Abstract
Regulatory circuits that control stem cell fate decisions can be identified and understood by manipulating individual regulatory elements genetically. While impractical in the rare somatic stem cells of primary tissue, this approach is feasible in embryonic stem cells differentiated in vitro into the somatic stem cell type of interest. We present an improved highly efficient targeting system allowing genes to be integrated into a predetermined, doxycycline-inducible locus, and corresponding inducible embryonic stem cell lines to be generated rapidly. We apply this system to evaluate a key hematopoietic progenitor cell regulatory element, HoxB4, and its mammalian paralogs, whose effects have not yet been tested in this context. We show that all Hox paralog group 4 members, A4, B4, C4, and D4, have similar effects on hematopoietic stem and progenitor self-renewal in vitro, and thus classify Hox paralog group 4 as promoting self-renewal. Each paralog group 4 member both promotes proliferation and inhibits differentiation, enabling the exponential expansion of hematopoietic progenitors from the c-kit+/CD41+ cell fraction of day 6 murine embryoid bodies. By evaluating a set of deletion mutants we show that sequences in addition to the homeodomain and hexapeptide motif are required for this activity. These results highlight the utility of this expression system to perform functional and structural analyses of genetic regulators of cell fate decisions.
Introduction
Although a number of signaling molecules have been identified as important regulators of hematopoietic stem cell (HSC) homeostasis [1–6], and pathways through which they act are well understood, the regulatory circuits formed by transcription factors on which these signals impinge are not well understood. Many transcription factors are known to be expressed in the HSC [7–9] but the role that the vast majority of these play is obscure. The classic loss of function genetic approach to evaluating gene activity, generating a knockout, gives insight into necessity but is commonly confounded by genetic and circuit redundancy. Furthermore, because a circuit made of many interconnecting components can have multiple stable states, the role of individual regulators cannot be determined solely by their complete removal, but also requires testing each regulator at varying levels of expression and observing the output of the circuit as a whole. Therefore, gain of function studies, preferably employing conditional and variable expression, are essential. We have developed an improved embryonic stem (ES) cell–based gene targeting and conditional expression system for performing such gain of function analyses and applied it to interrogating the function of one set of genetic regulators of HSC homeostasis, the paralog group 4 Hox genes.
There are 39 mammalian Hox genes which can be ordered into 13 paralogous groups, based on the sequence of the DNA-binding homeodomain encoded by each. Although first discovered through their early role in establishing the embryonic anterior-posterior axis [10], Hox genes are also expressed in proliferating tissues of the adult, and presumably have a role in tissue homeostasis. The misexpression of Hox genes in leukemia, through activation by MLL fusion proteins [11] or by chromosomal translocations [12] suggests a role in regulating the proliferation and differentiation of hematopoietic progenitors. Several Hox genes have been studied by retroviral transduction into adult bone marrow, followed by transplantation. Retroviral overexpression often results in leukemia, and is generally accompanied by defects in the differentiation of specific hematopoietic lineages [13–16]. The exception is HoxB4, which although it significantly enhances HSC self-renewal, does not commonly lead to leukemia. Furthermore, HoxB4 is compatible with differentiation of all hematopoietic lineages [17,18], although lymphoid differentiation is inhibited at high levels of expression [19–21]. We have shown that expressing HOXB4 conditionally allows the expansion of embryonic hematopoietic progenitors derived from ES cells differentiated in vitro [19]. HoxB4 has three paralogs, HoxA4, C4, and D4. Each member of paralog group 4 encodes a protein with a highly conserved homeodomain, a conserved upstream sequence adjacent to the homeodomain, which includes the presumptive Pbx-binding hexapeptide, and an N-terminal conserved sequence of unknown function (Fig. 1A). When compared with Dfd (Deformed, the Drosophila homologue), the homeodomain and upstream hexapeptide-containing sequences are strongly conserved, the N-terminal sequence is moderately conserved, and sequence conservation throughout the remainder of the protein is difficult to detect.
FIG. 1.
Conserved domains in paralog group 4 members. Conserved residues are highlighted in reverse face. (A) The N-terminus of each paralog group 4 member, containing a conserved domain of 21 amino acids. (B) The homeodomain and upstream Pbx-binding hexapeptide motif.
Although large elements of protein sequence may be unconserved between paralogs, elegant substitution studies in paralog group 3 have shown that when one sequence is swapped into the locus of another, it compliments the knockout phenotype perfectly [22]. The paralog group 4 members are expressed in broadly similar domains with anterior boundaries in the paraxial mesoderm at the cervical level. However the precise rostral boundary differs slightly: HoxB4 and HoxD4 in the prevertebrae 1, HoxA4 at prevertebrae 2 and HoxC4 at prevertebrae 4 [23,24]. Targeted deletion of the HoxA4, HoxB4, and HoxD4 show anterior homeotic transformations at the rostral boundary of expression while in the HoxC4 deletion, the most anterior transformation is seen in the first thoracic segment, which is caudal of its rostral boundary of expression [25–28]. The HoxB4 knockout has a very subtle hematopoietic defect, apparent only upon transplantation [29]; the other paralog group 4 mutants have not been analyzed at this level. Based on these data, it is not obvious whether HoxA4, C4, and D4 should function similarly or differently in the regulation of proliferation of hematopoietic progentiors.
Using the gene-targeting system described above, we have generated a panel of ES cell lines representing each member of paralog group 4, and studied the effect of overexpressing these genes on self-renewal of hematopoietic progenitor cells derived in vitro from these ES cell lines. We find that all paralog group 4 Hox genes promote proliferation and inhibit differentiation, enabling the exponential expansion of undifferentiated hematopoietic progenitors in vitro. This establishes a uniform self-renewal activity for Hox paralog group 4 in hematopoiesis. Finally, we show that this activity requires sequences in addition to the conserved homeodomain and hexapeptide motif.
Materials and Methods
Generation of A2Lox ES cells
The hypoxanthine-guanine phosphoribosyltransferase 5′ repair/targeting plasmid [30] carrying the TRE-loxP-Δneo inducible target locus [19] was modified by inserting lox2272 into the BamH1 site using primer linkers encoding the following sequence: 5′-GGATCCATAACTTCGTATAGGATACTTTATACGAAGTTATGGATCC-3′. Homologous recombination followed by selection in HAT medium was then performed as previously described [19].
Derivation of inducible HoxA4, C4, and D4 ES cell lines
p2Lox was generated from pLox by digesting with XbaI/NotI to remove loxP, the stuffer, and the multiple cloning site, and primer linkers were inserted encoding the following sequence: 5′-CCATGGTGTCGATAACTTCGTATAGCATACATTATACGAAGTTATCGATAACTTCGTATAGGATACTTTATACGAAGTTATCTCGAGGGTACCAA TCACTAGTGAATTCGCGGCCGC-3′. The derivative plasmid was then digested with XhoI/NotI, and the enhanced green fluorescent protein (EGFP) stuffer was inserted as an XhoI/NotI fragment (from pBluescript-EGFP, in which EGFP is inserted at the EcoRI site).
pGEM containing mouse HoxA4 (the generous gift of Deborah Wolgemuth, encoding UniprotKB/swiss-prot ID P06798) was digested with HindIII and XbaI and cloned into pBS at these restriction sites. Then the fragment was excised by digestion of pBS with HindIII and NotI and cloned into HindIII / NotI-digested p2lox. Mouse HoxC4 was amplified from total embryonic RNA using the following primers: C4F 5′-GTCGACCATGATCATGAGCTCGTATTTG-3′ and C4R 5′-TTATTACCTGGTGATGTCCT-3′. It was then TA cloned into pGEM-Teasy (Promega) and subcloned int p2Lox as a SalI/EcoRI fragment. After sequencing we determined that the clone corresponds to the UniprotKB/swiss-prot ID Q08624. Mouse HoxD4 was cloned in the same way using the primers D4F 5′-GTCGACCATGGCCATGAGTTCGTATATG-3′ and D4R 5′-CTATTAGGTCGTCAGGTCCGT-3′. After sequencing we determined that the clone corresponded to the UniprotKB/swiss-prot ID P10628. Twenty micrograms of p2Lox-HoxA4, or −HoxC4, or –HoxD4 were coelectroporated with 20 μg of cre expression plasmid, into the A2Lox-targeting cells, and cells were selected on 300 μg/mL of G418, as previously described [19].
ES cell culture and in vitro differentiation
ES cells were cultured on mouse embryonic fibroblasts (MEFs) in Dulbecco's modified eagle's medium (DME) supplemented with 15% fetal bovine serum (FBS), 0.1 mM nonessential amino acids (GIBCO), 2 mM glutamax (Invitrogen), penicillin/streptomycin (Gibco), 0.1 mM β-mercaptoethanol, and 1,000 U/mL LIF (Millipore), at 37°C in 5% CO2, and differentiated as EBs by preplating for 40 min to remove MEFs and suspension in hanging drops (100 cells/10 μL drop) in embryoid body differentiation (EBD) medium: Iscove's modified Dulbecco's medium (IMDM) supplemented with 15% FBS, 200 μg/mL ironsaturated transferrin (Sigma), 4.5 mM monothiolglycerol (Sigma), 50 μg/mL ascorbic acid (Sigma), penicillin/streptomycin (Gibco), and 2 mM glutamax at 37°C in 5% CO2, 5% O2. After 48 h, EBs were harvested from hanging drops and plated in nonadherent 10-cm dishes on a swirling rotator in EBD medium EBs were fed after 48 h by exchanging 50% of spent medium for fresh EBD medium.
Transcriptional analysis
EBs were treated with 1 μg/mL of doxycycline for different time points, and RNA was extracted at day 6 of differentiation by Trizol (invitrogen). RT was performed with Thermoscript (Invitrogen). Specific probes to detect human HoxB4, murine HoxB4, HoxA4, HoxC4 were obtained from Applied Biosystems. HoxD4 levels were monitored by using the following primers set F: 5′-TGTGGTCTACCCTTGGA TGAAG-3′ and R: 5′-TAGTTGGGGTTCACCGAATTCACG-3′. Embryonic globin was monitored from OP9 coculture–induced samples by using the following primer/probe sets: β-major F, AGGGCACCTTTGCCAGC; β-major R, GGCAGCCTCTGCAGCG; β-major probe, 6FAM-CGTGATTGTGCTGGGCCACCACCT-TAMRA. β-H1 F, CCTCAAGGAGACCTTTGCTCAT; β-H1R, CAGGCAGCCTGCACCTCT; β-H1 probe, 6FAM-CAACATGTTGGTGATTGTCCTTTCT-TAMRA.
OP9 cocultures
100,000 c-Kit+/CD41+ cells from day 6 EBs were purified by flow cytometry and plated on OP9 monolayers (50,000 OP9 cells per well in six-well dishes plated 1 day before) in IMDM supplemented with 10% FBS, 5 ng/mL vascular endothelial growth factor, 40 ng/mL TPO, 40 ng/mL Flt-3 ligand, penicillin/streptomycin (Gibco), 2 mM glutamax, and 1.0 μg/mL doxycycline at 37°C in 5% CO2, 5% O2. Semiadherant cells were passaged by trypsinization every 4–5 days and replated at a density of 50,000 cells/well in six-well dishes.
Antibody staining and flow cytometry
The following antibodies were used: c-Kit-APC, CD41-FITC, CD45-PE, Gr-1-PE, Ter119-PE (all from eBiosciences). Cells were analyzed and sorted on a FACS Aria (BD Biosciences).
Colony assays
For each colony assay, 50,000 cells were plated into 1.5 mL methylcellulose medium supplemented with interleukin-3 (IL-3), IL-6, Epo, and SCF (M3434, StemCell Technologies), and where indicated 1 μg/mL doxycycline. Primitive erythroid colonies were counted after 6 days, other colonies after 10 days.
Results
An improved gene targeting/inducible expression system
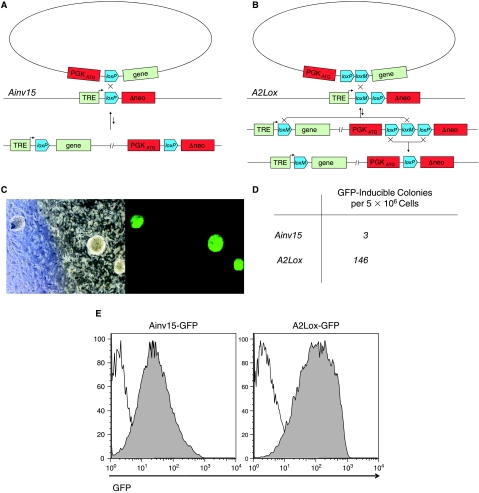
We have previously generated an ES cell line with a conditional variable locus on the X chromosome into which a gene of interest can be inserted by cre/lox recombination [19]. In this system, a circular plasmid carrying a single LoxP site integrates at a LoxP site on the X chromosome. A nonfunctional neo gene (lacking a promoter and start codon) resides downstream of the chromosomal LoxP site. Recombination is promoted by transient expression of cre recombinase through coelectroporation of an expression construct. Proper integration repairs the neo gene by placing a promoter and inframe start codon upstream, enabling efficient selection. Random integration events are rare (because the plasmid is not linearized) and do not result in G418 resistance, therefore they are not recovered [31]. A tetracycline-responsive promoter resides upstream of the chromosomal loxP site and precise integration places a gene of interest downstream of this promoter, such that it can be expressed in the presence of doxycycline. The reverse tetracycline transactivator is expressed from the constitutive Rosa26 promoter, enabling inducible expression in all cell types. Although selection is extremely effective, resulting in essentially all derivative ES cell lines being correctly targeted, the frequency of such clones is extremely low in any given experiment, making the system unsuitable for high-throughput or large-scale production of cell lines. A key problem with this system is that the great majority of recombination events are quickly reverted. After integration, the plasmid is flanked by identical loxP sites, and can therefore be excised by cre recombinase which, because its expression plasmid is coelectroporated, persists for at least as long as the integrating plasmid. Integration follows second order kinetics while excision is essentially first order, therefore the excision reaction proceeds with a much higher rate than the integration reaction (Fig. 2A). To solve this problem, we redesigned the targeting aspect of this system making use of a mutant version of loxP (lox2272 [32], referred to as loxM in Fig. 2B), which self-recombines, but does not cross-recombine with wild-type loxP. Topologically, the integration is a cassette exchange, with almost the entire plasmid being exchanged for the few nucleotides in between the chromosomal loxP and loxM sites. The final product is identical to that generated by the 1-Lox system, with the exception of the two base pair changes distinguishing loxM from loxP. Compared with the previous configuration, the efficiency of recovering recombinants in the 2-Lox system is improved by nearly two orders of magnitude (Fig. 2C and D).
FIG. 2.
2-Lox targeting system. (A) Gene targeting in the original 1-Lox system (Ainv15 ES cells). (B) Gene targeting by cassette exchange in the A2Lox cells. (C) Colonies arising from selection 10 days after targeting A2Lox cells with p2Lox-EGFP. The plate was exposed to doxycycline (1 μg/mL) 24 h prior to photographing under bright field (left panel) or fluorescence (right panel). (D) Comparison of targeting efficiency between the two systems. (E) Expression of GFP upon dox treatement of day 6 embryoid body differentiation. Induction was achieved from Day 0 to Day 6. Left Ainv15-GFP (one lox), right A2Lox-GFP (two lox).
To evaluate the efficiency of induction, we generated an inducible GFP ES cell line by using the A2Lox cells (2-Lox system) and compared the level of GFP expression during embryoid body differentiation, to iGFP cells, produced previously using 1-Lox targeting in Ainv15 cells [19]. As shown in Fig. 2E the new system can efficiently drive GFP in all cells of the day 6 embryoid body, and is comparable to the previously published system in terms of background leakiness and locus silencing.
Paralog group 4 members promote proliferation and inhibit differentiation of multipotent progenitors
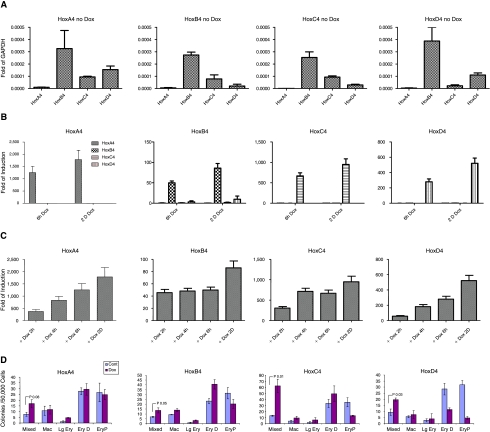
We then used the targeting system to derive new ES cell lines in which mouse HoxA4, C4, and D4, were integrated into the same inducible locus, and could thus be compared directly to HOXB4. To compare the regulation of these genes, we first evaluated the baseline levels of paralog group 4 members in each cell line at day 6 of embryoid body differentiation in the absence of doxycycline (Fig. 3A). All four cell lines gave similar results: HOXB4 was predominant, HoxC4 and D4 were expressed at lower levels, and HoxA4 expression was very low. We then induced gene expression with doxycycline for various time periods prior to harvesting on day 6, and observed that induced levels dwarf endogenous levels (Fig. 3B). We also observed that expression of one paralog does not induce expression of others, at least not to levels comparable to doxycycline-induced levels, a result that allows us to ascribe specific effects to specific paralog group 4 genes. Since induced levels were comparable, fold induction was highest for HoxA4 and lowest for HOXB4. Fold induction increased with longer induction times, as expected (Fig. 3C).
FIG. 3.
Induction of paralog group 4 members during EB differentiation. (A) Baseline level of expression for each paralog group 4 gene in uninduced EBs. (B) Level of expression of each paralog group 4 gene expressed as fold of baseline, when one paralog group 4 gene is induced. None of the genes induces the expression of its paralogs. (C) Fold induction for each paralog group 4 member after 2, 4, and 6 h and 2 days of induction, determined at Day 6. (D) Colony-forming cell assays from Day 6 embryoid bodies. 50,000 cells were plated into methylcellulose assay medium, either supplemented or not supplemented with 1 μg/mL doxycycline. Right bars indicate colonies from doxycycline-treated group; left bars indicate colony number from control, untreated group. For each paralog group 4 member, doxycycline results in an increase of multipotent (Mixed) colonies. Abbreviations: Mac, macrophage; Lg Ery, large erythroid; Ery D, definitive erythroid; Ery P, primitive erythroid.
We then differentiated each cell line as embryoid bodies for 6 days, and assayed hematopoietic progenitor activity in methylcellulose colony assay medium in the presence and absence of doxycycline, an experiment designed to study the role of a given paralog on committed hematopoietic progenitors. In the presence of doxycycline to induce gene expression during colony formation, all paralog group 4 members significantly increased the colony-forming activity of multipotent progenitors, observed as a significant increase in numbers of mixed (erythroid-myeloid) colonies (Fig. 3D). In addition, mixed colonies were considerably larger in the doxycycline-treated arms of each respective experiment (not shown). We also observed an inhibition of erythroid differentiation, most evident in the primitive erythroid lineage.
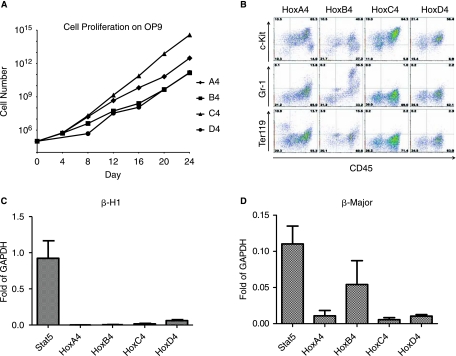
We have previously shown that hematopoietic progenitors derived from day 6 EBs can be expanded exponentially on OP9 stromal cells in the presence of HOXB4 expression [19]. We therefore sorted the hematopoietic progenitor fraction (c-Kit+/CD41+ cells) from day 6 EBs derived from each cell line, and compared the effect of expression of each paralog group 4 member on cells plated on OP9. In each case, doxycycline induced proliferation and exponential expansion of a hematopoietic population, which was sustained for over 1 month without exhaustion (Fig. 4A). These cultures were rich in hematopoietic colony-forming cells, particularly CFU-GEMM (not shown). We characterized the proliferating cells using a variety of lineage-specific markers, and observed that while undifferentiated hematopoietic progenitors dominated these cultures, there was a certain rate of ongoing differentiation in each case (Fig. 4B). This rate was somewhat higher for HoxA4 and B4 than it was for HoxC4 and D4, implying that HoxC4 and D4 are more potent inhibitors of differentiation. We have also previously shown that hematopoietic progenitors expanded under the influence of HOXB4 have altered globin gene expression patterns, indicative of primitive to definitive hemtopoietic switching, compared to hematopoietic progenitors expanded under mitogenic signaling, for example through activated Stat5 [33]. We therefore measured β-H1 (embryonic) and β-major (adult) globin levels in each culture. While all four paralogs showed greatly inhibited levels of β-H1 globin (Fig. 4C), switching was most apparent in the HOXB4 cultures, as these expressed significant levels of β-major globin (Fig. 4D). The other paralogs showed much lower levels of β-major globin, possibly due to the ability of these genes to inhibit differentiation more strongly than HOXB4.
FIG. 4.
Hematopoietic effects of paralog group 4 expression. (A) Growth curves of hematopoietic progenitors (derived from Day 6 EBs by c-Kit+/CD41+ sorting) on OP9 in the presence of doxycycline. In the absence of doxycycline, no growth was observed. (B) Representative figure of flow cytometric profile of cells expanded on OP9. CD45 is shown on the x-axis, lineage markers (c-Kit marking progenitors, Gr1 marking granulocytes, and Ter119 marking erythroblasts) are shown on the y-axis. Each paralog group 4 member promotes the outgrowth of c-Kit+ progenitors, with variable levels of ongoing differentiation. HoxA4 and B4 show more differentiation than HoxC4 and D4. (C) Level of expression of embryonic globin, β-H1. (D) Level of expression of definitive globin, β-major.
Domain analysis
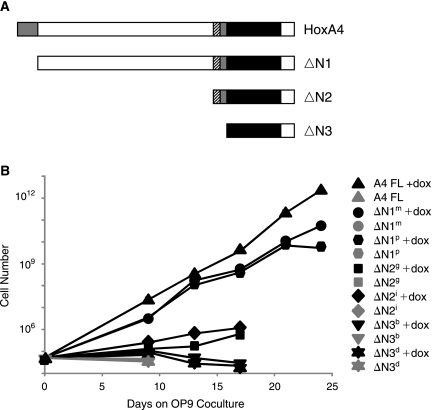
There exist two clearly obvious regions of similarity among the mammalian paralog group four members: part of the short N-terminal sequence and a C-terminal sequence comprising the paralog group 4 homeodomain and Pbx-interacting hexapeptide. To determine whether the homeodomain alone was sufficient, or if not, what N-terminal sequences were required for the hematopoietic progenitor expansion activity, we generated a nested set of N-terminal deletions comprising homeodomain-only, ΔN3, hexapeptide + homeodomain, ΔN2, and near-full length (lacking only the N-terminal conserved domain, ΔN1) versions of HoxA4, and compared the ability of these constructs to promote progenitor expansion on OP9 stromal cells to full length HoxA4. Neither the homeodomain alone nor the homeodomain + associated hexapeptide were capable of promoting growth of hematopoietic cells on OP9. The larger construct, lacking only the N-terminal conserved sequence, did promote growth, although not to the same extent as full length HoxA4. These results showed clearly that sequences in addition to the homeodomain and hexapeptide are required for progenitor expansion, and that the N-terminal conserved sequence, while not essential, is necessary for maximal activity (Figs. 5B).
FIG. 5.
Deletion analysis of HoxA4. (A) Deletion constructs tested. Conserved residues regions are highlighted gray, the hexapeptide motif is marked with stripes, and the homeodomain is shown in black. (B) Growth curves of hematopoietic progenitors from day 6 EBs, in which doxycycline was added or not added to induce expression of HoxA4 or deletion derivatives.
Discussion
Using a high-efficiency gene targeting and conditional expression system, we have generated a panel of ES cell lines representing each member of Hox paralog group 4, and compared their ability to promote self-renewal of hematopoietic stem and progenitor cells. Self-renewal requires both proliferation and inhibition of differentiation, and each paralog group 4 member has both activities. Because each construct was expressed from the same genetic location, variation due to promoter activity or chromosomal position, is minimized. Differential activity can be attributed to either RNA turnover or to protein sequence-intrinsic characteristics such as differential translation, stability, or activity of functional domains.
Understanding how multiple interacting regulators control a biological process requires manipulating each individually. While the loss-of-function approach is often preferred as it gives insight into necessity, for the study of complex genetic circuits the conditional variable gain of function system that we describe here has two distinct advantages. The first, not used in this study, is that it enables varying levels of expression. A complex circuit as a whole generally has multiple stable states, and individual elements may drive the system into different states depending on their level of expression. The ability to titrate levels of gene expression with doxycycline makes such an analysis possible. The second, used to advantage in our analysis of paralog group 4, is that being a gain of function assay, it is not affected by the redundancy that is often present in genetic systems. While each member of paralog group 4 has a similar gain of function effect on self-renewal, hematopoietic defects were not obvious when any paralog group 4 gene was deleted individually [25–28,34,35]. The appearance of a loss-of-function phenotype only after all or multiple paralogs have been deleted is typical for axial patterning phenotypes caused by Hox gene mutations. One of the most striking examples is the knockout of paralog group 10 (3 genes × 2 alleles). When all representatives of paralog group 10 were eliminated, the complete anterior transformation of all ribless posterior vertebrae to ribbed vertebrae occurred [36]. When any five of six representatives were knockout alleles, the single remaining wild-type allele was sufficient to suppress ectopic rib formation in the vertebrae of the tail. Our results showing that each paralog group 4 member has broadly similar effects on hematopoietic progenitor proliferation and differentiation indicate that loss of function studies involving HOXB4 ought to encompass multiple and preferably all paralog group 4 members. To date, hematopoiesis has not been characterized in compound mutants involving multiple paralog group 4 members.
In addition to redundancy within a paralog group, there are several examples of interparalog redundancies, usually involving neighboring paralog groups [37–39]. The loss of the entire B cluster did not inhibit FL-HSC activity [40], however the possibility of other Hox paralog groups having similar phenotypes in our simple gain of function assays (promotion of proliferation and inhibition of differentiation of ES-derived hematopoietic progenitors) is compelling, given that overexpression of numerous Hox genes can lead to leukemia. We are currently comparing the self-renewal activity of other paralog groups to that of paralog group 4.
The efficiency of gene targeting into the inducible locus of the A2Lox ES cell line allows the rapid generation of a large number of derivative inducible ES cell lines. This is ideal for the two applications that we demonstrate here: testing multiple genes for activity in a biological process, and testing mutants of a given gene to map that activity to particular sequence elements. Provided that a given biological process can be modeled in vitro with differentiating ES cells, this cell line facilitates mechanistic and genetic studies of a scope not possible with the conventional transgenic approach. The time-consuming nature of mammalian transgenesis is one reason that drives discovery phase experiments into other model organisms. As improvements in genetic tools combine with advances in differentiation culture, the ES in vitro differentiation system is poised to play a more important role.
Acknowledgments
We thank the Dr. Bob and Jean Smith Foundation for their generous support. This work was supported by the National Institutes of Health grant 1R01HL081186-01 and the March of Dimes 5-FY2006-272.
References
- 1.Bodine DM. Seidel NE. Zsebo KM. Orlic D. In vivo administration of stem cell factor to mice increases the absolute number of pluripotent hematopoietic stem cells. Blood. 1993;82:445–455. [PubMed] [Google Scholar]
- 2.Fleming WH. Alpern EJ. Uchida N. Ikuta K. Weissman IL. Steel factor influences the distribution and activity of murine hematopoietic stem cells in vivo. Proc Natl Acad Sci USA. 1993;90:3760–3764. doi: 10.1073/pnas.90.8.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sitnicka E. Lin N. Priestley GV. Fox N. Broudy VC. Wolf NS. Kaushansky K. The effect of thrombopoietin on the proliferation and differentiation of murine hematopoietic stem cells. Blood. 1996;87:4998–5005. [PubMed] [Google Scholar]
- 4.Yagi M. Ritchie KA. Sitnicka E. Storey C. Roth GJ. Bartelmez S. Sustained ex vivo expansion of hematopoietic stem cells mediated by thrombopoietin. Proc Natl Acad Sci USA. 1999;96:8126–8131. doi: 10.1073/pnas.96.14.8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arai F. Hirao A. Ohmura M. Sato H. Matsuoka S. Takubo K. Ito K. Koh GY. Suda T. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118:149–161. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Zhang CC. Kaba M. Ge G. Xie K. Tong W. Hug C. Lodish HF. Angiopoietin-like proteins stimulate ex vivo expansion of hematopoietic stem cells. Nat Med. 2006;12:240–245. doi: 10.1038/nm1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forsberg EC. Prohaska SS. Katzman S. Heffner GC. Stuart JM. Weissman IL. Differential expression of novel potential regulators in hematopoietic stem cells. PLoS Genet. 2005;1:e28. doi: 10.1371/journal.pgen.0010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivanova NB. Dimos JT. Schaniel C. Hackney JA. Moore KA. Lemischka IR. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 9.Taghon T. Thys K. De Smedt M. Weerkamp F. Staal FJ. Plum J. Leclercq G. Homeobox gene expression profile in human hematopoietic multipotent stem cells and T-cell progenitors: implications for human T-cell development. Leukemia. 2003;17:1157–1163. doi: 10.1038/sj.leu.2402947. [DOI] [PubMed] [Google Scholar]
- 10.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 11.Milne TA. Briggs SD. Brock HW. Martin ME. Gibbs D. Allis CD. Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura T. Largaespada DA. Lee MP. Johnson LA. Ohyashiki K. Toyama K. Chen SJ. Willman CL. Chen IM. Feinberg AP. Jenkins NA. Copeland NG. Shaughnessy JD., Jr Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat Genet. 1996;12:154–158. doi: 10.1038/ng0296-154. [DOI] [PubMed] [Google Scholar]
- 13.Perkins A. Kongsuwan K. Visvader J. Adams JM. Cory S. Homeobox gene expression plus autocrine growth factor production elicits myeloid leukemia. Proc Natl Acad Sci USA. 1990;87:8398–8402. doi: 10.1073/pnas.87.21.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sauvageau G. Thorsteinsdottir U. Hough MR. Hugo P. Lawrence HJ. Largman C. Humphries RK. Overexpression of HOXB3 in hematopoietic cells causes defective lymphoid development and progressive myeloproliferation. Immunity. 1997;6:13–22. doi: 10.1016/s1074-7613(00)80238-1. [DOI] [PubMed] [Google Scholar]
- 15.Thorsteinsdottir U. Sauvageau G. Hough MR. Dragowska W. Lansdorp PM. Lawrence HJ. Largman C. Humphries RK. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol Cell Biol. 1997;17:495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thorsteinsdottir U. Mamo A. Kroon E. Jerome L. Bijl J. Lawrence HJ. Humphries K. Sauvageau G. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood. 2002;99:121–129. doi: 10.1182/blood.v99.1.121. [DOI] [PubMed] [Google Scholar]
- 17.Sauvageau G. Thorsteinsdottir U. Eaves CJ. Lawrence HJ. Largman C. Lansdorp PM. Humphries RK. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev. 1995;9:1753–1765. doi: 10.1101/gad.9.14.1753. [DOI] [PubMed] [Google Scholar]
- 18.Antonchuk J. Sauvageau G. Humphries RK. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell. 2002;109:39–45. doi: 10.1016/s0092-8674(02)00697-9. [DOI] [PubMed] [Google Scholar]
- 19.Kyba M. Perlingeiro RCR. Daley GQ. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002;109:29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- 20.Rideout WM. Hochedlinger K. Kyba M. Daley GQ. Jaenisch R. Correction of a genetic defect by nuclear transplantation and combined cell and gene therapy. Cell. 2002;109:17–27. doi: 10.1016/s0092-8674(02)00681-5. [DOI] [PubMed] [Google Scholar]
- 21.Pilat S. Carotta S. Schiedlmeier B. Modlich U. Kamino K. Will E. Mairhofer A. Steinlein P. Ostertag W. Beug H. Baum C. Klump H. HOXB4 Enforces Equivalent Fates of ES-Cell-Derived and Adult Hematopoietic Cells. Blood. 2004;104:144a. doi: 10.1073/pnas.0505624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greer JM. Puetz J. Thomas KR. Capecchi MR. Maintenance of functional equivalence during paralogous Hox gene evolution. Nature. 2000;403:661–665. doi: 10.1038/35001077. [DOI] [PubMed] [Google Scholar]
- 23.Gaunt SJ. Krumlauf R. Duboule D. Mouse homeogenes within a subfamily, Hox-1.4, −2.6 and −5.1, display similar anteroposterior domains of expression in the embryo, but show stage- and tissue-dependent differences in their regulation. Development. 1989;107:131–141. doi: 10.1242/dev.107.1.131. [DOI] [PubMed] [Google Scholar]
- 24.Geada AM. Gaunt SJ. Azzawi M. Shimeld SM. Pearce J. Sharpe PT. Sequence and embryonic expression of the murine Hox-3.5 gene. Development. 1992;116:497–506. doi: 10.1242/dev.116.2.497. [DOI] [PubMed] [Google Scholar]
- 25.Ramirez-Solis R. Zheng H. Whiting J. Krumlauf R. Bradley A. Hoxb-4 (Hox-2.6) mutant mice show homeotic transformation of a cervical vertebra and defects in the closure of the sternal rudiments. Cell. 1993;73:279–294. doi: 10.1016/0092-8674(93)90229-j. [DOI] [PubMed] [Google Scholar]
- 26.Kostic D. Capecchi MR. Targeted disruptions of the murine Hoxa-4 and Hoxa-6 genes result in homeotic transformations of components of the vertebral column. Mech Dev. 1994;46:231–247. doi: 10.1016/0925-4773(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 27.Horan GS. Wu K. Wolgemuth DJ. Behringer RR. Homeotic transformation of cervical vertebrae in Hoxa-4 mutant mice. Proc Natl Acad Sci USA. 1994;91:12644–12648. doi: 10.1073/pnas.91.26.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boulet AM. Capecchi MR. Targeted disruption of hoxc-4 causes esophageal defects and vertebral transformations. Dev Biol. 1996;177:232–249. doi: 10.1006/dbio.1996.0159. [DOI] [PubMed] [Google Scholar]
- 29.Brun AC. Bjornsson JM. Magnusson M. Larsson N. Leveen P. Ehinger M. Nilsson E. Karlsson S. Hoxb4-deficient mice undergo normal hematopoietic development but exhibit a mild proliferation defect in hematopoietic stem cells. Blood. 2004;103:4126–4133. doi: 10.1182/blood-2003-10-3557. [DOI] [PubMed] [Google Scholar]
- 30.Bronson SK. Plaehn EG. Kluckman KD. Hagaman JR. Maeda N. Smithies O. Single-copy transgenic mice with chosen-site integration. Proc Natl Acad Sci USA. 1996;93:9067–9072. doi: 10.1073/pnas.93.17.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukushige S. Sauer B. Genomic targeting with a positive-selection lox integration vector allows highly reproducible gene expression in mammalian cells. Proc Natl Acad Sci USA. 1992;89:7905–7909. doi: 10.1073/pnas.89.17.7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee G. Saito I. Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene. 1998;216:55–65. doi: 10.1016/s0378-1119(98)00325-4. [DOI] [PubMed] [Google Scholar]
- 33.Kyba M. Perlingeiro RC. Hoover RR. Lu CW. Pierce J. Daley GQ. Enhanced hematopoietic differentiation of embryonic stem cells conditionally expressing Stat5. Proc Natl Acad Sci USA. 2003;100(Suppl 1):11904–11910. doi: 10.1073/pnas.1734140100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horan GS. Kovacs EN. Behringer RR. Featherstone MS. Mutations in paralogous Hox genes result in overlapping homeotic transformations of the axial skeleton: evidence for unique and redundant function. Dev Biol. 1995;169:359–372. doi: 10.1006/dbio.1995.1150. [DOI] [PubMed] [Google Scholar]
- 35.Saegusa H. Takahashi N. Noguchi S. Suemori H. Targeted disruption in the mouse Hoxc-4 locus results in axial skeleton homeosis and malformation of the xiphoid process. Dev Biol. 1996;174:55–64. doi: 10.1006/dbio.1996.0051. [DOI] [PubMed] [Google Scholar]
- 36.Wellik DM. Capecchi MR. Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science. 2003;301:363–367. doi: 10.1126/science.1085672. [DOI] [PubMed] [Google Scholar]
- 37.Rancourt DE. Tsuzuki T. Capecchi MR. Genetic interaction between hoxb-5 and hoxb-6 is revealed by nonallelic non-complementation. Genes Dev. 1995;9:108–122. doi: 10.1101/gad.9.1.108. [DOI] [PubMed] [Google Scholar]
- 38.Davis AP. Capecchi MR. A mutational analysis of the 5′ HoxD genes: dissection of genetic interactions during limb development in the mouse. Development. 1996;122:1175–1185. doi: 10.1242/dev.122.4.1175. [DOI] [PubMed] [Google Scholar]
- 39.de la Cruz CC. Der-Avakian A. Spyropoulos DD. Tieu DD. Carpenter EM. Targeted disruption of Hoxd9 and Hoxd10 alters locomotor behavior, vertebral identity, and peripheral nervous system development. Dev Biol. 1999;216:595–610. doi: 10.1006/dbio.1999.9528. [DOI] [PubMed] [Google Scholar]
- 40.Bijl J. Thompson A. Ramirez-Solis R. Krosl J. Grier DG. Lawrence HJ. Sauvageau G. Analysis of HSC activity and compensatory Hox gene expression profile in Hoxb cluster mutant fetal liver cells. Blood. 2006;108:116–122. doi: 10.1182/blood-2005-06-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]