the Mpa indicated a separately folded domain, ~150 amino acids long, from Pro-97 to Glu-245, that we termed the inter-domain (Mpa-ID), in a region between the N-terminal coiled coil domain and the C-terminal AAA domain. This domain appears to be unique to proteasomal ATPases (Fig. 2A) (Zhang et al., 2004). This stably-folded domain was identified by limited trypsin digestion and mass spectroscopy (data not shown). Mpa-ID eluted from the gel filtration column at a volume corresponding to ~100 kD (Fig. 2B), suggesting that Mpa-ID oligomerized in solution as a hexamer. The purified Mpa-ID migrated at ~20 kD, near the predicted 16.6 kD in the Coomassie-stained SDS-PAGE (Fig. 2C). We inspected the purified and His6-tag removed Mpa-ID by negative stain electron microscopy, and found that Mpa-ID indeed formed a ring-like structure with an apparent six-fold symmetry (Fig. 2D). The ring is about 65 Å in diameter with a ~ 20 Å central pore. Based on these observations, we conclude that Mpa-ID on its own is capable of hexamerization, and consequently, this domain may contribute to the stability of the full-length Mpa hexamer that we previously visualized by negative stain EM (Darwin et al., 2005).
Fig. 2.
Mpa has a multi-domain architecture. (A) Overview of the domain organization of the full-length Mpa. (B) The Superdex-200 gel filtration profiles of the purified full-length Mpa in black, Mpa_N97Δ in blue, and Mpa-ID in red. Each of the three proteins is eluted at the volume corresponding to their respective hexameric form. (C) The Coomassie blue stained SDS-PAGE of the purified Mpa-ID (lane 2), Mpa_N97Δ (lane 3), and Mpa (lane 4). Lane 1 is the molecular weight marker in kD. (D) The negatively stain EM image of the purified Mpa-ID and the inserted six 2D class averages reveal a hexameric structure about 65 Å in diameter with a ~20 Å central low density channel.
3. The Mpa-ID has two sub-domains, each with a five-stranded β-barrel
We determined the atomic structure of Mpa-ID at 2.0 Å resolution by X-ray crystallography (See Experimental Procedures; Table 1 and 2, and Fig. S2A for the quality of the electron density map). The refined structure has an Rfree of 26% and Rwork of 21%, and includes all residues except in a long loop region between Leu-188 and Gly-217. This loop can be fully traced only in one monomer (Fig. S2B); in the other monomers, it is partially disordered and has a gap between Ile-193 and Leu-214.
Table 1.
Data reduction & structure determination statistics
| Dataset | Native I (1°-360°) | Native II (1°-270°) | Hg2+ derivative (1°-360°) |
|---|---|---|---|
| Oscillation | 1° | 1° | 1° |
| Space group | P21 | P21 | P21 |
| Unite cell parameters (Å) | a = 74.86 | a = 75.39 | a = 75.03 |
| b = 74.99 | b = 75.92 | b = 75.59 | |
| c = 200.85 | c = 192.74 | c = 192.37 | |
| α = 90 | α = 90 | α = 90 | |
| β = 90.319 | β = 90.015 | β = 90.179 | |
| γ = 90° | γ = 90° | γ = 90° | |
| Resolution (Å) | 20~2.0 | 20–2.6 | 20~2.8 |
| Wavelength (Å) | 1.000 | 1.000 | 1.0066 |
| Rmerge*, ** | 0.21 (0.55) *** | 0.06 (0.60) | 0.06 (0.49) |
| Completeness** | 0.98 (0.89) | 0.97 (0.93) | 0.91 (0.82) |
| Mean of redundancy | 7.2 | 4.0 | 3.2 |
| I/σ (I)** | 13.0 (1.9) | 22.1 (1.9) | 14.9 (1.8) |
| f′ (Hg) | −12.98 | ||
| f″(Hg) | 9.74 | ||
| Measured reflections | 1,064,345 | 287,826 | 328,382 |
| Unique reflections | 147,655 | 66,845 | 98,072 |
| Heavy atom sites | 17 |
Rmerge = Σ((I − <I>)2)/Σ (I2)
Values in parentheses refer to the highest resolution shell.
High Rmerge value maybe due to the presence of twinning or a small crack.
Table 2.
Refinement statistics of the Mpa-ID crystal structure
| Resolution | 20-2.0 (Å) |
|---|---|
| No. of reflections used | 140,155 |
| No. of non-hydrogen atoms | 13,735 |
| No of water | 866 |
| Rwork* | 0.213 |
| Rfree* | 0.266 |
| Mean B value (Å2) | 33.4 |
| R.m.s. deviation of bond angle (°) | 1.682 |
| R.m.s. deviation of bond length (Å) | 0.018 |
| Mean figure of merit | 0.83 |
Rwork = Σ (|Fo − Fc|)/Σ |Fo|; Rfree is the same as Rwork calculated with a test set of 5% random selected subset of the reflections that was not included in refinement calculations.
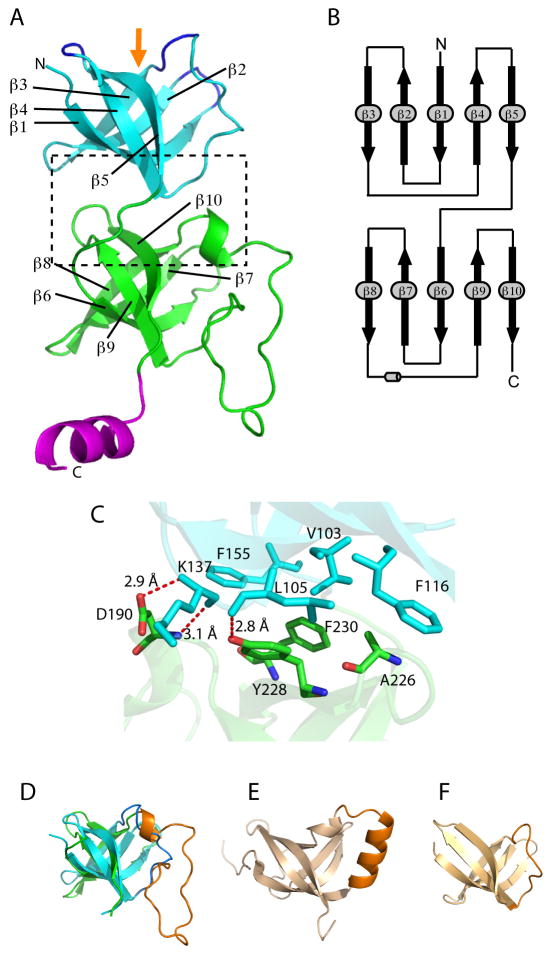
The Mpa-ID contains two sub-domains (Fig. 3A). Each sub-domain is a five-stranded β-barrel. Unexpectedly, the two β-barrels are arranged in tandem and connected by a 6-amino acid loop between Thr154 – Gly159. In each sub-domain, the five β-strands form a g−b type fold containing a left-handed Greek key motif, where g refers to Greek key motif, and b is β5 (Fig. 3B) (Zhang and Kim, 2000). The interaction between the two sub-domains is mainly of hydrophobic nature, involving four large hydrophobic residues (Phe-116 and Phe-155 in subdomain 1 and Tyr-228 and Phe-230 in subdomain 2) and three smaller ones (Val-103, Leu-105, Ala-226) (Fig. 3C). In addition, there are three hydrogen bonds that further stabilize the interface (two H-bonds between Lys-137 and Asp-190; and one H-bond between Try-228 and Leu-105). Although no sequence repeat can be observed within the Mpa-ID, the two β-barrels can be superimposed with a root mean square deviation (RMSD) of 1.9 Å (Fig. 3D). The only significant difference is the connecting loop between the third and the fourth β-strands in each β-barrel, as highlighted in blue and orange, respectively (Fig. 3D). The Mpa-ID tandem β-barrels are followed by a C-terminal α-helix (Fig. 3A). This helix is a linker between the ID and the following AAA domain, because the predicted α/β subdomain of AAA domain starts immediately after the α-helix.
Fig. 3.
The crystal structure of the Mpa-ID. (A) Mpa-ID contains two five-stranded β-barrel subdomains connected by a loop. The C-terminal helix might be a linker between the interdomain and the following AAA domain. The orange arrow indicates the substrate recognition/binding region at the top as defined by the three β-hairpin loops highlighted in blue. (B) The topology of Mpa-ID contains two Greek key motifs formed by the first four β-strands in each sub-domain. (C) An enlarged view of the interface between the two tandem β-barrels as boxed in (A), showing the hydrophobic and H-bond interactions. (D) The two β-barrel sub-domains have a similar fold; they can be overlapped with an RMSD of 1.9Å. (E) and (F) are the OB-fold structures of the vaccinia virus K3L (D, PDB ID 1LUZ, RMSD = 2.5 Å) and the transcription factor 5A (E, PDB ID 1EIF, RMSD = 1.5 Å, sequence identity = 21%), respectively, aligned with the first β-barrel sub-domain in Mpa-ID.
The five-stranded β-barrel with the Greek key motif as observed in Mpa-ID is also called the oligonucleotide and oligosaccharide binding-fold (OB-fold), due to the nature of its binding substrates (Murzin, 1993). The first β-barrel of Mpa-ID can be superimposed with an RMSD of 2.5 Å with the OB-fold structure found in the vaccinia virus protein K3L that is involved in substrate recognition and subversion of host immune defense (Fig. 3E) (Dar and Sicheri, 2002), and with an RMSD of 1.5 Å with the OB-fold in the transcription factor 5A (TF5A) (Fig. 3E) (Kloks et al., 2002). Interestingly, TF5A has 21% sequence identity with Mpa-ID. In a canonical OB-fold, an α-helix links the β3 and β4 strands and consequently caps the β-barrel (Fig. 3E). However, in the first β-barrel of Mpa-ID, a 14-amino acid loop (Thr-125 to Gly-138), instead of an α-helix, connects the β3 and β4 strands, and in the second β-barrel, a long (30 amino acids) and partially disordered loop (Leu 188 ~ Gly 217), connects the two corresponding β-strands (Fig. 3D). In the OB-fold proteins, the nucleotide or sugar binding sites are clustered in a similar region: on the side surface of the β-barrel where three loops, the loop between β1 and β2, the beginning section of the long loop between β3 and β4, and the loop between β4 and β5, come together (Murzin, 1993). In Fig. 3A, these loops in the first β-barrel are highlighted in blue and the region is labeled by an orange arrow. The corresponding region in the first β-barrel roughly translates to the top surface in the Mpa-ID hexamer (Fig. 3A and Fig. 4). This region in the Mpa-ID second β-barrel is shielded by intramolecular interaction. Given the role of Mpa in protein degradation, this analysis suggests that the top surface of the Mpa-ID hexamer is involved in protein-substrate recognition and binding.
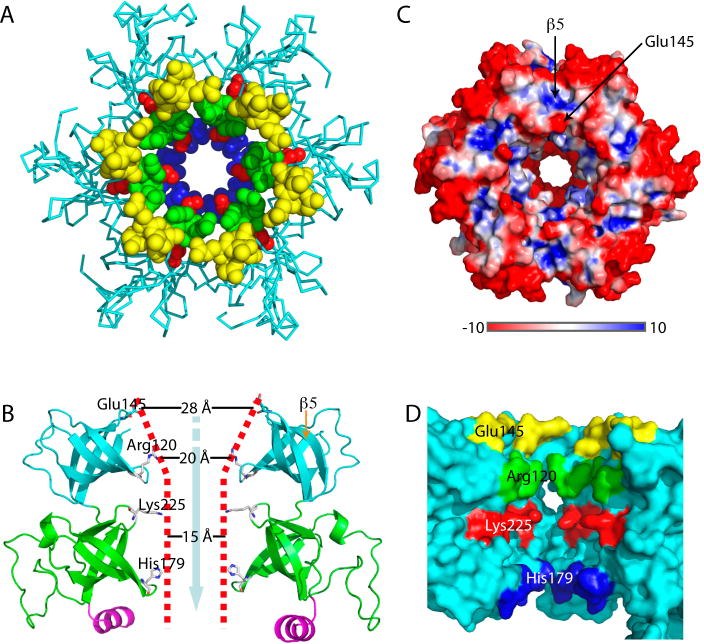
Fig. 4.
Crystal structure of the Mpa-ID hexamer. (A) The top view of the hexamer, with residues of the four β-hairpins lining the central channel shown as spheres in different colors. (B) Side view showing only two diagonal subunits. The four residues lining the funnel-like central channel are highlighted in the ball-and-stick presentation. The potential peptide translocation path is illustrated by a cyan arrow. (C) Surface-charge presentation of the Mpa-ID hexamer in the top view. Arrows indicate the substrate-binding site in the OB-fold above the β5 strand, and the negatively charged Glu-145 at the opening of the channel, respectively. (D) Surface rendered side view of the Mpa-ID hexamer with the front two subunits removed to show the interior channel. Charged residues lining the channels are labeled and shown in different colors.
4. The Mpa-ID hexamer structure has a negatively charged outside surface and a positively charged central channel
Mpa-ID formed hexamers in the crystal structure (Fig. 4A). In fact, two hexamers stacked end-to-end, forming a dodecamer as the packing unit in the crystal structure. The interface between the two hexamers in the crystals is maintained by three hydrophobic residues in the C-terminal helix (Fig. S3). The Mpa-ID hexamer is ~70 Å in diameter and ~60 Å high, comparable to the dimensions observed by EM (Fig. 2D). Intriguingly, the Mpa-ID hexamer has a highly negatively charged side surface, but several positively charged patches at the top, which we have described above as contributing to a potential substrate-binding site (Fig. 4A and 4C). The hexamer has a central channel. At the opening of the channel on the top surface, the six Glu-145 of the 6 β-hairpin loops (β4-β5) define a large and negatively-charged pore (28 Å) (Fig. 4B, 4C, and 4D). The interior of the channel is, however, lined by highly positively charged residues in three layers formed by β-hairpins from top to bottom: six Arg-120 of β2-β3 at the first layer define a 20-Å pore, six Lys-225 of β9-β10 at the middle layer define a 15-Å pore, and six His-179 of β7-β8 at the bottom layer also form a 15 Å pore (Fig. 4B, 4D). The central channel of Mpa-ID is funnel-shaped, constricting gradually from the top to the bottom by a factor of 2, from 28 Å to 15 Å (Fig. 4B).
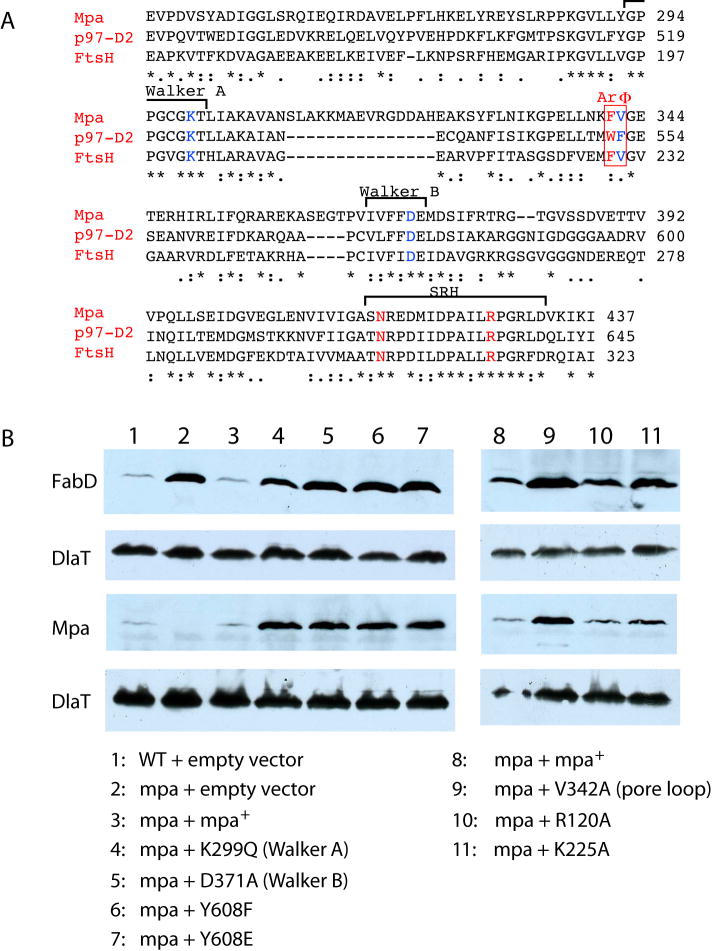
We tested the possible role of these positively charged amino acids on the recognition of a known proteasomal degradation substrate, malonyl coA-acyl carrier protein transacylase (FabD). FabD is targeted for degradation by Pup, which is highly negatively charged (Pearce 2008). Point mutations in Mpa-ID were made at either Arg-120 or Lys-225 and introduced into an mpa null mutant to determine if they could restore degradation of FabD. The predicted α/β sub-domain, but not the α-helical sub-domain, of the Mpa AAA domain can be aligned with mammalian p97 (43% identity, 61% similarity) and shares 38% identity with AAA domain of the bacterial membrane associated protease FtsH (Fig. 5A). The D2 domain of p97 was recently implicated in protein substrate binding and possibly unfoldase activity (DeLaBarre et al., 2006). ATPases involved in protein substrate unfolding and translocation contain a conserved Ar-Φ pore loop that was proposed to pull the unfolded peptide through the central channel (Martin et al., 2008a, b). The Ar-Φ pore loop motif, containing single aromatic residue and a hydrophobic amino acid, is also conserved in Mpa (Fig. 5A). Therefore, we also mutated the Φ residue Val-342. In addition, we analyzed several previously reported mutants that disrupt the in vitro ATPase activity or the penultimate tyrosine (Tyr) that is highly conserved in most proteasome-associated ATPases (Darwin et al., 2005).
Fig. 5.
Site-directed mutations in Mpa disrupt protein degradation. (A) Sequence alignment of Mpa with p97/VCP and FtsH reveals unexpected high identity at the α/β sub-domain of the AAA domain with p97 D2 domain at 46% and FtsH at 38.5%. The conserved aromatic and hydrophobic residues in the pore-loop region of AAA domain are highlighted by a red box (ArΦ). The Walker A/B motifs, and the conserved sensor-1 residue (N) and the arginine finger (R) in the second region of homology (SRH) are also marked. (B) Immunoblot analysis of total Mtb lysates of equivalent cell numbers of wild type, mpa mutant or mpa mutant Mtb transformed with plasmids encoding wild type or mutated mpa. Samples were analyzed with either polyclonal antibodies to FabD or Mpa. The same blots were stripped and re-analyzed with antibodies to dihydrolipoamide dehydrogenase (DlaT), the loading control.
Under steady state conditions, mutations in the Walker boxes (required for ATPase activity), the penultimate Tyr (Tyr-608), or the pore loop dramatically abrogated FabD degradation (Fig. 5B, lanes 1–7, 9). Importantly, Mpa is itself a target of proteasomal degradation (Pearce 2006), thus these mutations also increased the steady state levels of Mpa, but not a control protein, DlaT. In contrast, Arg-120 or Lys-225, which we hypothesized may interact with pupylated substrate, did not appear to dramatically affect the degradation of FabD, (Fig. 5B, lanes 10, 11). This indicates that the central channel in the OB-ring might not be directly involved in Pup recognition. However it is still possible that these amino acids are important for efficient degradation under different growth conditions, or of other proteins.
5. The second β-barrel ring in Mpa-ID contributes to the stability of the Mpa hexamer
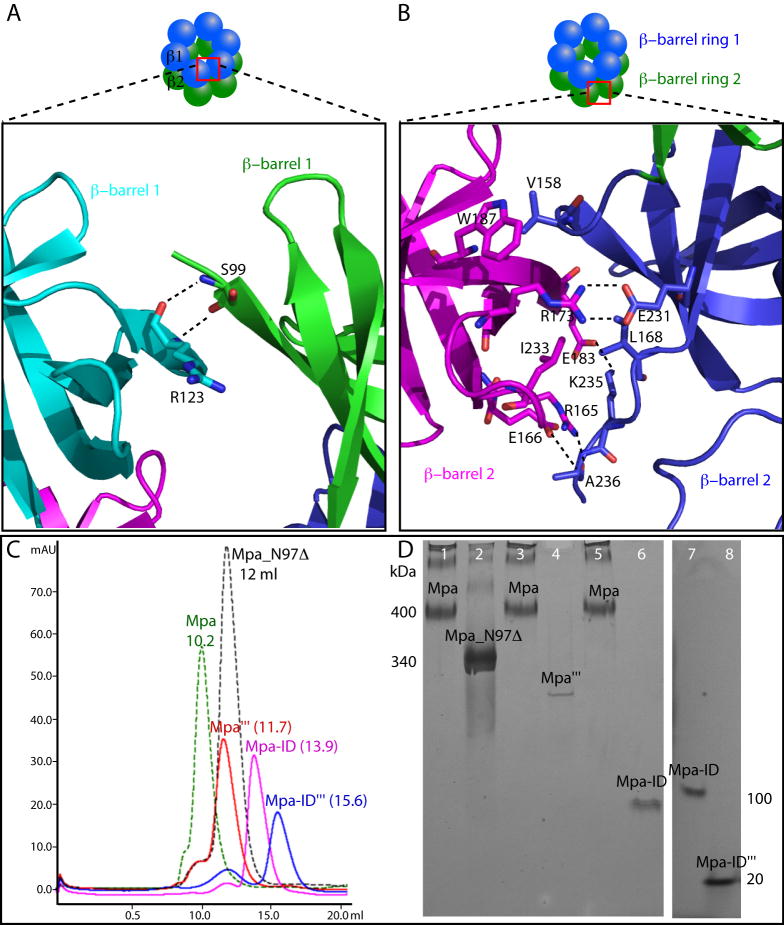
The interactions between the monomers within the hexamer are quite different for the two sub-domains: in the first or top β-barrel ring, interactions between the two neighboring β-barrels are minimal, involving only main chain hydrogen bonds (3.1 Å and 3.2 Å) between Ser-99 and Arg-123 (Fig. 6A). In contrast, in the second β-barrel ring, interactions between the two neighboring β-barrels are extensive, involving two salt bridges (between Glu-231 and Arg-173, 2.9 Å; Lys-235 and Glu-183, 2.7 Å), two hydrophobic interactions (between Val-158 and Trp-187; Ile-233 and Leu-168), and two hydrogen bonds (between carboxyl O of Ala-236 with the NH2 of Arg-165; 3.3Å; between the main chain NH of Ala-236 with the side chain carboxyl O of Glu-166, 2.73 Å) (Fig. 6B). Therefore, the second β-barrel ring of Mpa-ID appears to be in a position to contribute to the stability of the full-length Mpa hexamer. Mutagenesis was employed to investigate this possibility. We found that disruption of the H-bonds between Ala-236 and Arg-165 by deleting the Ala-236 did not abolish hexamerization of Mpa-ID (data not shown). Subsequently, we designed a triple mutant (R173E; W187A; K235E) that disrupts the salt bridges and the hydrophobic interaction. We found by gel filtration (Fig. 6C) and blue native gel electrophoresis (Fig. 6D) that the triple mutant Mpa-ID (denoted Mpa-ID‴) was monomeric. Furthermore, no ring-like hexameric particles of Mpa-ID‴ were observed by negative stain EM (data not shown). The same triple mutations when introduced into the full-length Mpa significantly reduced the production of the recombinant protein in E. coli, and markedly altered the migration of Mpa in both gel filtration profile and blue native gel electrophoresis (Fig. 6C and 6D). The mutant Mpa (Mpa‴) eluted from the sizing column and migrated in the native gel between the hexameric Mpa (400 kD) and the hexameric Mpa-ID (100 kD), and appeared to form loosely-associated trimeric particles (predicted mass 200 kD) in negatively stained electron micrographs, with an appearance drastically different from that of the native hexameric Mpa (Fig. S4). These data indicate an important role of the second β-barrel of Mpa-ID for stabilizing the functional Mpa hexamer.
Fig. 6.
Subunit-subunit interactions in the Mpa-ID hexamer. (A) The interface between two neighboring β-barrel one is limited, involving only two main chain H-bonds between the Arg-123 and Ser-99. (B) The interactions between the two neighboring β-barrel two are extensive. At the top of the interface, Trp-187 and Val-158 form a hydrophobic interaction; at the middle section, the interface involves two salt bridges/H-bonds (between Arg-173 and Glu-231, and between Glu-183 and Lys-235), and a pair of hydrophobic residues (between Ile-233 and Leu-168); and at the bottom section, Arg-165 and Glu-166 form two H-bonds with the Ala-236. (C). Gel filtration profiles of the native full-length Mpa, Mpa_N97Δ, the full-length Mpa triple mutant (Mpa‴), Mpa-ID, and Mpa-ID with the triple mutations (Mpa-ID‴). (D) The Blue Native gel of the corresponding Mpa constructs purified as shown in (C). The right panel (lane 7 and 8) was run separately for a shorter period of time than the left panel. This was to prevent the small Mpa-ID‴ (~ 20 kD) from running out of the gel.
DISCUSSION
Weak in vitro interaction between Mtb 20S and Mpa
To our knowledge, this is the first demonstration that a bacterial proteasome can directly interact with its cognate ATPase. Previously, Smith et al reported that PAN can bind to a non-cognate proteasome and stimulate protein degradation (Smith et al., 2007). Mpa was previously only circumstantially linked to the Mtb proteasome by genetic studies (Darwin et al., 2003; Pearce et al., 2006). We found that the in vitro interaction between Mpa and 20SOG was not stable to gel filtration (data not shown), and must thus be weak. In electron micrographs, only 15% of Mtb 20S core particles were bound to the Mpa_N97Δ, although the Mpa ATPases were incubated at high molar ratio (4:1) with 20S CP. It is currently unclear why Mpa_N97Δ binds better than the full-length Mpa to 20S CP. We only observed Mpa binding to one end of 20S CP. This is likely due to the low in vitro affinity of “apo” Mpa for 20S CP. Binding of PAN with the proteasome CP was also weak, undetectable by native PAGE, gel filtration, ultracentrifugation, or immunoprecipitation (Forster et al., 2003), and was only demonstrated by highly sensitive techniques such as surface plasmon resonance and EM (Smith et al., 2005). The off-axis and flexible binding of Mpa on 20S CP was also similar to the binding of PAN on 20S CP. Unlike PAN, we were unable to detect Mpa-enhanced 20S protein degradation in vitro with either peptides or β-casein as substrates (data not shown). These observations underscore the fact that other factors such Pup are required for protein degradation by Mpa-20S system in Mtb (Pearce et al., 2008; Striebel et al., 2009).
Functional implications of Mpa-ID hexamer with the OB fold
Most recently, the archaeal proteasome-activating nucleotidase (PAN) from Methanocaldococcus jannaschii was shown to contain a single OB-fold structure (Zhang et al., 2009), and the AAA ATPase forming ring shaped complex (ARC) from the bacterium Rhodococcus erythropolis contained the tandem double OB-motifs (Djuranovic et al., 2009). All these OB domains, like Mpa-ID, formed hexamers. Thus the hexameric OB-ring appears to be a unique feature of all proteasomal ATPases (Djuranovic et al., 2009). The OB-fold has been found in more than 80 other unrelated proteins. The OB-fold is in general highly stable. For example, the OB-fold structures in three unrelated proteins, the staphylococcal nuclease SN, the E coli anticodon binding domain of Lys-tRNA synthetase and the cold shock protein CspA, were shown by mutagenesis and hydrogen exchange experiments to be resistant to unfolding (Alexandrescu et al., 1995; Alexandrescu et al., 1999; Jaravine et al., 2000). We found that Mpa-ID is also stable and resistant to prolonged protease treatment. Therefore, the hexameric Mpa-ID with a total of 12 OB-motifs might be mechanically strong. Given its high structural stability, we propose that the Mpa-ID hexamer might provide a rigid platform upon which the mechanical force exerted from the Mpa AAA domain unfolds the substrate protein. The Mpa AAA domain alone is unstable, and does not form hexamer (data not shown). Therefore, the Mpa OB-ring is also the major stabilizing factor of the Mpa hexamer.
Thus, the OB-ring as the core of Mpa serves multiple functions: the OB-ring is a main factor that holds the Mpa hexamer together, provides the initial binding site for the substrate in conjunction with the coiled coil domain, an entrance point into the ATPase central channel, and a rigid platform against which the protein substrate can be pulled apart for translocation and ultimate degradation in the proteasome core. Our in vitro and in vivo mutagenesis studies support these functions.
Mpa facilitated protein degradation by the Mtb proteasome
The system for protein tagging and degradation in Mtb consists at least of Pup-Mpa-20SCP, the organization of which is reminiscent of the prokaryotic SsrA-ClpX-ClpP and eukaryotic ubiquitin-19S-20SCP systems. Each uses a protease complex that requires an oligomeric ATPase for unfolding and translocating the protein substrate into the protease core for degradation. In the ClpXP system, the protein substrate unfolding and translocation involves the Ar-Φ pore loop of the ClpX ATPase (Martin et al., 2008a, b). We found that the conserved Ar-Φ pore loop in Mpa is also essential for substrate degradation in Mtb (Fig. 5B), suggesting that Mpa might share a similar protein translocation mechanism.
In summary, our demonstration of the physical interaction between Mpa and the Mtb proteasome and the crystallographic revelation of the OB-fold structure of Mpa-ID, in conjunction with our previous genetic and functional evidence (Darwin et al., 2003; Darwin et al., 2005; Pearce et al., 2006), firmly establish Mpa as the bona fide Mtb proteasomal ATPase. Because the virulence of Mtb relies on proteasome function (Darwin et al., 2003; Darwin et al., 2005; Gandotra et al., 2007; Pearce et al., 2006), and the function of the Mtb proteasome apparently depends on Mpa, it is possible to take advantage of the unique structure to design inhibitors for Mpa. Such an approach potentially circumvents the difficulty in developing species-specific proteasome inhibitors (Lin et al., 2008), given the high conservation of proteasome structure across all domains of life.
EXPERIMENTAL PROCEDURES
1. Purification of Mpa
Purification of the full-length Mpa was as described previously (Darwin et al., 2005).
2. Purification of Mpa-ID and Mpa_N97Δ, and the triple mutant forms of Mpa and Mpa-ID
For all cloning, Pfu Ultra (Stratagene) was used for the polymerase chain reactions and the amplified products were cloned into pET24b(+) (Novagen) using enzymes from New England Biolabs. The inserts were sequenced by GENEWIZ (South Plainfield, NJ). Plasmids were transformed into DH5a (Gibco, BRL) and ER2566 (Chong and Garcia, 1994) for expression. Bacteria producing various Mpa constructs were grown in Luria-Bertani medium. Gene expression was induced by IPTG overnight at 20°C. The supernatant of the lysed bacteria was purifed by a 5 ml Qiagen Ni2+ affinity column. The imidazole eluate was buffer exchanged to 1× PBS and treated by thrombin for His tag removal. A second Ni2+ affinity chromatography was performed and the flow-through was collected and further purified by the Resource Q ion exchange column and the Superdex-200 gel filtration column. The purified proteins were dialyzed in buffer (10 mM Tris, pH 8.0, 10 mM NaCl, 1 mM β-ME), and concentrated with a Vivaspin tube (WMCO 10 kD). The Blue Native Gel (4~15% gradient Ready Gel) was purchased from Bio-Rad, run with the standard Tris-HCl Gel method, and stained by Coomassie blue. See Supplementary information for details.
3. Mtb Wild Type 20S Proteasome and the Open Gate 20S Mutant Expression and Purification
The wild type and the open gate 20S are expressed and purified as described (Lin et al., 2006).
4. Electron Microscopy of Mpa-20SOG complex
The purified Mpa ATPases and proteasome samples were mixed at a molar ratio of 4:1, in a buffer containing ATPγS, incubated on ice briefly and then used directly for negative stain EM grids preparation. Microscopy was carried out in a JEOL-1200EX TEM. See Supplementary Methods.
5. 3D Crystallization of Mpa-ID and Diffraction Data Collection
The protein was prone to aggregation, so the detergent cyclohexylpropanoyl-N-hydroxyethylglucamide (C-HEGA-9) was essential for crystallization. The hanging drop vapor diffusion method was used for crystallization of Mpa-ID. Briefly, a 2-μl droplet of Mpa-ID at 10.0 mg/ml in 10 mM Tris, pH 8.0, 10 mM NaCl, 1 mM β-ME was mixed with 2 μl of reservoir solution consisting of 0.1 M MES, pH 6.0, 50 mM MgCl2, 50 mM NaCl, 20% PEG-2000, 108 mM C-HEGA-9. We screened an array of heavy atom derivatives by native gel electrophoresis of the purified Mpa-ID, and found that mercury acetate (Hg(CH3CO2)2) induced a clear upward shift of the protein on the native gel, so Hg(CH3CO2)2 was chosen for derivatizing the crystals. The cryoprotectants used for flash freezing heavy atom derivatized crystals were 10% (v/v) glycerol and 10% (v/v) ethylene glycerol. Crystal screening and dataset collection were carried out at X25 and X29 beam lines at NSLS, Brookhaven National Laboratory. See Supplementary Experimental Procedures for more details.
6. Data Reduction and Structure Determination
7. Mpa mutant analysis in Mtb
Mutations were introduced into pMV-mpa using overlap extension (“sewing”) PCR or QuikChange Multi Site-Directed Mutagenesis kit (Statagene). Plasmids were introduced into mpa null mutant Mtb as described elsewhere (Darwin, 2003). Equivalent Mtb cell numbers, determined by culture OD580, were harvested for each strain, and washed in 0.05% Tween 80 in phosphate buffered saline (PBS). Bacteria were resuspended in 250 μl of lysis buffer (100 mM Tris-Cl, 100 mM KCl, 1 mM EDTA 5 mM MgCl2 pH 8) and transferred to bead beating tubes with 100–200 μl of zirconia silica beads (BioSpec Products, Inc). Each tube was bead beaten in a BioSpec Mini Bead Beater for 30 s, 3 times (tubes were chilled on ice between beatings). Lysates were not filtered due to significant loss of Mpa from the samples during filtration. Samples were boiled for 5 m prior to separation by 10% SDS-PAGE. For loading controls, immunoblots were stripped and incubated with anti-DlaT. SuperSignal West Pico or Femto Chemiluminescent Substrate (ThermoScientific) was used for detection.
Supplementary Material
Acknowledgments
We thank Drs. A. Héroux and S. Eswaramoorthy for their expert advice on X-ray crystallography. We thank staff of Beam lines X25 and X29 at the National Synchrotron Light Source, Brookhaven National Laboratory for technical assistance in data collection. This work is supported by NIH grant AI070285 and Brookhaven National Laboratory LDRD grant 06-60 to H.L., and by NIH grants AI065437 and HL092774 to K.H.D., and AI064768 to C.N.
Footnotes
ACCESSION NUMBER
The Mpa-ID X-ray coordinates have been deposited in the Protein Data Bank (accession code 3FP9).
Supplemental Data include Supplementary Experimental Procedures, Supplemental References, four figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexandrescu AT, Gittis AG, Abeygunawardana C, Shortle D. NMR structure of a stable “OB-fold” sub-domain isolated from staphylococcal nuclease. J Mol Biol. 1995;250:134–143. doi: 10.1006/jmbi.1995.0365. [DOI] [PubMed] [Google Scholar]
- Alexandrescu AT, Jaravine VA, Dames SA, Lamour FP. NMR hydrogen exchange of the OB-fold protein LysN as a function of denaturant: the most conserved elements of structure are the most stable to unfolding. J Mol Biol. 1999;289:1041–1054. doi: 10.1006/jmbi.1999.2813. [DOI] [PubMed] [Google Scholar]
- Baker TA, Sauer RT. ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem Sci. 2006;31:647–653. doi: 10.1016/j.tibs.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaroudj N, Zwickl P, Seemuller E, Baumeister W, Goldberg AL. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol Cell. 2003;11:69–78. doi: 10.1016/s1097-2765(02)00775-x. [DOI] [PubMed] [Google Scholar]
- Borissenko L, Groll M. 20S proteasome and its inhibitors: crystallographic knowledge for drug development. Chem Rev. 2007;107:687–717. doi: 10.1021/cr0502504. [DOI] [PubMed] [Google Scholar]
- Butler SM, Festa RA, Pearce MJ, Darwin KH. Self-compartmentalized bacterial proteases and pathogenesis. Mol Microbiol. 2006;60:553–562. doi: 10.1111/j.1365-2958.2006.05128.x. [DOI] [PubMed] [Google Scholar]
- Chong S, Garcia GA. A versatile and general prokaryotic expression vector, pLACT7. Biotechniques . 1994;17:686–688. 690–681. [PubMed] [Google Scholar]
- Dar AC, Sicheri F. X-ray crystal structure and functional analysis of vaccinia virus K3L reveals molecular determinants for PKR subversion and substrate recognition. Mol Cell. 2002;10:295–305. doi: 10.1016/s1097-2765(02)00590-7. [DOI] [PubMed] [Google Scholar]
- Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- Darwin KH, Lin G, Chen Z, Li H, Nathan CF. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol Microbiol. 2005;55:561–571. doi: 10.1111/j.1365-2958.2004.04403.x. [DOI] [PubMed] [Google Scholar]
- DeLaBarre B, Christianson JC, Kopito RR, Brunger AT. Central pore residues mediate the p97/VCP activity required for ERAD. Mol Cell. 2006;22:451–462. doi: 10.1016/j.molcel.2006.03.036. [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Hartmann MD, Habeck M, Ursinus A, Zwickl P, Martin J, Lupas AN, Zeth K. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol Cell. 2009;34:580–590. doi: 10.1016/j.molcel.2009.04.030. [DOI] [PubMed] [Google Scholar]
- Festa RA, Pearce MJ, Darwin KH. Characterization of the proteasome accessory factor (paf) operon in Mycobacterium tuberculosis. J Bacteriol. 2007;189:3044–3050. doi: 10.1128/JB.01597-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A, Whitby FG, Hill CP. The pore of activated 20S proteasomes has an ordered 7-fold symmetric conformation. Embo J. 2003;22:4356–4364. doi: 10.1093/emboj/cdg436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandotra S, Schnappinger D, Monteleone M, Hillen W, Ehrt S. In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat Med. 2007;13:1515–1520. doi: 10.1038/nm1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AL. Functions of the proteasome: from protein degradation and immune surveillance to cancer therapy. Biochem Soc Trans. 2007;35:12–17. doi: 10.1042/BST0350012. [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hu G, Lin G, Wang M, Dick L, Xu RM, Nathan C, Li H. Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate. Mol Microbiol. 2006;59:1417–1428. doi: 10.1111/j.1365-2958.2005.05036.x. [DOI] [PubMed] [Google Scholar]
- Jaravine VA, Rathgeb-Szabo K, Alexandrescu AT. Microscopic stability of cold shock protein A examined by NMR native state hydrogen exchange as a function of urea and trimethylamine N-oxide. Protein Sci. 2000;9:290–301. doi: 10.1110/ps.9.2.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloks CP, Spronk CA, Lasonder E, Hoffmann A, Vuister GW, Grzesiek S, Hilbers CW. The solution structure and DNA-binding properties of the cold-shock domain of the human Y-box protein YB-1. J Mol Biol. 2002;316:317–326. doi: 10.1006/jmbi.2001.5334. [DOI] [PubMed] [Google Scholar]
- Lin G, Hu G, Tsu C, Kunes YZ, Li H, Dick L, Parsons T, Li P, Chen Z, Zwickl P, et al. Mycobacterium tuberculosis prcBA genes encode a gated proteasome with broad oligopeptide specificity. Mol Microbiol. 2006;59:1405–1416. doi: 10.1111/j.1365-2958.2005.05035.x. [DOI] [PubMed] [Google Scholar]
- Lin G, Tsu C, Dick L, Zhou XK, Nathan C. Distinct specificities of Mycobacterium tuberculosis and mammalian proteasomes for N-acetyl tripeptide substrates. J Biol Chem. 2008;283:34423–34431. doi: 10.1074/jbc.M805324200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. Mol Cell. 2008a;29:441–450. doi: 10.1016/j.molcel.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat Struct Mol Biol. 2008b;15:1147–1151. doi: 10.1038/nsmb.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medalia N, Beer A, Zwickl P, Mihalache O, Beck M, Medalia O, Navon A. Architecture and molecular mechanism of PAN, the archaeal proteasome regulatory ATPase. J Biol Chem. 2009 doi: 10.1074/jbc.M809643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. Embo J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy I, Tamura T, Vanderleyden J, Baumeister W, De Mot R. The 20S proteasome of Streptomyces coelicolor. J Bacteriol. 1998;180:5448–5453. doi: 10.1128/jb.180.20.5448-5453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C, Gold B, Lin G, Stegman M, de Carvalho LP, Vandal O, Venugopal A, Bryk R. A philosophy of anti-infectives as a guide in the search for new drugs for tuberculosis. Tuberculosis (Edinb) 2008;88(Suppl 1):S25–33. doi: 10.1016/S1472-9792(08)70034-9. [DOI] [PubMed] [Google Scholar]
- Pearce MJ, Arora P, Festa RA, Butler-Wu SM, Gokhale RS, Darwin KH. Identification of substrates of the Mycobacterium tuberculosis proteasome. Embo J. 2006;25:5423–5432. doi: 10.1038/sj.emboj.7601405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Hanna J, Elsasser S, Finley D. Proteasome-associated proteins: regulation of a proteolytic machine. Biol Chem. 2005;386:725–737. doi: 10.1515/BC.2005.085. [DOI] [PubMed] [Google Scholar]
- Smith DM, Benaroudj N, Goldberg A. Proteasomes and their associated ATPases: a destructive combination. J Struct Biol. 2006;156:72–83. doi: 10.1016/j.jsb.2006.04.012. [DOI] [PubMed] [Google Scholar]
- Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DM, Kafri G, Cheng Y, Ng D, Walz T, Goldberg AL. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and traslocation of unfolded polypeptides. Molecular Cell. 2005;20:1–12. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Striebel F, Imkamp F, Sutter M, Steiner M, Mamedov A, Weber-Ban E. Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1597. [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Wolf S, Nagy I, Lupas A, Pfeifer G, Cejka Z, Muller SA, Engel A, De Mot R, Baumeister W. Characterization of ARC, a divergent member of the AAA ATPase family from Rhodococcus erythropolis. J Mol Biol. 1998;277:13–25. doi: 10.1006/jmbi.1997.1589. [DOI] [PubMed] [Google Scholar]
- Zhang C, Kim SH. A comprehensive analysis of the Greek key motifs in protein beta-barrels and beta-sandwiches. Proteins. 2000;40:409–419. doi: 10.1002/1097-0134(20000815)40:3<409::aid-prot60>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Zhang F, Hu M, Tian G, Zhang P, Finley D, Jeffrey PD, Shi Y. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009;34:473–484. doi: 10.1016/j.molcel.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Stoffels K, Wurzbacher S, Schoofs G, Pfeifer G, Banerjee T, Parret AH, Baumeister W, De Mot R, Zwickl P. The N-terminal coiled coil of the Rhodococcus erythropolis ARC AAA ATPase is neither necessary for oligomerization nor nucleotide hydrolysis. J Struct Biol. 2004;146:155–165. doi: 10.1016/j.jsb.2003.10.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.