Abstract
Many biotrophic fungal and oomycete plant pathogens deliver effector proteins directly into host cells during infection. Recent advances are revealing the extensive effector repertoires of these pathogens and are beginning to shed light on how they manipulate host cells to establish a parasitic relationship. Surprisingly, oomycete effectors seem to share a common uptake system with those from the human malaria pathogen. The current explosion of information is opening new research avenues in molecular plant pathology and is providing new opportunities to limit the impact of plant disease on food production.
A new fungal strain causing stem rust disease of wheat emerged recently in East Africa and is now spreading through the Near East, causing worldwide concern (www.globalrust.org/) (1). This highly virulent Ug99 strain of the wheat stem rust fungus Puccinia graminis tritici overcomes wheat lines that are widely used and have heretofore shown durable resistance to fungal infection. Ug99 is already well established in Kenya, where it causes losses of up to 80% of the wheat in farmers’ fields. P. graminis represents a class of destructive plant pathogens that share a biotrophic life-style; that is, they rely entirely on living host tissue for the completion of their life cycle (2). Elaborate parasitic relationships allow these biotrophic fungi to feed from their host plants. During infection, they establish their lifeline through the formation of specialized infection structures (haustoria) that penetrate the plant cell wall and allow nutrient uptake (Fig. 1). These structures are shared with some of the equally destructive oomycete pathogens, which resemble fungi but are more related to brown algae. For instance, the pathogen responsible for the Irish potato famine, the oomycete Phytophthora infestans, forms haustoria during its initial biotrophic stage of infection.
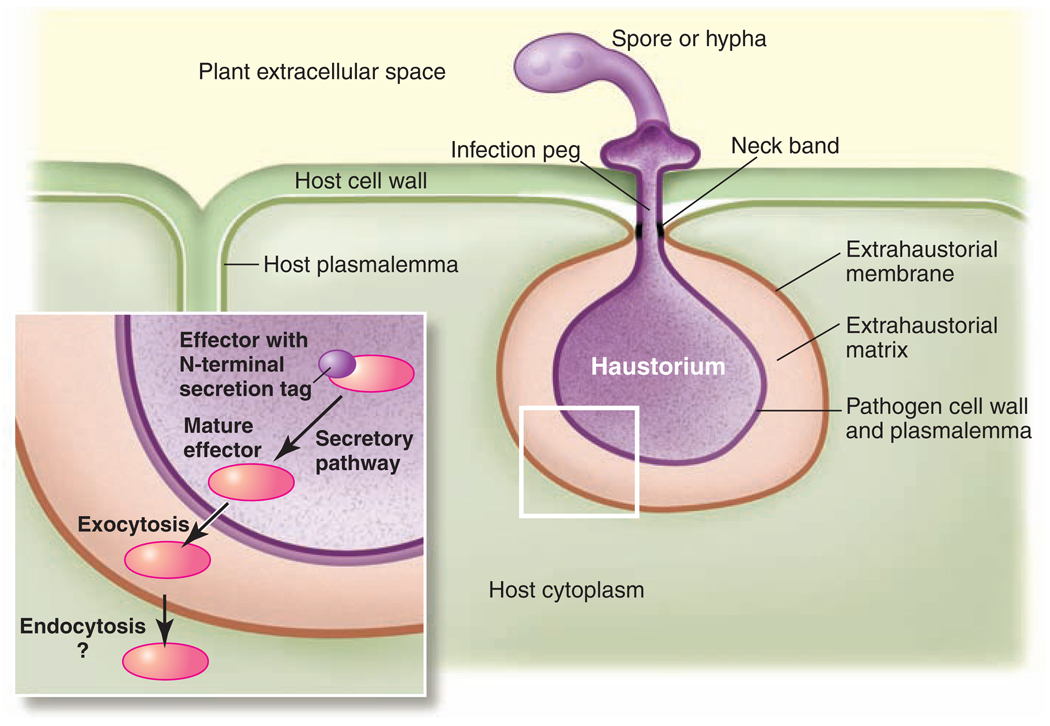
Fig. 1.
Organization of haustoria and the presumed route of effector delivery. Haustoria emerge from extracellular pathogen infection structures (such as spores or hyphae) and enter plant cells through the infection peg by invaginating the host plasma membrane. Haustoria are surrounded by the pathogen plasmalemma and cell wall as well as the modified host plasma membrane (extrahaustorial membrane). These membranes enclose the extrahaustorial matrix, an interface zone that is sealed by the neck band. The inset illustrates the delivery of a pathogen effector (red) with an N-terminal secretion tag (purple) through the secretory pathway and into the host cell.
Plant strains that are able to fend off these attacks do so in part by means of molecular recognition of proteins (effectors) delivered from haustoria into host cells during infection (3). However, haustoria are separated from the host cells by three distinct physical barriers: the pathogen plasma membrane, the cell wall, and the extrahaustorial membrane, which is derived from the plant plasma membrane but is molecularly distinct (Fig. 1). Exciting new research is now shedding light on the intriguing and as-yet unanswered question of how molecules are shuttled between the host and the invader.
Effector Delivery
Many bacterial pathogens of animals and plants possess a syringe-like apparatus, such as the one produced by prokaryotic (type III) secretion systems, which injects pathogenicity effector proteins into host cells. Effector proteins from fungal and oomycete parasites, on the other hand, seem to travel by way of the eukaryotic (type II) secretory pathway, which involves exocytosis of Golgi-derived secretory vesicles. Most known effectors from these parasites harbor a canonical N-terminal type II secretion signal (2, 4–7), which suffices to get the effectors past the first two barriers. However, although some effectors function in the extracellular environment, many others clearly do actually get into the plant cell because they are recognized by intracellular plant proteins [the resistance (R) proteins] that are part of the plant innate immune system (5, 7–9). Direct evidence of the transfer of effector proteins during infection came from the immunocytological detection of a rust effector (10) and the detection of a tagged oomycete effector (11) inside infected host cells.
Many oomycete effectors harbor a bipartite amino acid motif [RXLR–(D)EER], located downstream of the signal peptide, that is dispensable for exocytosis but required for plant access (11, 12). The RXLR domain is necessary and sufficient to mediate entry of effectors or reporter proteins into host cells in the absence of the oomycete, which indicates that the uptake occurs independently of any pathogen-derived machinery (12). The identification of a number of plant proteins that also contain the RXLR motif, several of them associated with membrane trafficking (13), suggests that oomycetes may exploit the plant endocytic pathway for host cell entry. Some fungal effectors can also enter plant cells in the absence of the pathogen, although they lack an RXLR domain and do not share other conserved peptide motifs (8, 14). Indeed, in plant-rust interactions, ultrastructural studies have visualized tubular extensions of the extrahaustorial membrane with associated budding vesicles that reach into the host cell cytoplasm and can form close contact with host endoplasmic reticulum and dictyosomes (15). In an alternative scenario, effector uptake could be mediated by a parasite-derived protein channel (16). The oomycete RXLR sequence motif is reminiscent of a similar domain in effectors of the protist Plasmodium falciparum, the malaria parasite that invades mammalian erythrocytes (17). The oomycete and Plasmodium delivery motifs appear to function interchangeably in either pathogen and in either host, which suggests that they target the same host admission route (18, 19). Because oomycetes and Plasmodium are members of the Chromalveolata, this peptide domain may have a common origin in these plant and animal pathogens, although convergent evolution is another possibility.
Biotroph effectors were initially identified based on their function as avirulence proteins that are recognized by plant immune receptors (7, 8). However, with the use of sequence signatures from the conserved N-terminal secretion signal and, for oomycetes, the RXLR motif, a more extensive suite of effectors can now be predicted from genome and transcriptome analyses (11, 20–23). Through these biocomputational means, the genomes of several fungal and oomycete phytopathogens unexpectedly were found each to encode several hundred effector candidates. In the case of Phytophthora species, many of these probably evolved from a common ancestor through rapid duplication and divergence (23). Nevertheless, only a few RXLR-type effectors are known to be detected by the plant defense machinery. However, this biocomputational approach may still overlook effectors that engage alternative trafficking routes, such those currently known from powdery mildew fungi, which lack canonical eukaryotic secretion tags (24).
Toward Effector Functions
The next challenge is to understand how these effector proteins turn the host cell to their own purposes. Although the biocomputational approach promises riches of new proteins to work with, the sheer number of effector candidates renders functional analysis a formidable task. Most candidates lack informative homologies other than the signal domain. Functional studies are hindered by the inaccessible genetics of many of these pathogens. Alternate means are thus being explored for the expression of candidate effector proteins inside plant cells or the heterologous delivery of effectors. The bacterial type III secretion system can be engineered to deliver these proteins (25), and certain hemibiotrophic phyto-pathogens that are more amenable to genetic manipulation may be useful transfer vehicles as well. Some effectors have been shown to heighten pathogen virulence (24, 25) or suppress host immune responses (25–27). Mutational analysis of Ustilago maydis suggests a high degree of redundancy in effector functions (22), and there is also considerable redundancy in oomycete effector repertoires, with the suppression of plant cell death processes being a common function of many effectors (26). The actual biochemical functions and host targets of identified effectors also need to be characterized. Protein x-ray crystallography has revealed that the structure of some bacterial effectors resembles known host proteins despite the absence of any sequence homology (28). However, the intensive analysis of protein crystallography is difficult to scale up to encompass whole-genome effector repertoires. Bioassays are therefore needed to probe the functions of numerous effector candidates simultaneously. Conceivable possibilities include the search for cellular targets on the basis of protein-protein interactions or experimental settings that explore the capacity of effectors to interfere with host defense or cell death. Alternatively, activity-based enzyme profiling might be employed to test the interference of effectors with host enzyme activities (29).
Most effectors seem to be specific to individual pathogen species, and even closely related pathogens reveal little overlap in their effector complements (21). This probably reflects a very rapid evolution of these proteins, which is driven by both the proteins’ need to target divergent host proteins and to escape recognition by host immune receptors. It will be intriguing to learn whether the effector suites from different parasites are of similar size and targeted to similar biochemical host functions. So far, few effectors with nuclear localization have been described (10), although the transcriptional reprogramming of host cells is a plausible effector task. Other effectors localizing to defined host compartments have not been identified yet and possibly await discovery.
Given that pathogenesis of obligate biotrophs is a multistep process that is characterized by a series of morphogenetic changes, it seems unlikely that all effectors are released simultaneously and from the same location into host cells. It is rather conceivable that sets of effectors are delivered in a coordinated manner as pathogenesis progresses, perhaps first from extracellular infection structures and then from haustoria or intracellular infection hyphae. It will be informative in this respect to find out whether the spatially linked effector genes in the genome of U. maydis (22) are transcriptionally co-regulated. Unravelling the spatial and temporal order of effector delivery as well as untangling the regulatory networks governing this proposed hierarchical process will surely be prominent topics in future research.
Although recent experimental efforts have mainly been focused on proteinaceous effectors, biotrophic pathogens also use nonproteinaceous metabolites to manipulate their host’s cells. In the phytopathogenic bacterium Pseudomonas syringae, the low–molecular-weight compounds coronatine (a mimic of the plant defense–signaling molecule jasmonic acid) and syringolin (a protea-some inhibitor) help to divert plant defense responses. Secondary metabolites may also be revealed as powerful nonproteinaceous effectors of fungal and oomycete biotrophs (30). It seems, however, that this effector category would be even more difficult to analyze than their proteinaceous cousins.
Future Perspectives
Secreted effectors are emerging as the prime weapons of plant parasites and also as targets for host recognition and immunity. As such, the current focus on pathogen effectors promises to reveal fascinating insights into the molecular basis of the biotrophic life-style and plant-microbe coevolution and probably will lead to innovative disease-control measures. It will be intriguing to learn whether biotroph effectors actively divert host metabolism or whether nutrient flow to the pathogen is an indirect consequence of disarmed host cells. It will also be fascinating to compare the effector suites of closely related pathogen species to find out whether and how effector diversity contributes to host range restriction and parasite speciation. Molecular finger-printing of pathogen isolates on the basis of the natural variation in their effector repertoires has the potential to serve as a valuable diagnostic tool in epidemiological surveys and will shed light on how new pathogen strains, such as the wheat stem rust strain Ug99, evolve to overcome host resistance.
Plant resistance genes that recognize effectors with important and nonredundant virulence functions are more likely to prove durable in the field because this will constrain the evolution of the pathogen. New resistance genes may be found in wild relatives of crop plants, a strategy that has already proven successful in defending against the Irish potato famine pathogen P. infestans (31).
Several biotechnological resistance strategies can now be envisaged, including the blocking of the effector delivery mechanism, the modifying of host targets in order to elude effector function, and the rational design of synthetic plant immune receptors in order to detect as-yet unrecognized parasite effectors. The pathogens’ effectors may be the source of their own undoing if research into their biochemical activity generates tools to selectively manipulate plant cellular functions. The current devastating impact of Ug99 stem rust on wheat production in Africa high-lights the critical importance of developing durable and effective strategies to protect agricultural production from disease threats.
References and Notes
- 1.Singh RP, et al. Adv. Agron. 2008;98:271. [Google Scholar]
- 2.O’Connell RJ, Panstruga R. New Phytol. 2006;171:699. doi: 10.1111/j.1469-8137.2006.01829.x. [DOI] [PubMed] [Google Scholar]
- 3.Ellis JG, Dodds PN, Lawrence GJ. Curr. Opin. Microbiol. 2007;10:326. doi: 10.1016/j.mib.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 4.Catanzariti AM, Dodds PN, Ellis JG. FEMS Microbiol. Lett. 2007;269:181. doi: 10.1111/j.1574-6968.2007.00684.x. [DOI] [PubMed] [Google Scholar]
- 5.Rehmany AP, et al. Plant Cell. 2005;17:1839. doi: 10.1105/tpc.105.031807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen RL, et al. Science. 2004;306:1957. doi: 10.1126/science.1104022. [DOI] [PubMed] [Google Scholar]
- 7.Dodds PN, Lawrence GJ, Catanzariti AM, Ayliffe MA, Ellis JG. Plant Cell. 2004;16:755. doi: 10.1105/tpc.020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catanzariti AM, Dodds PN, Lawrence GJ, Ayliffe MA, Ellis JG. Plant Cell. 2006;18:243. doi: 10.1105/tpc.105.035980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Armstrong MR, et al. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7766. [Google Scholar]
- 10.Kemen E, et al. Mol. Plant Microbe Interact. 2005;18:1130. doi: 10.1094/MPMI-18-1130. [DOI] [PubMed] [Google Scholar]
- 11.Whisson SC, et al. Nature. 2007;450:115. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- 12.Dou DL, et al. Plant Cell. 2008;20:1930. doi: 10.1105/tpc.107.056093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birch PRJ, et al. Curr. Opin. Plant Biol. 2008;11:373. doi: 10.1016/j.pbi.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Manning VA, Ciufetti LM. Plant Cell. 2005;17:3203. doi: 10.1105/tpc.105.035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mims CW, Rodriguez-Lother C, Richardson EA. Protoplasma. 2002;219:221. doi: 10.1007/s007090200023. [DOI] [PubMed] [Google Scholar]
- 16.Morgan W, Kamoun S. Curr. Opin. Microbiol. 2007;10:332. doi: 10.1016/j.mib.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Marti M, Good RT, Rug M, Knuepfer E, Cowman AF. Science. 2004;306:1930. doi: 10.1126/science.1102452. [DOI] [PubMed] [Google Scholar]
- 18.Bhattacharjee S, et al. PLoS Pathog. 2006;2:453. doi: 10.1371/journal.ppat.0020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grouffaud S, van West P, Avrova AO, Birch PRJ, Whisson SC. Microbiology. 2008;154:3743. doi: 10.1099/mic.0.2008/021964-0. [DOI] [PubMed] [Google Scholar]
- 20.Dean RA, et al. Nature. 2005;434:980. doi: 10.1038/nature03449. [DOI] [PubMed] [Google Scholar]
- 21.Tyler BM, et al. Science. 2006;313:1261. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 22.Kämper J, et al. Nature. 2006;444:97. doi: 10.1038/nature05248. [DOI] [PubMed] [Google Scholar]
- 23.Jiang RHY, Tripathy S, Govers F, Tyler BM. Proc. Natl. Acad. Sci. U.S.A. 2008;105:4874. doi: 10.1073/pnas.0709303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ridout CJ, et al. Plant Cell. 2006;18:2402. doi: 10.1105/tpc.106.043307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sohn KH, Lei R, Nemri A, Jones JDG. Plant Cell. 2007;19:4077. doi: 10.1105/tpc.107.054262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dou DL, et al. Plant Cell. 2008;20:1118. doi: 10.1105/tpc.107.057067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bos JIB, et al. Plant J. 2006;48:165. doi: 10.1111/j.1365-313X.2006.02866.x. [DOI] [PubMed] [Google Scholar]
- 28.Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE. Science. 2006;311:222. doi: 10.1126/science.1120131. [DOI] [PubMed] [Google Scholar]
- 29.van der Hoorn RAL, Leeuwenburgh MA, Bogyo M, Joosten M, Peck SC. Plant Physiol. 2004;135:1170. doi: 10.1104/pp.104.041467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bohnert HU, et al. Plant Cell. 2004;16:2499. doi: 10.1105/tpc.104.022715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vleeshouwers VGAA, et al. PLoS One. 2008;3:e2875. doi: 10.1371/journal.pone.0002875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.We thank S. Kamoun, J. Ellis, and S. Schmidt for inspiring discussions. Work in the lab of R.P. is supported by funds of the Max-Planck Society and the Deutsche Forschungsgemeinschaft (DFG; SFB670). Work in the lab of P.D. is supported by funding from the Australian Research Council, the Australian Grains Research and Development Corporation, and NIH (grant GM074265-01A2).