Abstract
Background & Aims
MicroRNAs (miRNAs) are non-protein coding RNAs that regulate gene expression. The tumor suppressor miRNA let-7a has been reported to be inhibited post-transcriptionally in embryonic stem cells and in human cancers. The microtubule-associated kinase DCAMKL-1 is a putative intestinal stem cell marker that is expressed in ApcMin/+ adenomas. We investigated the role of DCAMKL-1 on expression of let-7a miRNA and the oncogene c-Myc and in tumorigenesis.
Methods
Immunostaining for DCAMKL-1 was performed on human tissue microarray slides. HCT116 and SW480 cells were transfected with DCAMKL-1 small interfering (si)RNA and analyzed for DCAMKL-1, c-Myc (using immunoblot and real-time RT-PCR) and pri-let-7a miRNA (using real-time RT-PCR) levels. A liposomal preparation of DCAMKL-1 siRNA was administered into HCT116 xenografts in nude mice and tumor volumes were measured. A luciferase reporter assay, with a plasmid containing a let-7a binding site at the 3’ UTR, was utilized to measure let-7a in cell lines. Cells were isolated from normal mouse intestine using DCAMKL-1 and fluorescence-activated cell sorting (FACS) and subjected to pri-let-7a miRNA analysis.
Results
Expression of DCAMKL-1 was increased in human colorectal cancers. siRNA-mediated blockade of DCAMKL-1 resulted in HCT116 tumor xenograft growth arrest, increased levels of pri-let-7a miRNA, a corresponding decrease in luciferase activity, as well as decreased expression of the oncogene c-Myc. DCAMKL-1+ cells isolated by FACS demonstrated a significant decrease in pri-let-7a miRNA, compared to more-differentiated cells.
Conclusion
DCAMKL-1 is a negative regulator of let-7a miRNA biogenesis in intestinal stem and colorectal cancer cells; it could represent a novel target for anti-cancer stem cell-based strategies.
Keywords: DCAMKL-1, Let-7a, microRNA, siRNA, c-Myc, HCT116, SW480, tumor xenograft, stem cell
Introduction
MicroRNAs (miRNAs) are small, non-coding RNAs that regulate gene expression in animal and plant systems 1, 2. miRNAs have emerged as important developmental regulators and control critical processes such as cell fate determination and cell death 3. There is increasing evidence that several miRNAs are mutated or poorly expressed in human cancers and may act as tumor suppressors or oncogenes 4, 5. Gene expression is regulated by miRNAs through complementary elements in the 3’ untranslated regions (3’UTRs) of their target messenger RNAs (mRNAs) 6. lethal-7 (let-7), a founding member of the miRNA family, is required for timing of cell fate determination in C. elegans 7. In humans, various let-7 genes have been reported to map to regions that are deleted in human cancers 8. In addition, let-7 is poorly expressed in lung cancers 9, suggesting that let-7 miRNAs may be tumor suppressors. In support of this, overexpression of let-7 inhibited cell growth of a lung cancer cell line in vitro 9.
Mature miRNAs are produced from primary miRNA transcripts (pri-miRNAs) through sequential cleavages by the Microprocessor complex, comprising the ribonuclease III Drosha component and the double-stranded RNA (dsRNA) binding protein DGCR8 10 and Dicer 11. This coordinated enzyme complex results in the release of pri-miRNA and mature miRNA species. Posttranscriptional control of miRNA expression has been reported to occur in a tissue-specific 12 and developmentally regulated fashion 13, 14. In mouse embryonic stem (ES) cells and in mouse embryonal carcinoma (EC) cells, the magnitude of the Microprocessor processing block is greatest for members of the let-7 family of miRNAs; although it is quite possible that the processing of all miRNAs may be regulated at the Microprocessor step 13, 14. It has been recently discovered that in many cancers, the miRNA profile is altered when compared to normal tissue 15. It is becoming increasingly recognized that most cancers have a stem-cell-like compartment that is responsible for inciting and sustaining tumorigenesis 15, 16. One might hypothesize that miRNA profiles are altered in cancer stem cells (CSCs) within a particular tumor. Moreover, it is quite possible that such alterations are key factors in the initiation of the CSC. Recent evidence suggests that several miRNAs may be responsible for maintaining stem-cell-like characteristics 17, 18.
Furthermore, miRNA profiling of human and mouse ES cells reveals high levels of miRNAs expression, previously associated with oncogenesis and cell-cycle control 19, 20. Moreover, lack of let-7 miRNA expression was observed as an indicator for “stemness” in epithelial progenitor cells. Recent studies have also demonstrated that let-7 expression is absent in certain tumor cell lines, and that re-introduction of let-7 into these cells causes differentiation and reduction in proliferation and tumor-forming ability 22–24. The regulatory mechanisms that control the maturation process of miRNA are unclear and the regulatory factors that control let-7 miRNA levels, particularly in epithelial stem/progenitor cells, are completely unknown. The study of epithelial stem cell biology has been hampered by the lack of reliable stem cell markers that distinctly define and distinguish between stem and progenitor cell populations. There has been an accelerated interest however, in defining these populations as it is becoming increasingly clear that many important diseases including cancers are likely driven by effects on stem and/or progenitor cells.
We have recently demonstrated that the novel putative intestinal stem cell marker DCAMKL-1, a microtubule associated kinase expressed in post mitotic neurons 21 and in the stomach 22, is expressed in the intestine, colon and ApcMin/+ adenomas 23. Given the importance of stem cells in mucosal regeneration and neoplasia, we sought to determine whether DCAMKL-1 played a functional role in tumorigenesis and whether these effects were mediated through regulation of let-7a miRNA.
Materials and Methods
Cell culture
HCT116, HCT116 p21−/− and SW480 human colon adenocarcinoma cell lines were obtained from the American Type Culture Collection (ATCC) and grown in Dulbecco’s modified eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 100 U/mL penicillin-streptomycin in a humidified chamber at 37°C with 5% CO2.
Silencer RNA
DCAMKL-1 siRNA (si-DCAMKL-1) sequence targeting the coding region of DCAMKL-1 (Accession # NM_004734) (GGGAGUGAGAACAAUCUACtt) and scrambled control siRNAs (si-Scr) not matching any of the human genes were obtained (Ambion Inc., Austin, TX) and transfected using Transfectol™ (Ambion Inc.).
Real-time reverse transcription-PCR analyses
Total RNA isolated either from cells or from human colon cancer cell tumor xenograft samples was subjected to reverse transcription with Superscript™ II RNase H - Reverse Transcriptase and random hexanucleotide primers (Invitrogen, Carlsbad, CA). The cDNA was subsequently used to perform Real-time PCR by SYBR chemistry (SYBR® Green I; Molecular Probes) for specific transcripts using gene specific primers and Jumpstart Taq DNA polymerase (Sigma-Aldrich, St. Louis, MO). The crossing threshold value assessed by Real-time PCR was noted for the transcripts and normalized with β-actin mRNA. The changes in mRNA were expressed as fold change relative to control with ± SEM value.
Human primers used are:
| β-actin: | Forward: 5’-GGTGATCCACATCTGCTGGAA-3’ |
| Reverse:5’-ATCATTGCTCCTCCTCAGGG-3’ | |
| DCAMKL-1: | Forward: 5’-AGTCTTCCGATTCCGAGTTGAG-3’ |
| Reverse: 5’-CAGCAACCAGGAATGTATTGGA-3’ | |
| c-Myc: | Forward: 5’-CACACATCAGCACAACTACGCA-3’ |
| Reverse: 5’-TTGACCCTCTTGGCAGCAG-3’ | |
| Mouse primers used are: | |
| DCAMKL-1: | Forward: 5’-CAGCCTGGACGAGCTGGTGG-3’ |
| Reverse: 5’-TGACCAGTTGGGGTTCACAT-3’ | |
miRNA analysis
Total miRNA was isolated using mirVana™ miRNA isolation kit (Ambion Inc.,). Total miRNA isolated either from cells or from human colon cancer cell tumor xenograft samples were subjected to reverse transcription with Superscript™ II RNase H - Reverse Transcriptase and random hexanucleotide primers (Invitrogen, Carlsbad, CA). The cDNA was subsequently used to perform Real-time PCR by SYBR chemistry (SYBR® Green I; Molecular Probes) for pri-let-7a transcript using specific primers and Jumpstart Taq DNA polymerase (Sigma-Aldrich, St. Louis, MO). The crossing threshold value assessed by Realtime PCR was noted for pri-let-7a miRNA and normalized with U6 pri-miRNA.
The changes in pri-miRNA were expressed as fold change relative to control with ± SEM value. Primers used are:
| pri-U6: | Forward: 5’-CTCGCTTCGGCAGCACA-3’ |
| Reverse: 5’-AACGCTTCACGAATTTGCGT-3’ | |
| pri-let-7a: | Forward: 5’-GAGGTAGTAGGTTGTATAGTTTAGAA-3’ |
| Reverse: 5’-AAAGCTAGGAGGCTGTACA-3’ |
Western Blot analysis
HCT116 and SW480 cells were cultured in a 6 well plates to 40% confluency and were transfected with si-DCAMKL-1 or si-Scr for 72 h. Cells or the tumor xenograft samples were lysed and the concentration of protein was determined by BCA protein assay kit (Pierce Biotechnology Inc., Rockford, IL). Forty µg of the protein was size separated in a 7.5–15% SDS polyacrylamide gel and transferred onto a nitrocellulose membrane with a semidry transfer apparatus (Amersham-Pharmacia, Piscataway, NJ). The membrane was blocked in 5% non-fat dry milk for 1 h and probed overnight with a rabbit anti-DCAMKL-1 antibody (Abcam Inc., Cambridge, MA) or with rabbit anti-c-Myc antibody (Santa Cruz). Subsequently, the membrane was incubated with anti-rabbit or anti-goat IgG horseradish peroxidase-conjugated antibodies (Amersham-Pharmacia) for 1 h at room temperature. The 82 kDa DCAMKL-1 and 49 kDa c-Myc proteins were detected using ECL™ Western Blotting detection reagents (Amersham-Pharmacia). Actin (43 kDa), used as loading control was identified using a goat polyclonal IgG (Santa Cruz Biotechnology Inc., Santa Cruz, CA).
Immunohistochemistry
Heat Induced Epitope Retrieval was performed on 4 µm formalin-fixed paraffin-embedded sections utilizing a pressurized Decloaking Chamber (Biocare Medical) in citrate buffer (pH 6.0) at 99°C for 18 min. (a) Brightfield: Slides were incubated in 3% hydrogen peroxide, then normal serum and BSA at room temperature for 20 min. After incubation with primary antibody [DCAMKL-1 C-terminal (Abcam), anti-c-Myc (Santa Cruz Biotechnologies), L-FABP (Santa Cruz Biotechnologies)], the slides were incubated in peroxidase-conjugated EnVision™+ polymer detection kit (DAKO). Slides were developed with Diaminobenzidine (Sigma). (b) Fluorescence: Slides were first incubated in Image-iT FX signal enhancer (Invitrogen), followed by normal serum and BSA at room temperature for 20 min. After incubation with primary antibody [L-FABP (Santa Cruz Biotechnologies)], slides were incubated in appropriate Alexa Fluor® conjugated secondary [488 (green)].
Microscopic Examination
Slides were examined utilizing the Nikon 80i microscope and DXM1200C camera for brightfield. Fluorescent images were taken with PlanFluoro objectives, utilizing CoolSnap ES2 camera (Photometrics). Images were captured utilizing NIS-Elements software (Nikon).
Stem cell isolation
Based on protocols developed in intestinal stem cell biology 24, 25, we isolated stem cells from mouse intestine. The intestine was chopped into small strips, washed and incubated with 1 mM DTT (Sigma) for 30 min at room temperature. It was further incubated with 30 mmol/L EDTA (Sigma) for 10 min at 37° C. The strips were shaken vigorously in fresh HBSS (Cellgro) and filtered through 400 µm mesh (Spectrum Labs) to separate the detached intestinal crypt epithelial cells from the tissue. The filtrate was passed through 80 µm mesh (BD Falcon) to retain the crypts and washed. The crypts were digested at 37°C to create a single cell suspension.
FACS
The cells isolated from mouse intestine were incubated with 1:100 dilution of Alexa Fluor® 568 (Invitrogen) conjugated DCAMKL-1 antibody (Abcam) for 30 min. The cells were washed twice with HBSS containing 10% serum and sorted using Influx-V cell sorter (Cytopeia). DCAMKL-1 positively and negatively sorted cells were collected and subjected to total mRNA and miRNA isolation. mRNA was reverse transcribed and subjected to real-time RT-PCR for DCAMKL-1. Total miRNA was subjected to real-time RT-PCR for pri-let-7a miRNA.
Xenograft tumor model. (a) Liposomal preparation
siRNA was administered into the xenografts after incorporation into 1, 2-Dioleoyl-sn-Glycero-3-Phosphocholine (DOPC) (Avanti Polar Lipids, Alabaster, AL). DOPC and siRNA were mixed in the presence of excess tertiary butanol at a ratio of 1:10 (w/w) (siRNA/DOPC). Tween 20 (Sigma-Aldrich) was added to the mixture at a ratio of 1:19 (Tween 20 to siRNA/DOPC). The mixture was vortexed and frozen in an acetone/dry ice bath and lyophilized. Before administration, the siRNA preparation was reconstituted in 0.9% sterile saline and injected at a dose of 50 µl (5 µM) per injection. (b) Tumor therapy: Female athymic nude mice (NCr-nu) were purchased from the National Cancer Institute-Frederick Cancer Research and Development Center (Frederick, MD) and housed in specific pathogen-free conditions. They were cared for in accordance with guidelines set forth by the American Association for Accreditation of Laboratory Animal Care and the USPHS “Policy on Human Care and Use of Laboratory Animals,” and all studies were approved and supervised by the Institutional Animal Care and Use Committee. HCT116 cells (6 × 106) were injected subcutaneously into the flanks of 4–6 week-old female athymic nude mice (5 mice per group). Tumors were measured with calipers and the volume was calculated as (length X width2) X 0.5. The tumors reached 1000 mm3 after 15 days of injection of cells. These tumors were injected with 50 µl (5 µM) of siRNA preparation on every third day from day 15 for a total of 5 doses.
Luciferase reporter gene assay
pLet7a–Luc reporter vector contains a let-7a miRNA specific binding site at the 3’UTR of the firefly (Photinus pyralis) luciferase gene obtained from Signosis Inc (Supplementary Figure 2). HCT116 and SW480 cells were transfected with the pLet7a–Luc reporter vector, Renilla luciferase expressing plasmid pRL-TK (Promega) along with DCAMKL-1 or scrambled siRNA using Transfectol™ (Ambion Inc.). Luciferase activity was determined as per the manufacturer’s instructions (Dual-Luciferase Reporter Assay System; Promega) using a Monolight 2010 luminometer (Analytical Luminescence Laboratory, San Diego, CA) as described earlier 27. The activity, normalized to Renilla Luciferase activity, is presented as relative luciferase units relative to control with ± SEM values. Assays were performed in triplicate wells and experiments were repeated 3 times.
Statistical analysis
All the experiments were performed in triplicate. The data was analyzed by Student’s t-test. Where indicated, the data is presented as mean ± SEM. A p value of <0.01 was considered statistically significant.
Results
DCAMKL-1 is overexpressed in cancer
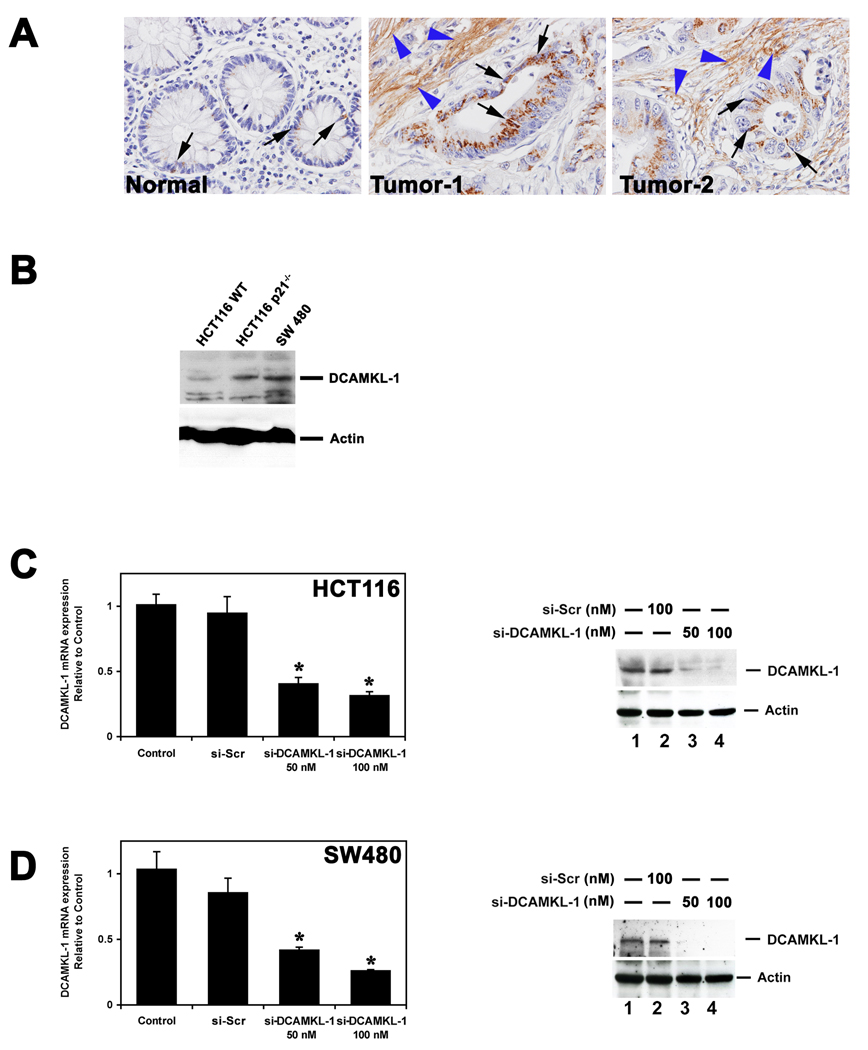
To determine whether DCAMKL-1 was expressed in human colorectal cancers, we performed immunohistochemical analysis on human cancer tissue microarrays (Tissue Array Network and National Cancer Institute – Tissue Array Research Program). Staining revealed increased DCAMKL-1 protein (Figure 1A; brown – indicated by black arrows) in human colorectal cancers specimens, compared to normal colonic mucosa. In tumors, the staining pattern was particularly impressive in the stroma surrounding malignant crypts (brown – indicated by blue arrow heads). Representative images of normal mucosa and two different human colorectal cancer specimens are shown in figure 1A. Similarly, we observed DCAMKL-1 expression in a variety of human colon cancer cell lines (Figure 1B). HCT116 and SW480 cells were transfected with DCAMKL-1 and scrambled siRNA; then total RNA was isolated and subjected to real-time RT-PCR. We noted a >70% reduction in DCAMKL-1 mRNA expression in DCAMKL-1 siRNA (si-DCAMKL-1) treated cells (Figure 1C and D). A reduction in DCAMKL-1 protein was also observed following si-DCAMKL-1 transfection (Figure 1C and D). Scrambled siRNA (si-Scr) did not affect the expression of DCAMKL-1 mRNA or protein (Figure 1C and D).
Figure 1.
DCAMKL-1 is overexpressed in colorectal cancer. (A) Immunohistochemistry for DCAMKL-1 (brown) in normal (left panel) and two different colon cancer tissues (middle and right panels). Black arrow indicates representative epithelial cells positive for DCAMKL-1. Blue arrow head indicates the presence of DCAMKL-1 in the stromal compartment. (B) Western blot demonstrating the expression of DCAMKL-1 in three different colon cancer cell lines. Actin serves as control. (C) DCAMKL-1 specific siRNA (si-DCAMKL-1) decreases DCAMKL-1 mRNA (left panel) and protein expression (right panel) in HCT116 colon cancer cells compared to controls. (D) Similar decrease in DCAMKL-1 mRNA (left panel) and protein (right panel) observed following si-DCAMKL-1 transfection in SW480 colon cancer cells. For C and D, values in the bar graphs are given as average ± SEM and * denote statistically significant differences (*p<0.01) compared to control. All the experiments were performed in triplicates and were repeated 3 times.
siRNA mediated knockdown of DCAMKL-1 leads to tumor growth arrest
Given the increased DCAMKL-1 expression in human colorectal tumors (Figure 1A) and in ApcMin/+ adenomas 23, we wanted to determine its role in tumor progression. Tumor xenografts were generated by injecting HCT116 cells (6×106) subcutaneously into the flanks of athymic nude mice. After 15 days, si-DCAMKL-1 and si-Scr were injected into the xenografts. Tumor volumes were measured using calipers at various time points before sacrifice and weights were determined after sacrifice 26, 27. Administration of si-DCAMKL-1 resulted in a statistically significant reduction (p <0.01) in tumor size compared to the control or the si-Scr treated tumors (Figure 2A and B). Thus inhibition of DCAMKL-1 arrested HCT116 tumor xenograft growth. Total RNA isolated from these tumors was subjected to real-time RT-PCR and demonstrated a significant downregulation (55%) (p <0.01) of DCAMKL-1 mRNA expression in the si-DCAMKL-1-treated tumors compared to control and si-Scr treated tumors (Figure 2C). This downregulation was associated with reduced expression of DCAMKL-1 protein in those tumors by Western blot analyses (Figure 2D).
Figure 2.
DCAMKL-1 is essential for tumor growth. (A) HCT116 cells were injected into the flanks of athymic nude mice (n=5 per group) to generate tumors. At day 15 siRNAs (si-DCAMKL-1 and si-Scr) were injected directly into the tumors and followed by injections every third day (inset). After 5 injections, tumors were excised at day 28 and are represented above. Tumor sizes with standard error are shown from data collected at the time of every injection. (B) si- DCAMKL-1 treatment resulted in significantly decreased tumor weight when compared to controls. (C) The expression of DCAMKL-1 mRNA in the tumors quantitated by real-time RT-PCR. (D) Western blot analysis for DCAMKL-1 was performed on tumors samples as indicated. For A – C, values are given as average ± SEM and * denote statistically significant differences (*p<0.01) compared to control.
Knockdown of DCAMKL-1 induces pri-let-7a miRNA
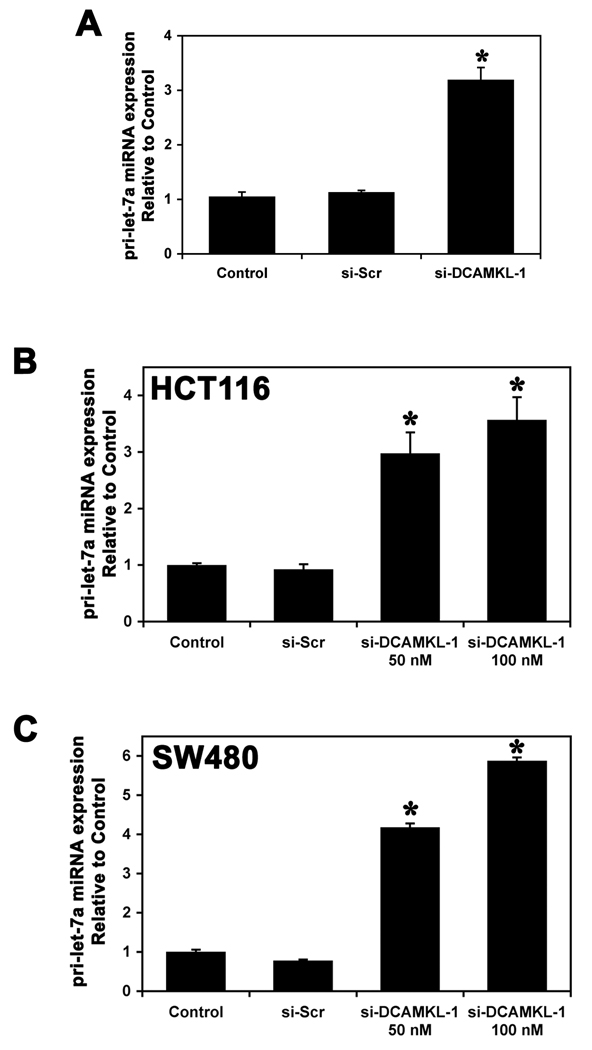
To determine the role of DCAMKL-1 mediated regulation of pri-let-7a miRNA, control and siRNA treated HCT116 tumor xenografts were analyzed for pri-miRNA expression by real-time RT-PCR. Compared to control and si-Scr treated tumors, there was a >3-fold increase in pri-let-7a miRNA expression in DCAMKL-1 siRNA treated tumors (Figure 3A). We next analyzed the effects of siRNA-mediated knockdown of DCAMKL-1 on pri-let-7a miRNA expression in HCT116 and SW480 cells. Real-time RT-PCR analysis revealed a 4-fold increase in pri-let-7a miRNA, compared to controls (Figure 3B and C). These data suggest that DCAMKL-1 negatively regulates pri-let-7a miRNA in human colon cancer cells.
Figure 3.
Knockdown of DCAMKL-1 induces pri-let-7a miRNA. (A) Quantitative real-time RT-PCR analysis for pri-let-7a miRNA in tumor xenografts. siRNA mediated knockdown of DCAMKL-1 results in increased expression of pri-let-7a miRNA. (B) si-DCAMKL-1 treated HCT116 cells demonstrate increased expression of pri-let-7a miRNA. (C) Similar induction of pri-let-7a miRNA was observed in SW480 cells. For A – C, values are given as average ± SEM and * denote statistically significant differences *p<0.01) compared to control.
DCAMKL-1 negatively regulates let-7a miRNA
As stated earlier, lack of let-7 miRNA is an indicator of “stemness” in epithelial progenitor cells 28–30. To determine whether pri-let-7a miRNA was expressed in stem cells, we utilized FACS based sorting to isolate DCAMKL-1 positive and negative cells, which were analyzed for pri-let-7a miRNA. The antibody used for FACS was directed against the c-terminal extracellular domain of DCAMKL-1 31 32 and conjugated to the Alexa Fluor® 568 fluorochrome. Following FACS, both sorted cell populations were examined by fluorescence microscopy. The positively sorted cells demonstrated the presence of DCAMKL-1 antibody staining, whereas the negatively sorted cells did not (Supplementary Figure 1A and B). Furthermore, DCAMKL-1 positive cells did not express L-type fatty acid binding protein (L-FABP), a marker of enterocyte lineage known to be expressed in differentiated intestinal epithelia 35, 36, indicating a less differentiated state (Supplementary Figure 1C and E). In contrast, L-FABP was found to be expressed in DCAMKL-1 negative cells (Supplementary Figure 1D and F), indicating that these cells are more differentiated compared to DCAMKL-1 positive cells.
Total miRNA isolated from DCAMKL-1 positive and DCAMKL-1 negative cells were subjected to pri-let-7a miRNA expression by real-time RT-PCR and normalized using pri-U6 miRNA. A 65% reduction in pri-let-7a miRNA was observed in DCAMKL-1 positive sorted “stem” cells relative to DCAMKL-1 negative cells (Figure 4A). To confirm sorting specificity, total RNA isolated from the cells was subjected to real-time RT-PCR for DCAMKL-1 mRNA expression (Figure 4B). These data suggests that DCAMKL-1 negatively regulates pri-let-7a miRNA in putative intestinal stem/progenitor cells.
Figure 4.
DCAMKL-1 inhibits let-7a miRNA. (A) Intestinal stem cells (DCAMKL-1 +) isolated from normal mouse intestine demonstrate decreased pri-let-7a compared to more differentiated cells (DCAMKL-1 -). (B) Real-time RT-PCR data demonstrate an increased expression of DCAMKL-1 mRNA in DCAMKL-1+ sorted stem cells compared to more differentiated (DCAMKL-1 -) cells. siRNA mediated knockdown of DCAMKL-1 decreases luciferase activity (Relative Luciferase Units – RLU) following transfection with plasmid encoding luciferase containing let-7a binding site in HCT116 (C) and SW480 cells (D). For A – D,values are given as average ± SEM and * denote statistically significant differences (*p<0.01) compared to control.
To determine quantitatively the effect of siRNA-mediated knockdown of DCAMKL-1 on let-7a miRNA, we performed a luciferase reporter gene assay. HCT116 and SW480 cells were transfected with a plasmid containing firefly luciferase gene with a complementary let-7a binding site at the 3’UTR (Supplementary Figure 2). A dose dependent reduction in luciferase activity was observed following the knockdown of DCAMKL-1 (Figure 4C and D). This suggests that DCAMKL-1 may be a posttranscriptional regulator of let-7a miRNA downstream targets. However we cannot rule out other alternative mechanisms for DCAMKL-1 such as acting as a transcriptional regulator of let-7a or as a posttranscriptional regulator of let-7a maturation.
Knockdown of DCAMKL-1 inhibits c-Myc
HCT116 tumor xenografts were evaluated for expression of the let-7a miRNA downstream oncogenic target c-Myc, following siRNA-mediated knockdown of DCAMKL-1 as described earlier. We observed a 45% reduction in c-Myc mRNA in si-DCAMKL-1 treated tumors compared to controls (Figure 5A). An even more striking reduction of c-Myc protein was seen by Western blot and immunohistochemical analyses (Figure 5B and C) of siDCAMKL-1 treated tumors. A significant reduction in c-Myc mRNA and protein was also observed in siDCAMKL-1 treated HCT116 (Figure 5D and E) and SW480 cells (Figure 5D and F). These data suggest that knockdown of DCAMKL-1 results in a reduced expression of c-Myc by a let-7a dependent mechanism.
Figure 5.
Downregulation of DCAMKL-1 results in decreased expression of a let-7a downstream target. A decreased expression of c-Myc mRNA (A) and protein (B) was observed in HCT116 tumor xenografts following the knockdown of DCAMKL-1. (C) Decreased c-Myc expression (brown) in si-DCAMKL-1 treated tumors compared to controls by immunohistochemica l analysis. siRNA mediated knockdown of DCAMKL-1 results in decreased c-Myc mRNA (D) and protein (E) in HCT116 cells. (D and F) Similar decrease was observed in SW480 cells. For bar graph in A and D, values are given as average ± SEM and * denote statistically significant differences (*p<0.01) compared to control.
Discussion
miRNAs play important gene-regulatory roles by pairing to the mRNAs of protein-coding genes to direct their posttranscriptional repression 37. The involvement of miRNAs in human cancer has been recently described 15; with several reports indicating that miRNAs might be used as future diagnostic and therapeutic targets 33. Furthermore, characteristic miRNA expression signatures in various cancers that can profoundly affect cancer cell behavior has been reported 15. miRNAs have been shown to play an important role in regulating stem cell self-renewal and differentiation by repressing the translation of selected mRNAs in stem cells and differentiating daughter cells. Let-7a is a tumor suppressor miRNA that is blocked posttranscriptionally in ES cells and in several human cancers 14, 15, 19. The regulatory factors that control miRNA expression, maturation and function in adult stem cells and cancers are just beginning to be explored.
This study demonstrates that the novel putative intestinal stem cell marker DCAMKL-1 is a negative regulator of let-7a miRNA expression/function. Here we demonstrate that DCAMKL-1 expression is increased in human colorectal cancers compared to normal uninvolved tissues. While we have previously shown DCAMKL-1 expression in ApcMin/+ adenomas, to our knowledge, this is the first demonstration of DCAMKL-1 in human colorectal cancer. In addition to the increased epithelial expression of DCAMKL-1 seen within the colorectal tumors examined, strong staining was also observed in the stroma surrounding malignant crypts. Given the importance of epithelial–mesenchymal cell interactions in cancer 34 and the role of the niche in epithelial stem cell fate 35, we speculate that stromal DCAMKL-1 may participate in tumor progression.
Using a tumor xenograft model generated from HCT116 human colorectal cancer cells, we demonstrate near complete tumor growth arrest following siRNA-mediated knockdown of DCAMKL-1. These data strongly implicate a functional role for DCAMKL-1 in the regulation of tumor growth. Given the potential roles of let-7a miRNA in the regulation of gene expression in stem cells and cancer, we next assayed the tumor xenografts for pri-let-7a miRNA expression. We found a significant increase in pri-let-7a miRNA in the tumors following siRNA-mediated inhibition of DCAMKL-1. These data confirm that pri-let-7a miRNA is indeed a tumor suppressor miRNA, which is regulated by DCAMKL-1 in colorectal cancer cells.
Cellular transformation and tumorigenesis are driven by activation of oncogenes and/or inactivation of tumor suppressors. Oncogenic c-Myc overexpression is observed in many cancers along with enhanced cell proliferation 36. Furthermore, transcripts encoding both c-Myc and Kras are known to contain target sites for the let-7 miRNA in their 3’UTR 37. Such findings led us to speculate that DCAMKL-1 may affect c-Myc expression in colon cancer via a let-7a dependent mechanism. Indeed, we found a 45% reduction in c-Myc mRNA and significant decrease in protein levels in the tumors following the inhibition of DCAMKL-1. These findings were confirmed in vitro in human colorectal cancer cell lines where knockdown of DCAMKL-1 resulted in increased pri-let-7a miRNA, which corresponded with a significant reduction of c-Myc. These data taken together strongly suggests that DCAMKL-1 negatively regulates the tumor suppressor miRNA let-7a resulting in reduced expression of its downstream target oncogene c-Myc.
In order to determine the effects of DCAMKL-1 knockdown on let-7a miRNA-dependent gene silencing of let-7a downstream targets, we performed a luciferase gene reporter assay containing a specific let-7a miRNA binding site at its 3’UTR. We found a significant dose-dependent reduction in luciferase activity following knockdown of DCAMKL-1. This provides a potential explanation and mechanism where inhibition of DCAMKL-1 results in decreased c-Myc and possibly other let-7a downstream targets.
In this report, we have demonstrated that DCAMKL-1, a protein expressed in both normal stem cells and in cancer, likely promotes tumorigenesis through the regulation of pri-let-7a miRNA and c-Myc. The presence of let-7a binding sites in the c-Myc 3’UTR leads us to speculate that DCAMKL-1 is regulating c-Myc posttranscriptionally. However, we cannot rule out other alternatives, such as direct transcriptional regulation. Nevertheless, the knockdown of DCAMKL-1 resulted in a marked reduction in c-Myc mRNA and protein in vitro and in vivo. Moreover, several other oncogenes contain let-7a binding sites in their 3’UTRs, thus it is quite possible that DCAMKL-1 may have similar effects on other oncogenic targets including Kras.
miRNAs are known to contribute to the preservation of ‘stemness’ and associated with self-renewal and differentiation in ES cells 38. Previous studies have also shown an overall reduction in miRNA expression in embryonic and tissue stem cells 39. We analyzed intestinal epithelial cells following FACS based sorting using DCAMKL-1 for pri-let-7a miRNA. A marked reduction in pri-let-7a miRNA was observed in DCAMKL-1 positively sorted “stem” cells relative to DCAMKL-1 negative cells. These data suggests that intestinal stem cells, like ES cells, express low levels of let-7a.
We cannot rule out the possibility that DCAMKL-1, alone or in concert with other factors, may block other pri-miRNAs in different physiological contexts. Furthermore, additional factors may be discovered that regulate miRNA processing. Future studies targeting the putative stem cell marker DCAMKL-1 may reveal the mechanisms by which miRNA processing contributes to oncogenesis. The findings presented in this report suggest that regulation of miRNAs may represent an exciting new strategy to combat tumorigenesis, particularly in cancers originating from cancer stem cells.
Supplementary Material
DCAMKL-1 positive cells are less differentiated. A representative image of Alexa Fluor® 568 conjugated DCAMKL-1 positively sorted cells (A) (red) and negatively sorted cells (B) following FACS. (C) Brightfield image of L-FABP immunostaining. DCAMKL-1 positive cells do not express L-FABP. (D) DCAMKL-1 negative cells express L-FABP (brown – arrows). (E) Fluorescent image of L-FABP immunostaining. DCAMKL-1 positive cells do not express L-FABP. (F) L-FABP was found in DCAMKL-1 negative cells (green).
Nuclei in A, B, E and F are stained blue with Hoechst 33342 DNA dye post-sorting.
Map of pLet7a–Luc Reporter Vector (LR-0037) (Signosis, Inc. CA) demonstrating the presence of the let7a binding site at the 3’UTR of Luciferase gene.
Acknowledgements
This work was supported by National Institute of Health Grants DK-065887, DK-002822 and Veteran Administration Merit award to C.W.H. We thank Dr. Michael Bronze, University of Oklahoma Health Sciences Center for his support. We also thank Dr. Lurdes Queimado, University of Oklahoma Health Sciences Center for her valuable input, Nguyet Hoang, Oklahoma University Advanced Immunohistochemistry & Morphology Core and Jim Henthorn of The Flow and Image Cytometry Core Laboratory.
Grant Support: National Institute of Health Grants DK-066161, DK-002822 and Veteran Administration Merit award to CWH
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
All the authors have no conflicts of interest.
References
- 1.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 2.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.McManus MT. MicroRNAs and cancer. Semin Cancer Biol. 2003;13:253–258. doi: 10.1016/s1044-579x(03)00038-5. [DOI] [PubMed] [Google Scholar]
- 5.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 6.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3[prime] UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reinhart B. The 21 nucleotide let-7 RNA regulates C. elegans developmental timing. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 8.Calin GA. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takamizawa J. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 10.Gregory RI. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 11.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. Rna. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 16.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 19.Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, Kim VN, Kim KS. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 20.Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer’s role in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin PT, Gleeson JG, Corbo JC, Flanagan L, Walsh CA. DCAMKL1 encodes a protein kinase with homology to doublecortin that regulates microtubule polymerization. J Neurosci. 2000;20:9152–9161. doi: 10.1523/JNEUROSCI.20-24-09152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giannakis M, Stappenbeck TS, Mills JC, Leip DG, Lovett M, Clifton SW, Ippolito JE, Glasscock JI, Arumugam M, Brent MR, Gordon JI. Molecular properties of adult mouse gastric and intestinal epithelial progenitors in their niches. J Biol Chem. 2006;281:11292–11300. doi: 10.1074/jbc.M512118200. [DOI] [PubMed] [Google Scholar]
- 23.May R, Riehl TE, Hunt C, Sureban SM, Anant S, Houchen CW. Identification of a novel putative gastrointestinal stem cell and adenoma stem cell marker, doublecortin and CaM kinase-like-1, following radiation injury and in adenomatous polyposis coli/multiple intestinal neoplasia mice. Stem Cells. 2008;26:630–637. doi: 10.1634/stemcells.2007-0621. [DOI] [PubMed] [Google Scholar]
- 24.Dekaney CM, Rodriguez JM, Graul MC, Henning SJ. Isolation and characterization of a putative intestinal stem cell fraction from mouse jejunum. Gastroenterology. 2005;129:1567–1580. doi: 10.1053/j.gastro.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 25.Grossmann J, Walther K, Artinger M, Kiessling S, Steinkamp M, Schmautz WK, Stadler F, Bataille F, Schultz M, Scholmerich J, Rogler G. Progress on isolation and short-term ex-vivo culture of highly purified non-apoptotic human intestinal epithelial cells (IEC) Eur J Cell Biol. 2003;82:262–270. doi: 10.1078/0171-9335-00312. [DOI] [PubMed] [Google Scholar]
- 26.Sureban SM, May R, George RJ, Dieckgraefe BK, McLeod HL, Ramalingam S, Bishnupuri KS, Natarajan G, Anant S, Houchen CW. Knockdown of RNA Binding Protein Musashi-1 Leads to Tumor Regression In Vivo. Gastroenterology. 2008;134:1448–1458. doi: 10.1053/j.gastro.2008.02.057. e2. [DOI] [PubMed] [Google Scholar]
- 27.Sureban SM, Ramalingam S, Natarajan G, May R, Subramaniam D, Bishnupuri KS, Morrison AR, Dieckgraefe BK, Brackett DJ, Postier RG, Houchen CW, Anant S. Translation regulatory factor RBM3 is a proto-oncogene that prevents mitotic catastrophe. Oncogene. 2008;27:4544–4556. doi: 10.1038/onc.2008.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ibarra I, Erlich Y, Muthuswamy SK, Sachidanandam R, Hannon GJ. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 2007;21:3238–3243. doi: 10.1101/gad.1616307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 30.Stadler BM, Ruohola-Baker H. Small RNAs: keeping stem cells in line. Cell. 2008;132:563–566. doi: 10.1016/j.cell.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sossey-Alaoui K, Srivastava AK. DCAMKL1, a brain-specific transmembrane protein on 13q12.3 that is similar to doublecortin (DCX) Genomics. 1999;56:121–126. doi: 10.1006/geno.1998.5718. [DOI] [PubMed] [Google Scholar]
- 32.Kim MH, Cierpicki T, Derewenda U, Krowarsch D, Feng Y, Devedjiev Y, Dauter Z, Walsh CA, Otlewski J, Bushweller JH, Derewenda ZS. The DCX-domain tandems of doublecortin and doublecortin-like kinase. Nat Struct Biol. 2003;10:324–333. doi: 10.1038/nsb918. [DOI] [PubMed] [Google Scholar]
- 33.Tricoli JV, Jacobson JW. MicroRNA: Potential for Cancer Detection, Diagnosis, and Prognosis. Cancer Res. 2007;67:4553–4555. doi: 10.1158/0008-5472.CAN-07-0563. [DOI] [PubMed] [Google Scholar]
- 34.Arias AM. Epithelial mesenchymal interactions in cancer and development. Cell. 2001;105:425–431. doi: 10.1016/s0092-8674(01)00365-8. [DOI] [PubMed] [Google Scholar]
- 35.Rizvi AZ, Wong MH. Epithelial stem cells and their niche: there’s no place like home. Stem Cells. 2005;23:150–165. doi: 10.1634/stemcells.2004-0096. [DOI] [PubMed] [Google Scholar]
- 36.Smith DR, Goh HS. Overexpression of the c-myc proto-oncogene in colorectal carcinoma is associated with a reduced mortality that is abrogated by point mutation of the p53 tumor suppressor gene. Clin Cancer Res. 1996;2:1049–1053. [PubMed] [Google Scholar]
- 37.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 38.Shcherbata HR, Hatfield S, Ward EJ, Reynolds S, Fischer KA, Ruohola-Baker H. The MicroRNA pathway plays a regulatory role in stem cell division. Cell Cycle. 2006;5:172–175. doi: 10.4161/cc.5.2.2343. [DOI] [PubMed] [Google Scholar]
- 39.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DCAMKL-1 positive cells are less differentiated. A representative image of Alexa Fluor® 568 conjugated DCAMKL-1 positively sorted cells (A) (red) and negatively sorted cells (B) following FACS. (C) Brightfield image of L-FABP immunostaining. DCAMKL-1 positive cells do not express L-FABP. (D) DCAMKL-1 negative cells express L-FABP (brown – arrows). (E) Fluorescent image of L-FABP immunostaining. DCAMKL-1 positive cells do not express L-FABP. (F) L-FABP was found in DCAMKL-1 negative cells (green).
Nuclei in A, B, E and F are stained blue with Hoechst 33342 DNA dye post-sorting.
Map of pLet7a–Luc Reporter Vector (LR-0037) (Signosis, Inc. CA) demonstrating the presence of the let7a binding site at the 3’UTR of Luciferase gene.