Abstract
The full complement of homeobox transcription factor sequences, including genes and pseudogenes, was determined from the analysis of 10 complete genomes from flowering plants, moss, Selaginella, unicellular green algae, and red algae. Our exhaustive genome-wide searches resulted in the discovery in each class of a greater number of homeobox genes than previously reported. All homeobox genes can be unambiguously classified by sequence evolutionary analysis into 14 distinct classes also characterized by conserved intron–exon structure and by unique codomain architectures. We identified many new genes belonging to previously defined classes (HD-ZIP I to IV, BEL, KNOX, PLINC, WOX). Other newly identified genes allowed us to characterize PHD, DDT, NDX, and LD genes as members of four new evolutionary classes and to define two additional classes, which we named SAWADEE and PINTOX. Our comprehensive analysis allowed us to identify several newly characterized conserved motifs, including novel zinc finger motifs in SAWADEE and DDT. Members of the BEL and KNOX classes were found in Chlorobionta (green plants) and in Rhodophyta. We found representatives of the DDT, WOX, and PINTOX classes only in green plants, including unicellular green algae, moss, and vascular plants. All 14 homeobox gene classes were represented in flowering plants, Selaginella, and moss, suggesting that they had already differentiated in the last common ancestor of moss and vascular plants.
Keywords: homeobox, homeodomain, transcription factor, plant
Introduction
Homeobox genes encode a typical DNA-binding domain of 60 amino acids, known as homeodomain (HD), that characterizes a large family of transcription factors. The homeodomain folds into a characteristic 3D structure containing three alpha-helices, of which the second and third form a helix-turn-helix motif. The first homeobox genes were isolated from the fruit fly Drosophila melanogaster and were subsequently found to be involved in many aspects of development (for review, see Gehring et al. 1994; Bürglin 2005). Many more homeobox genes have been subsequently identified from all major eukaryotic lineages (Derelle et al. 2007). Based on sequence differentiation and fusion with characteristic codomain sequences, animal homeodomain proteins have been classified into several distinct classes, including TALE, Antp, PRD, SIX, LIM, POU, ZF, CUT, HNF, and PROS (Bürglin 1994, 2005; Holland et al. 2007; Takatori et al. 2008). A distinction has been made between “typical” homeodomains, characterized by a length of 60 amino acids, versus “atypical” ones of different lengths (Bürglin 1994). The latter include a group characterized by homeodomains of 63 aa, with three extra residues inserted between helix 1 and 2 (Bürglin 1995), that have been named TALE (Three Amino acid Loop Extension) homeobox genes (Bertolino et al. 1995; Chen et al. 2003). Both TALE and typical homeobox genes were found to be present in all major eukaryotic lineages including plant, fungi, and animals, suggesting that these two types of homeobox were present in the eukaryote ancestor (Bürglin 1995; Bharathan et al. 1997; Bürglin 1997, 1998a; Derelle et al. 2007). Besides TALE, other homeobox genes of noncanonical length have emerged from the analysis of animal sequences (Bürglin 1997; Bürglin and Cassata 2002). These can be clustered into separate classes of atypical homeobox genes characterized by unique homeodomain insertions and by class-specific codomain architectures, making it apparent that insertions in homeodomain loops have independently occurred multiple times in evolution.
Plant homeodomain proteins have been classified in the literature into various groups based on sequence similarity of their homeodomains and on the presence of characteristic codomains. Bharathan et al. (1997) classified them into seven classes: KNOX and BEL, belonging to the TALE superclass (Bürglin 1997), ZM-HOX, HAT1, HAT2, ATHB8, and GL2. The HAT1, HAT2, ATHB8, and GL2 genes are all characterized by a leucine-zipper motif downstream of the homeodomain (Ruberti et al. 1991) and have been successively renamed HD-ZIP I, HD-ZIP II, HD-ZIP III, and HD-ZIP IV, respectively (Bharathan et al. 1997; Meijer et al. 1997; Aso et al. 1999; Sakakibara et al. 2001). Chan et al. (1998) proposed an alternative classification into five groups (HD-ZIP, GLABRA, KNOTTED, PHD, and BEL).
Although many homeobox genes have been reported from plants, a complete survey and classification of all homeobox genes in plant species from disparate evolutionary groups is lacking. The completion of several high-quality plant genome sequencing projects provided us with the unique opportunity to make a complete assessment and thorough comparative analysis of the homeodomain proteins encoded in plants. The analysis of the full set of homeobox genes in genomes from diverse species allows for a definitive classification of plant homeodomain proteins and an assessment of their origins, evolutionary relations, patterns of differentiation, and proliferation in the various phylogenetic groups. We are interested in finding answers to the following questions: 1) What are the evolutionary relations among plant homeodomain proteins?; 2) How many classes of plant homeodomain proteins can we distinguish in the complete collections from multiple genomes?; 3) When did each class appear in evolutionary time?; 4) What classes are present in which plant groups?; 5) How did each class proliferate in the different plant groups?; 6) Can we classify plant homeodomain sequences based on a characteristic pattern of insertions in homeodomain loops?; 7) Can we characterize homeodomain evolution based on intron–exon structure?; 8) What is the relation of plant homeodomain sequence evolution with the evolution of their codomain architecture?; 9) Can we relate homeobox gene and class presence and differentiation within each genome with the developmental properties of the species bearing them?
Our thorough analyses provide a comprehensive classification scheme and suggest a reconstruction of the evolutionary history of plant homeodomain protein codomain architecture applicable to all plant homeodomain proteins. The results of our study provide a clear picture of the patterns of origin and differentiation of homeodomain classes in different plant groups and suggest interpretations linking their appearance to organism differentiation and increasing developmental complexity. We recognize appearance of all present-day classes of plant homeodomain proteins early in land plant evolution and their differential proliferation into variable numbers of paralogs within each class in different plant groups. We identified and classified in this study a higher number of homeobox genes than previously reported, allowing us to identify several subclasses based on phylogenetic clustering and presence of subclass-specific motifs. We found that, compared with animal homeobox genes, insertions in the homeodomain loops are quite common among plant homeobox genes. We newly characterized several conserved codomain motifs, specific to each of the classes, and predicted their functionality based on sequence similarity to proteins available in databases. The resulting rich data set of plant homeodomain proteins, comprising over 1,000 sequences, is available for download.
Methods
Database Searches and Retrieval of Protein Sequences
Thorough TBlastN searches with several divergent homeodomain proteins of plants and animals were performed to retrieve homeobox genes of Arabidopsis through the TAIR database server (http://www.arabidopsis.org) and of rice through the Gramene database server (http://www.gramene.org). Likewise, analogous searches were performed for the maize genome, through the TAIR maize genome server (http://tigrblast.tigr.org/tgi_maize/) as well as the Plant Genome Database server (http://www.plantgdb.org/PlantGDB-cgi/blast/PlantGDBblast). The plant genome database server was also used to retrieve homeodomain sequences from other plants. The JGI Blast server (http://genome.jgi-psf.org/Poptr1/) was used to retrieve all the Selaginella, moss, and poplar homeobox genes using TBlastN searches against the genomic sequence. Similarly, the TIGR Medicago genome server (http://tigrblast.tigr.org/er-blast/index.cgi?project=mtbe) was used to retrieve Medicago homeodomain sequences. A local database was created using Filemaker Pro 5 (Filemaker, Inc.) to store the retrieved sequences, annotations, accession numbers, expressed sequence tags (ESTs), and chromosomal location. The National Center for Biotechnology Information (NCBI) version of Mac Os X Blast (ftp.ncbi.nih.gov) was installed on a PowerMacG4 computer. The protein sequences in the Filemaker Pro 5 database were exported and reformatted for the local Blast. In order to remove the redundancy in the database, each sequence in the database was blasted against the database, and redundant entries were consolidated. New rounds of BlastP and TBlastN searches of the nr protein and GenBank databases at NCBI restricted to Arabidopsis thaliana using default values were carried out using representative homeodomains of different classes from plants and animals as a query (e.g., POU, LIM, CUT, Antp, HD-ZIP, BEL [Bürglin 1994]). Hits from these searches were checked against data in the local Filemaker database using the “Search” feature (e.g., accession numbers, or N-terminal protein sequences), or a local Blast search of a retrieved hit was performed against the local database. New sequences identified in this fashion were added to the database. A preliminary Neighbor-Joining tree was generated, and PSI-Blast searches with a member of each evolutionary group used as a query were carried out against the nr protein database at NCBI, and iterations were stopped when no new sequences were detected. Every hit for the BlastP, TBlastN, and PSI-Blast searches was manually examined for its potential to be a homeodomain, according to the criteria outlined in the text. No particular e value was taken as an automatic cut-off point, as the goal was to detect as many homeobox genes as possible. In a few instances, Blast matrix and expected value were relaxed to include additional sequences, which were then checked manually for their potential as homeodomain sequences. Arabidopsis thaliana ESTs at NCBI were searched using each entry of our local database as a query using TBlastN. Searches of newly discovered conserved domains/motifs linked to homeobox genes were carried out using BlastP with default values and without species restriction, unless the results would lead to too many hits (e.g., PHD, DDT). All A. thaliana homeodomain protein entries (mislabeled as Hox) in the SMART database were also checked against the local database.
For rice, maize, and poplar, the genomic sequences of the few thousand nucleotides upstream and downstream of the homeodomain were analyzed to predict the complete homeodomain protein sequences using either the MIT Genescan server or the softberry FGENESH+ server, using the most similar plant gene from the blast searches as a guide. In the multiple sequence alignment, if the homeodomain showed obvious misalignment, the most similar plant sequence was used as a guide to correct the homeodomain following a three-frame translation of the sequence and manual determination of potential intron and exon boundaries. A similar procedure was used in some sequences that showed obvious gaps and misalignments within conserved domains upon sequence alignment. In those cases, the genomic sequence was examined, and it was compared either as three-frame translation with a closely related protein sequence or as DNA against a closely related DNA sequence, using the dot matrix program PPCMatrix (Bürglin 1998b). The genomic sequence was translated in all three frames in PPCMatrix and examined for splice sites in the regions where the sequence similarity terminated. In the case of maize, part of the homeodomain could be found in one contig and the other part in a different contig. In this case, manual contig assemblies were carried out and finally confirmed by Blast searches. In several cases, Arabidopsis homeobox genes were repredicted with FGENSH+ using the most similar gene ortholog as a guide to obtain the full-length protein.
The European Molecular Biology Open Software suite was used for pairwise alignment and for locating the homeodomain sequence within a protein sequence. Sequences were submitted also to the SMART and NCBI CDD (Schultz et al. 1998; Marchler-Bauer et al. 2003) to identify conserved domains. These databases did not contain or identify many of the conserved motifs and unusual homeodomain sequences.
The homeodomain DNA from representative members of each class were blasted against the JGI Chlamydomonas project Blast server (http://genome.jgi-psf.org/Chlre3/Chlre3.home.html) to retrieve the Chlamydomonas reinhardtii homeodomain sequences. Similarly, to recover the homeodomain protein sequences of Physcomitrella patens, TBlastN searches were performed at JGI (http://genome.jgi-psf.org/).
Multiple Sequence Alignment and Phylogenetic Analysis
The multiple sequence alignment tool MUSCLE (Edgar 2004) was used for multiple protein sequence alignment. Sequences were further edited and aligned manually, when necessary, using the “Seaview” multiple sequence editor (Galtier et al. 1996). For phylogenetic analyses of conserved domains, sequences were trimmed so that only the relevant protein domains remained in the alignment. Phylogenetic relationships were inferred using the maximum likelihood (ML) method as implemented in PHYML (Guindon and Gascuel 2003). For the ML trees, the JTT (Jones et al. 1992) substitution model was used and results were evaluated with 100 or 1,000 bootstrap replicates. Use of alternative substitution models (WAG, LG) did not affect the results in any of the cases tested (results not shown). The generated trees were displayed using TREEVIEW 1.6.6 (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) and NJPLOT by M. Gouy (http://pbil.univ-lyon1.fr/software/njplot.html).
Secondary Structure Prediction and Protein Homology Modeling
We predicted the secondary structures of several divergent homeodomain sequences using the PHD, JPRED, and Predict Protein servers (Rost and Sander 1993; Cuff et al. 1998). For homology modeling, the crystal structure of the engrailed homeodomain (PDB id: 1enh) obtained from Protein Data Bank (PDB) was used as a template. The aligned sequences were submitted to SWISS-MODEL (http://www.expasy.org/swissmod/) to obtain the 3D structure of some of the atypical homeodomains. The model was viewed in Swiss-PDB Viewer (Kaplan and Littlejohn 2001), and the quality of the model was judged by the phi–psi angle represented in Ramachandran Plots.
Results
We searched homeodomain sequences in several plant genomes (table 1) including the genomes of the red alga Cyanidioschyzon merolae (Rhodophyta), the unicellular green algae Ostreococcus lucimarinus, Ostreococcus tauri and Chl. reinhardtii (Chlorophyta), the moss P. patens (Bryophyta), the spikemoss Selaginella moellendorffii (Lycopodiophyta), and the flowering plants A. thaliana, poplar (eudicots), maize and rice (monocots), using the TBlastN (Gertz et al. 2006) search engine with a variety of homeodomain sequences as queries. In most cases, whenever significant similarity to a homeodomain sequence was identified, the genomic sequence was excised and homology-based gene predictions were performed using the most similar query as a guide. In many instances, regions of conservation were manually reconstructed by multiple sequence alignment of three-frame translations of the genomic sequence including the Blast hits. The results of our extensive database searches are summarized in table 2. In the genome of Arabidopsis (eudicot), we found 110 unique homeobox genes, and in the genome of rice (monocot), we found 117 sequences (110 genes and 7 pseudogenes). Searches in the genomes of poplar (eudicot) and maize (monocot) each yielded 149 homeobox sequences (148 genes and 1 pseudogene) (table 2). In the genome of Selaginella (Lycopodiophyta), we found 45 homeobox genes and 1 pseudogene. In the genome of moss P. patens (Bryophyta), we identified 66 homeobox genes and 1 pseudogene. Among unicellular green algae (Chlorophyta), we found five homeobox genes in the genome of Chl. reinhardtii and seven homeobox genes in the genomes of O. lucimarinus and O. tauri. We retrieved a similar number of homeobox genes (six genes) from the genome of the red alga (Rhodophyta) Cy. merolae. An alignment of HD sequence representatives from one monocot (rice), one dicot (Arabidopsis), moss, and Selaginella, showing sequence features unique to each class, is shown in fig. 1. The alignment of all sequences found in Arabidopsis, moss, Selaginella, unicellular algae, and red algae is shown in supplementary fig. 1 (Supplementary Material online). See supplementary table 1 (Supplementary Material online) for a complete catalog of all sequences identified in the plant genomes analyzed in this study. Several groups of homeodomains do not fit into the typical 60-residue pattern, as they have insertions between helix 1 and helix 2, and/or helix 2 and helix 3 (fig. 1). However, several criteria identify all these sequences as homeodomains: 1) The amino acid sequences fit the profile of amino acids established from 346 animal homeodomains (Bürglin 1994) when the loop regions are removed; 2) The pattern of conservation, that is, which positions are conserved and which ones are not, conforms to the profile established for the animal homeodomains (Bürglin 1994), with positions 16, 20, 48, 49, 51, and 53 highly conserved among plant and animal homeodomains; 3) Secondary structure prediction and homology modeling conform to the homeodomain structure; 4) Blast searches using as a query each newly identified homeodomain confirmed their highest similarity to homeodomain proteins from other plants. All plant homeodomain proteins that we retrieved conserve at least 36% identity with animal homeodomain proteins.
Table 1.
Plant Genomes Analyzed in This Study
| Species | Genome Size (Mbp) | Database | |
| Eudicots | Arabidopsis thaliana (thale cress) | 157 | TAIR |
| Populus trichocarpa (poplar) | 500 | JGI | |
| Monocots | Oryza sativa (rice) | 430 | Gramene |
| Zea mays (maize) | 2,400 | TAIR, PlantGDB | |
| Lycopodiophyta | Selaginella moellendorffii | 110 | JGI |
| Bryophyta (moss) | Physcomitrella patens | 500 | JGI |
| Chlorophyta | Chlamydomonas reinhardtii | 120 | JGI |
| Unicellular green algae | Ostreococcus lucimarinus | 12.5 | JGI |
| Ostreococcus tauri | 11.6 | JGI | |
| Rhodophyta (red algae) | Cyanodioschyzon merolae | 16.5 | Genome project |
Table 2.
Classification of All Homeobox Proteins Retrieved from Plant Genomes
| Eudicots |
Monocots |
Bryophyta | Lycopodiophyta | Unicellular Green Algae |
Red Algae | ||||||
| Super Class | Class | At | Poplar | Rice | Maize | Moss | Sm | Cr | Ol | Ot | Cm |
| HD-ZIP | HD-ZIP I | 17 | 22 | 14 | 24 | 17 | 4 | — | — | — | — |
| HD-ZIP II | 10 | 16 | 14 (2) | 17 | 7 | 2 | — | — | — | — | |
| HD-ZIP III | 5 | 8 | 7 (3) | 8 | 5 | 3 | — | — | — | — | |
| HD-ZIP IV | 16 | 15 | 12 | 21 | 4 | 4 | — | — | — | — | |
| PLINC | 14 | 17 | 11 | 17 | 11 (1) | 5 | — | — | — | — | |
| WOX | 16 | 19 | 15 | 16 | 3 | 6 | — | 1 | 1 | — | |
| TALE | KNOX | 8 | 15 | 12 (1) | 13 | 5 | 5 (1) | 1 | 1 | 1 | 1 |
| BEL | 13 | 19 | 14 | 17 (1) | 4 | 2 | 1 | 2 | 2 | 1 | |
| Uncharacterized | — | — | — | — | — | 6 | 1 | — | — | 2 | |
| DDT | 4 | 7 | 3 (1) | 4 | 3 | 2 | 1 | 1 | 1 | — | |
| PHD | 2 | 4 | 2 | 5 | 2 | 1 | — | — | — | — | |
| NDX | 1 | 2 | 1 | 1 | 2 | 1 | — | — | — | — | |
| LD | 1 | 2 | 1 | 1 | 1 | 1 | — | — | — | — | |
| PINTOX | 1 | 1 (1) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | — | |
| SAWADEE | 2 | 1 | 3 | 3 | 1 | 2 | — | — | — | — | |
| Uncharacterized | — | — | — | — | — | — | — | 1 | 1 | 2 | |
| Total | 110 | 148 (1) | 110 (7) | 148 (1) | 66 (1) | 45 (1) | 5 | 7 | 7 | 6 | |
NOTE.—Number of pseudogenes retrieved from each species is indicated in parentheses. At, Arabidopsis thaliana; Sm, Selaginella moellendorffii; Cr, Chlamydomonas reinhardtii; Ol, Ostreococcus lucimarinus; Ot, Ostreococcus tauri; Cm, Cyanodioschyzon merolae.
FIG. 1.—
Multiple sequence alignment of homeodomain sequence representatives from Arabidopsis thaliana (At), rice Oryza sativa (Os), moss Physcomitrella patens (Pp), and spikemoss Selaginella moellendorffii (Sm). Canonical homeodomain sequence numbering (excluding loops), the TALE class three-residue insertion (abc), and the position of the three homeodomain helices are indicated. The alignments presented in this and other figures were obtained using MUSCLE (Edgar 2004) and conserved amino acids of different physicochemical properties are highlighted in different shades of gray using the Clustal-Qt (Larkin et al. 2007) alignment-drawing software.
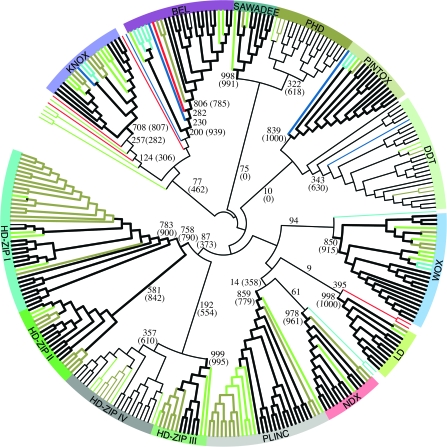
By phylogenetic analyses based on the ML procedure implemented in PHYML (Guindon and Gascuel 2003) and other approaches (see Methods), we found that all plant homeodomain proteins reliably group into 14 distinct classes with robust (generally 70% or more) bootstrap support (fig. 2 and supplementary figs. 2 and 3, Supplementary Material online). Furthermore, from the multiple sequence alignments of full-length homeodomain proteins belonging to the 14 classes, distinctive motifs were found conserved and uniquely associated with each class across monocots, eudicots, Selaginella, and moss and even among unicellular green algae or red algae (fig. 3). Finally, the intron positions within all the 110 homeodomain genes of Arabidopsis were strikingly conserved within each class (fig. 4). Based on previous classifications (see Introduction) and on the results of our analysis, we propose the following classification scheme for the plant homeobox genes.
FIG. 2.—
Circular representation of the evolutionary tree of plant homeodomain sequences (branch lengths not drawn to scale), based on all homeodomain sequences identified in Arabidopsis, Selaginella, moss, unicellular green algae, and red algae supplemented by selected sequences from other flowering plant species wherever only one protein from Arabidopsis was found (see supplementary table 1 [Supplementary Material online] for a complete list of species). The tree, which should be considered unrooted, was obtained with the ML procedure implemented in PHYML with the JTT substitution model using an alignment of homeodomain sequences as shown in supplementary fig. 1 (Supplementary Material online). Bootstrap support was based on 1,000 replicates and is indicated for relevant branches. Bootstrap support obtained after excluding all sequences from unicellular green algae and red algae is shown in parentheses. Clades supported by robust bootstrap values (70% or more) are shown with thicker lines. Bootstrap values for intraclass branches are omitted. Bootstrap values less than 5% for branches connecting different classes are not shown. All homeodomain classes are identified as separate clades in this analysis and are consistently supported by conserved, class-specific domain architecture (see fig. 3) and unique splice junctions (see fig. 4). Red-colored branches indicate sequences from red algae, blue-colored branches refer to the unicellular green alga Chlamydomans reinhardtii, the light-blue color identifies the unicellular green algae Ostreococcus lucimarinus and Ostreococcus tauri, gold-colored branches indicate moss proteins, and a light-green color is used to represent sequences from Selaginella.
FIG. 3.—
Schematic overview of the domain architecture of all 14 classes of plant homeodomain proteins. The following domains and motifs are indicated: HD, leucine-zipper (LZ), CPSCE motif, CESV motif, START domain, homeodomain-START associated domain (HD-SAD), MEKHLA domain, PLINC zinc finger, BEL domain (A & B), KNOX domain (A & B), ELK motif, DDT domain, WSD motif, D-TOX ZF, PEX-PHD, PHD, LUMI, conserved motifs in LD homeodomain proteins (LD1, LD2, LD4, AND LD5). Among DDT proteins, only D-TOX A is indicated with its full symbol; D-TOX B, D-TOX C, D-TOX D, D-TOX E, D-TOX F, and D-TOX G are indicated as B, C, D, E, F or G, respectively.
FIG. 4.—
Intron positions in the homeobox sequences of Arabidopsis thaliana. A consensus (majority) 60-residue homeodomain sequence is shown above a ruler where individual codon base positions are separated by tick marks. Intron positions are indicated by vertical lines labeled by the class names where each intron was found. The intron positions are generally conserved within each class. Exceptions are ATHB1, which has one intron, whereas other HD-ZIP I genes are single-exon genes; and At4g12750, a DDT class member, which has an extra intron at the beginning of the homeodomain. Gene classes not shown (HD-ZIP I, WOX, PLINC) do not have introns in the homeodomain. One of the splice sites in the NDX genes lies in the loop between helix 2 and helix 3 where it cannot be placed accurately on the consensus sequence. Its approximate position is marked by a crossbar at the end of the line.
The HD-ZIP Superclass (HD-ZIP I, HD-ZIP II, HD-ZIP III, HD-ZIP IV)
The HD-ZIP superclass is composed of four individual classes, HD-ZIP I, HD-ZIP II, HD-ZIP III, and HD-ZIP IV (Sessa et al. 1994), all of which are characterized by presence of a leucine-zipper adjacent to the C-terminus of the homeodomain (fig. 3). The HD-ZIP II class is distinguished by a “CPSCE” motif conserved downstream of the leucine-zipper (Chan et al. 1998). The HD-ZIP III and HD-ZIP IV classes were characterized by the START (STeroidogenic Acute Regulatory protein–related lipid Transfer) (Ponting and Aravind 1999) and HD-SAD (START associated conserved domain) (Schrick et al. 2004; Mukherjee and Bürglin 2006) domains. The HD-ZIP III class is distinguished from HD-ZIP IV by an additional conserved C-terminal domain, previously named MEKHLA domain (Mukherjee and Bürglin 2006). Our exhaustive database searches yielded the 48 leucine-zipper homeodomain proteins recently identified in the genome of Arabidopsis (Baima et al. 2001; Henriksson et al. 2005; Nakamura et al. 2006; Ciarbelli et al. 2008). We identified a similar number of leucine-zipper homeodomain proteins (47 genes) in the genome of rice. In the genomes of poplar, maize, Selaginella, and moss, we identified 61, 70, 13, and 33 leucine-zipper homeodomain proteins, respectively (table 2). Thus, the HD-ZIP superclass comprises from 40% to 50% of all homeobox genes of flowering plants and moss (table 2). The classification of HD-ZIP proteins was based on the evolutionary tree relations among HD sequences and was also supported by the association of these sequences with codomains characteristic of the different HD-ZIP classes (see above). From the alignment of all known and newly identified proteins we also discovered that HD-ZIP III proteins were distinguished from other HD-ZIP class genes by four extra residues inserted between helices 2 and 3 of the homeodomain (fig. 1). We propose this insertion as a diagnostic new feature for this class. We could not find any HD-ZIP gene in unicellular green algae or in red algae.
The PLINC Zinc Finger Class
A PLINC zing finger HD class protein was first identified in Flaveria trinervia (Asteraceae) (Windhovel et al. 2001), where it was named ZF-HD for two highly conserved zinc finger–like motifs upstream of the homeodomain (fig. 3). Subsequently 14 members of the same class have been characterized in Arabidopsis (Tan and Irish 2006). In each of the four flowering plant genomes that we analyzed, we identified between 11 and 17 gene sequences belonging to this class, along with 5 genes from Selaginella and 11 genes and 1 pseudogene in moss (table 2). The corresponding proteins were characterized by clustering in the ML tree reconstruction and by the presence of the two diagnostic putative “zinc finger motifs” upstream of the homeodomain (fig. 3). The first putative zinc finger has a consensus sequence C-X3-H-X9-D-X1-C, where H is not always conserved, and the second putative zinc finger has the conserved pattern C-X2-C-X1-C-H-X3-H. We propose to name this class and the corresponding domain “PLINC” (Plant Zinc Finger), to distinguish it from the unrelated zinc finger class of homeodomain proteins, called ZF, found in animals (Bürglin 1994). Most of these proteins were also characterized by the substitution of the usually conserved homeodomain residue F49 with a methionine residue and by insertion of one amino acid between helix 1 and helix 2, which we propose to be two strong diagnostic features of this class. Exceptions were two genes, one from A. thaliana (At1g14687) and one from rice (03g50920), which at the same position showed, respectively, 3 or 6 amino acid insertions (supplementary fig. 1, Supplementary Material online). We could not identify any gene belonging to this class in unicellular green algae or in red algae, suggesting that this class originated within the Streptophyta clade and that it was already present in the last common ancestor of moss and vascular plants.
The WOX Class
The homeodomain proteins of this class have one or two extra residues between helices 1 and 2, and four to five extra residues between helices 2 and 3 (fig. 1). Recently, Haecker et al. (2004), based on their genome-wide search, reported and annotated 14 WOX genes from Arabidopsis. We found in Arabidopsis 16 WOX genes (table 2), characterized by clustering in the evolutionary tree and by distinctive conserved motifs both upstream and downstream of the homeodomain, including the WUS Box (Haecker et al. 2004). The WOX class can be subdivided into several families (supplementary fig. 4, Supplementary Material online). Among these, we found that families such as WOX 11/12 and WOX 8/9 had a C-terminal motif of about 60 aa in length conserved between monocots and eudicots. We found six WOX genes in Selaginella, three in moss, and one WOX gene in the unicellular green algae O. lucimarinus and O. tauri (fig. 2 and supplementary fig. 1, Supplementary Material online) but none in Chl. reinhardtii, indicating that the WOX class of proteins was present in the last common ancestors of green plants (Chlorobionta) and was successively lost in the Chlamydomonas lineage.
The TALE Superclass (KNOX and BEL)
TALE homeodomain proteins are characterized by three extra residues between helix 1 and helix 2 (Bertolino et al. 1995; Bürglin 1997). In plants, TALE genes have been extensively studied and classified into the two classes KNOX and BEL. Among the homeobox gene classes, KNOX and BEL appeared to be the oldest classes, with members present in single-cell green algae and in red algae (fig. 2).
Members of the KNOX class have been previously classified into two families, KNOX I and KNOX II (Kerstetter et al. 1994). We found eight members of the KNOX class in A. thaliana, 15 in poplar, 12 (and one pseudogene) in rice, 13 in maize, 5 (and 1 pseudogene) in Selaginella, 5 in moss, and a single unambiguously classified gene in unicellular green and red algae (fig. 2). Five additional genes from Selaginella, one from Chlamydomonas, and two from the red alga clustered with the TALE class. However, they did not confidently cluster within the KNOX or BEL classes (fig. 2), and we could not recognize in their sequence a typical KNOX or BEL codomain architecture. From the analysis of the codomain structure of KNOX genes from flowering plants, we could further classify KNOX I genes into two subclasses, KNAT 2/6 and KNAT1, while the KNOX II class could be separated into the two subclasses KNAT7 and KNAT3/4/5 (fig. 5). Except for the STM subclass, which appeared to be dicot specific, all other subclasses had representatives from monocots and eudicots, suggesting that they were present in flowering plants prior to the separation of the two groups. KNOX gene representatives from both Selaginella and moss clustered within the KNOX I or the KNOX II subclasses (fig. 5), confirming the results of a recently published evolutionary analysis (Sakakibara et al. 2008) and suggesting that both KNOX I and KNOX II genes were present in the common ancestor of moss and vascular plants. Both KNOX families conserve a diagnostic KNOX domain upstream of the homeodomain, composed of two blocks (KNOX A and KNOX B) separated by a variable region (Bürglin 1997), as well as a shorter motif adjacent to the homeodomain, named ELK (Vollbrecht et al. 1991). The multiple sequence alignment of KNOX I and KNOX II proteins (supplementary fig. 5, Supplementary Material online) showed that in KNOX II proteins the KNOX B subdomain is characterized by a 30-aa insertion. In addition, we could newly identify in all KNOX II genes a highly conserved motif consisting of 20 amino acids that we named “KNOX II C” (supplementary fig. 5, Supplementary Material online).
FIG. 5.—
ML tree obtained using the homeodomain and codomain sequence of KNOX class proteins. KNOX class can be subdivided into two families: KNOX I and KNOX II. Each of these can be further subdivided into two subfamilies having members conserved in both monocots (light-green boxes) and eudicots (light-blue boxes). Selaginella (Sm) proteins are shown in yellowish green–colored boxes, whereas moss (Pp) proteins are shown inside the gold-colored boxes. Each subfamily harbors distinct signature motifs, schematically represented next to the subfamily name. The tree has been rooted with BEL class protein representatives.
The KNOX domain shares similarities with the MEIS and PBC domains of animal TALE homeobox genes, suggesting that these domains derived from a common ancestral domain that has been named MEINOX (Bürglin 1997, 1998a). The ELK motif, situated between the KNOX and HD domains of KNOX class proteins, is in a position analogous to the PBC-B motif of animal PBC class homeobox genes. From sequence analysis we found that the ELK domain showed similarities with the PBC-B motif, including highly conserved hydrophobic residues, and that both motifs are predicted to fold into helical conformations (fig. 6). This reinforces the notion that genes of the PBC class are also derived from an ancestral MEINOX TALE homeobox gene (Bürglin 1998a). We propose that the ELK domain, as well as the C-terminal end of the PBC-B domain, should be considered part of the MEINOX domain.
FIG. 6.—
Multiple sequence alignment of the ELK motif from KNOX proteins and of the C-terminal part of the PBC-B domain from PBC proteins showing the sequence similarity and likely homology of the two regions. Helices predicted from the alignments of PBC or KNOX sequences are shown above the alignment. At each position conserved amino acid types with similar physicochemical properties are highlighted in different shades of gray.
The BEL class of proteins is uniquely characterized by two conserved domains upstream of the homeodomain, called SKY and BEL (Bellaoui et al. 2001; Becker et al. 2002). We identified 13–19 BEL genes in flowering plants, 2 genes in Selaginella, 4 genes in moss, 1 or 2 genes in unicellular green algae, and 1 gene in red alga (table 2). By ML evolutionary tree reconstructions based on the homeodomain and codomain amino acid sequences and by similarities in codomain architecture, we could distinguish within the BEL class five newly classified subclasses, common to monocot and eudicot flowering plants (supplementary fig. 6, Supplementary Material online). In addition to SKY and BEL, we have detected in this class a third highly conserved 10-aa motif, which we named “ ZIBEL”, repeated at both the C-terminal and N-terminal ends of the BEL proteins (fig. 7, panel a). This motif was conserved in all BEL subclasses except at the C-terminal end of the ATH1 subclass, where the SKY domain was also missing (supplementary fig. 6, Supplementary Material online). Furthermore, we also found the ZIBEL motif at the N-terminal end of HD-ZIP II proteins (fig. 7, panel a).
FIG. 7.—
Multiple sequence alignment of (a) ZIBEL motif sequences identified at the N-terminus and C-terminus of BEL class homeodomain proteins and at the N-terminus of HD-ZIP II class proteins and (b) the BEL-A (SKY) region of plant BEL class proteins and the MEIS-A domain of animal MEIS class proteins. Secondary structure predictions of BEL-A and MEIS proteins are shown above and below the sequence alignment, respectively. At each position conserved amino acid types with similar physicochemical properties are highlighted in different shades of gray.
Considering that the KNOX, PBC, and MEIS domains are all bipartite, with a variable-length linker region connecting two conserved regions, we propose that a corresponding BEL domain should also be redefined as a bipartite domain composed of a conserved N-terminal domain (BEL-A), also including the SKY motif, and a conserved C-terminal domain (BEL-B). This definition is also consistent with the observation that both parts of the newly defined BEL domain are necessary for interaction with KNOX homeodomain proteins (Bellaoui et al. 2001; Smith et al. 2002). We also identified in all BEL subclasses except BLR a third conserved motif, that we named BEL-C, situated between the BEL-B and the homeodomain sequences (supplementary fig. 6, Supplementary Material online). No obvious conservation had been so far described between the BEL and MEINOX domains (Becker et al. 2002). However, when the redefined BEL domain was aligned with the MEIS domain, the BEL-A region aligned with the MEIS-A domain, with two adjacent leucine residues highly conserved in BEL-A and in the MEIS-A domain within a segment predicted in both classes to be helical (fig. 7, panel b).
New Classes of Plant Homeodomain Proteins
The DDT Class
One group of homeodomain proteins is characterized by the presence of the DDT domain (Doerks et al. 2001), located downstream of the homeodomain. Our extended searches and evolutionary analyses allowed us to newly define this group of proteins as a separate class of HD proteins. This group encompasses the largest plant homeodomain proteins, with sequences of lengths up to about 1,900 amino acids. We identified three to seven genes encoding a DDT domain in flowering plants, two genes in Selaginella, three genes in moss, and one gene in each genome of unicellular green algae (table 2). No DDT class genes were identified in red algae. In addition to the DDT domain, from the alignment of these proteins we identified seven other conserved motifs, distributed throughout the entire length of the protein, which we named D-TOX A to G (DDT Homeobox Class Domain, fig. 3). The newly identified D-TOX F motif is a zinc finger motif highly conserved between monocots and eudicots, characterized by the pattern C-X2-C-X10-C-X2-C. To distinguish it from other zinc finger domains, we propose to name the newly identified zinc finger motif “D-TOX ZF” (fig. 8, panel a). Using D-TOX E as a query in database searches, we identified other proteins from rice, Arabidopsis, and slime mold that did not contain the homeodomain but conserved the D-TOX E and the DDT domains, with 36–43% amino acid identity. We also identified similarity with the animal Williams-Beuren syndrome transcription factor proteins, which also contain the DDT domain, as well as PHD and bromo domains, with 25–28% sequence identity (fig. 8, panel b). We propose to uniquely identify the D-TOX E motif as the Williams-Beuren syndrome DDT (WSD) motif. We classified genes belonging to the DDT class by phylogenetic analyses and codomain architecture into three subclasses (D-TOX1, D-TOX2, D-TOX3), of which D-TOX1 and D-TOX2 are represented in both eudicots and monocots, whereas D-TOX3 is eudicot specific (fig. 9). The D-TOX3 subclass has lost all motifs characteristic of the DDT class of genes except for the D-TOX A motif. The DDT class of genes appears to have evolved early in green plant (Chlorobionta) evolution because members of this class were found in unicellular green algae and in land plants (fig. 2).
FIG. 8.—
Multiple sequence alignment of (a) the zinc finger motif (D-TOX ZF) uniquely found in the plant DDT class. Conserved cysteine residues are highlighted as “C.” (b) the WSD motif of the DDT class. At each position conserved amino acid types with similar physicochemical properties are highlighted in different shades of gray. Alignment of the WSD motif of the DDT class found in plant and animal sequences showing the regions of highest similarity (boxed residues).
FIG. 9.—
ML tree obtained using the homeodomain and associated signature codomains of DDT class proteins, outgrouped by PINTOX class proteins (At_At5g11270 and St_Pint1). Proteins from monocots are shown within light-green–colored boxes; from eudicots in light-blue–colored boxes; from moss in musky-green–colored boxes, and from Selaginella yellowish green–colored boxes. The DDT class can be subdivided into three families: D-TOX1, D-TOX2, and D-TOX3. D-TOX3 is eudicot specific and has secondarily lost all signature codomains found in D-TOX1 and D-TOX2 with the exception of the D-TOX A codomain. A DDT protein from Chlamydomonas reinhardtii (Cr_C460085), three proteins from moss (Pp_sca_419, Pp_sca_15b and Pp_sca_89), and two proteins from Selaginella (Sm_422900 and Sm_447019) cannot be classified with certainty within any of the three families.
The PHD Class
The pathogenesis-related homeobox gene A (PRHA) (Plesch et al. 1997) and HAT3.1 gene (Schindler et al. 1993) from Arabidopsis were characterized by the presence of the PHD codomain, located several hundred amino acids upstream of the homeodomain. We found from two to five PHD genes encoded in flowering plant genomes, one gene in Selaginella, and two genes in moss. We did not detect any PHD HD gene in unicellular green algae or in red algae. These genes formed a distinct clade in the ML tree reconstruction (fig. 2), which, together with the diagnostic presence of the PHD domain, allowed us to newly define the class of PHD homeodomain proteins. Adjacent to the PHD domain proteins of this class lies an additional conserved motif of about 90 aa, which we called PEX-PHD (fig. 3). This region, which we found only in plant PHD homeodomain proteins, is rich in charged residues. Other smaller conserved motifs are found mainly between the PHD domain and the homeodomain (fig. 3).
The ML evolutionary tree obtained from the alignment of the homeodomain and PHD sequence regions suggests that PHD proteins divide into two distinct subclasses (supplementary fig. 7, Supplementary Material online), named after the corresponding Arabidopsis genes: PRHA, with a high bootstrap support (98%), and HAT3.1, with a lower support (58%). Both subclasses are represented in all flowering plant genomes examined, but only proteins classified in the PRHA group are found in moss (two genes) and in Selaginella (one gene). The most parsimonious scenario is that the HAT3.1 subclass evolved in the flowering plant lineage after separation of moss and Selaginella. However, the topology of the rooted trees (fig. 2 and supplementary fig. 7, Supplementary Material online) suggests that both subclasses were already present before separation of the moss and Selaginella lineages and that HAT3.1 genes were successively lost from these genomes.
The NDX Class
Homeobox genes expressed in the nodule (Nodulin Homeobox genes, NDX) were first reported by Jørgensen et al. (1999) from soybean and from Lotus japonicus. We identified two NDX genes in the genomes of poplar and moss; one NDX gene in A. thaliana, rice, maize, and Selaginella; and none in unicellular green or red algae, suggesting that this class appeared early in the evolution of the land plant clade before separation of moss and vascular plants. We also searched NDX genes in several other unfinished genome sequences and EST collections of other plant species. A multiple sequence alignment of the corresponding proteins is presented in supplementary fig. 8 (Supplementary Material online). The homeodomains of the NDX class are atypical and highly divergent. We found that NDX proteins have a diagnostic insertion of six amino acids between helices 2 and 3 (fig. 1 and supplementary fig. 1, Supplementary Material online), conserved in moss, Selaginella, and flowering plants. From the alignment of moss and flowering plant sequences, we also newly identified two additional motifs, which we named NDX-A and NDX-B. NDX-A was a 540-aa-long domain located upstream of the homeodomain. NDX-B was an 80-aa-long domain located downstream of the homeodomain. Both domains are highly conserved among flowering plants and in moss (supplementary fig. 8, Supplementary Material online).
The LD Class
A homeobox gene called LD (Luminidependens) has been previously identified in Arabidopsis and in maize (van Nocke et al. 2000). In addition to the genes from these species, we found a single copy of LD genes in rice, maize, Selaginella, and moss and two copies of the gene in poplar. We did not find any LD gene in unicellular green algae or in red algae (table 2). The distribution of LD genes suggests that they were present in the last common ancestor of moss and vascular plants and that they evolved after the divergence of Chlorophyta and Streptophyta. The homeodomain of this class is of typical length, but it is characterized by several unusual substitutions of otherwise conserved residues, such as W48F and N51X (fig. 1). The sequence alignment of LD proteins revealed five conserved codomains, which we named LD1–LD5 (supplementary fig. 9, Supplementary Material online). PSI-Blast database searches revealed that the 80-aa-long LD3 region was also conserved within an unrelated group of plant transcription factors (fig. 10, panel a). We refer to this newly identified conserved domain as the “LUMI domain.”
FIG. 10.—
Multiple alignments of (a) the LUMI domain of the LD class; (b) the PINTOX domain of PINTOX class proteins; and (c) the SAWADEE domain of SAWADEE class proteins. The position of conserved cysteine and histidine residues is highlighted below the alignment. At each position conserved amino acid types with similar physicochemical properties are highlighted in different shades of gray.
The PINTOX Class
Homeodomain proteins of the PINTOX class formed another distinct clade with a strong bootstrap support of 84% (fig. 2). We named this class PINTOX from one of the genes represented in this class, the Plant Interactor Homeobox rice gene GF14c-int., which has been isolated and named for its interaction with GF14-c in a two-hybrid screen (Cooper et al. 2003). We newly identified at least one full-length gene as belonging to this class in all other examined green plant genomes, including Chl. reinhardtii and other unicellular green algae, indicating that PINTOX genes originated before the divergence of Cholorophyta and Streptophyta. PINTOX homeodomain proteins are characterized by the substitution N51D within the homeodomain region (fig. 1) and by a highly conserved basic domain of about 70 aa (named PINTOX domain) newly identified upstream of the homeodomain (fig. 10, panel b). Further upstream of the PINTOX domain is a conserved acidic domain, which we named “Acid Pint” (fig. 3), whereas the N-terminal region has conserved hydrophobic and basic residues.
The SAWADEE Class
In all examined genomes of flowering plants, in Selaginella and in moss we identified from one to three genes that clustered with high (99%) bootstrap support in the ML evolutionary tree (fig. 2) into a newly identified class that we named SAWADEE. These genes are characterized by diagnostic insertions of 10 aa between the second and third homeodomain helices (fig. 1), extending the second loop more so than in any other plant homeodomain protein. C-terminal to the homeodomain we identified a 130- to 140-aa-long conserved region, the SAWADEE domain (fig. 10, panel c). We found homologs of the SAWADEE domain also in non-HD genes conserved among monocots and eudicots (fig. 10, panel c). The SAWADEE domain is characterized by several conserved cysteine and histidine residues, suggesting that it may be involved in metal coordination.
Discussion
By exhaustive searches of complete genomes and other sequence data we have identified the full repertoire of homeodomain proteins found in diverse plant species, improving previous reports of homeobox genes in A. thaliana and in rice, and newly identifying the complete set of homeobox genes from the genomes of poplar and maize (eudicot and monocot flowering plants); spikemoss S. moellendorffii (Lycopodiophyta); moss P. patens (Bryophyta); the unicellular green algae O. lucimarinus, O. tauri, and Chl. reinhardtii (Chlorophyta); and the red alga Cy. merolae (Rhodophyta). From these analyses we obtained a comprehensive perspective of the evolutionary history of plant homeobox genes and gene classes and a reliable, exhaustive, nonredundant source catalog of plant homeodomain protein sequences. Our searches expanded the number of plant homeobox genes previously reported in the literature. In Arabidopsis we identified 110 genes versus the 87 homeobox genes previously reported (Shiu et al. 2005). In rice, the most recent report identified 107 homeobox genes (Jain et al. 2008). From this collection we excluded eight zinc finger domain proteins that did not contain a conserved homeodomain, and we added 11 other homeobox genes not previously reported. Among rice homeobox genes, 31 sequences have been recently identified as HD-ZIP genes (Agalou et al. 2008). In our analysis of the rice genome, we identified 47 HD-ZIP genes and 5 HD-ZIP pseudogenes. Although the genome of rice (400–430 Mb) is more than three times larger than the genome of Arabidopsis (125 Mb), the total number of its predicted genes (37,544 predicted genes) is only 34% more than the number estimated in Arabidopsis (28,000 predicted genes). We identified in both genomes 110 homeobox genes. To our knowledge, no data were previously available on the number of homeobox genes in the genomes of poplar and maize. Although poplar and maize have genomes of substantially different sizes (550 Mb and 2,900 Mb, respectively), they are thought to encode a similar total number of genes (58,000 and 54,000 predicted genes, respectively). Consistent with their greater total number of predicted genes compared with Arabidopsis or rice, we identified in both genomes 148 homeobox genes (vs. 110 genes in Arabidopsis and rice). However, in the genome of the moss P. patens (Bryophyta), with a genome size of 500 Mb and 30,000–35,000 estimated genes, we found only 66 homeobox genes, whereas we identified 45 homeobox genes in the 110-Mb genome of Selaginella. In unicellular algae we identified a much smaller number of homeobox genes. Although the unicellular green alga Chl. reinhardtii has a genome size similar to Arabidopsis at 121 Mb and about half as many predicted genes (15,000 genes), we identified in its genome only five homeobox genes. Similarly, the other unicellular green algae O. lucimarinus and O. tauri, with a genome size of about 13 Mb and 7,700–7,900 predicted genes, contained only seven homeobox genes each. Finally, six homeobox genes were found in the genome (of 16.5 Mb and 5,331 predicted genes) of the red alga Cy. merolae (Rhodophyta). Expression of about half of the homeobox genes of Arabidopsis has been experimentally verified. Searching EST databases we found expression data for 92 Arabidopsis homeobox genes (84%), and sequence conservation suggests that all 110 genes play a biological role.
The total number of homeobox genes found among plant genomes somewhat reflects the complex patterns of genome duplication and gene duplication or loss that have characterized the evolutionary histories of these species. Three genome duplications are recognized in the evolution of A. thaliana, poplar, and maize, two duplications in rice (see Adams and Wendel [2005] for a review), and one duplication in moss (Rensing et al. 2008). These duplications are likely to be related to the increased number of homeobox genes found in flowering plants compared with moss. However, conservation of a greater number of genes in flowering plants is also likely to be related to the increased developmental and organizational complexity of these species.
New Classes and Conserved Motifs
Based on our analysis, we propose that plant homeobox genes can be classified into 14 classes (HD-ZIP I to IV, PLINC, WOX, NDX, DDT, PHD, LD, PINTOX, SAWADEE, KNOX, and BEL), of which four are grouped into the HD-ZIP superclass (HD-ZIP I to IV) and two (KNOX and BEL) belong to the TALE superclass. All homeobox genes found in the genomes of flowering plants, Selaginella, and moss were unambiguously clustered by evolutionary tree reconstructions into 14 distinct groups (classes) with generally high bootstrap support (70% or more). Each class was additionally supported by distinctive signature motifs or codomains and by typical intron–exon structures. Among these, the classes PHD, DDT, NDX, LD, SAWADEE, and PINTOX are here for the first time defined. We discovered and characterized numerous new motifs, which appeared to be highly conserved in moss and flowering plants, and, whenever present, also in unicellular green and red algae, suggestive of a common, evolutionarily conserved functional role. Notable examples are the LUMI domain, the ZIBEL motif, the WSD motif, and the PINTOX domain. The ZIBEL motif is conserved at both the N-terminal and C-terminal ends of the BEL class of proteins and at the N-terminal end of the HD-ZIP II class of proteins. We speculate that through the ZIBEL motif the HD-ZIP II and BEL homeodomain proteins may interact with each other or may interact with the same target proteins. We found that the WSD motif of plant DDT proteins shows high sequence similarities with a protein that causes Williams-Beuren syndrome (WSTF) in humans (Cus et al. 2006), suggesting that the WSD motif was already present in the common ancestor of plants and animals. The WSTF gene was found to play an important role in chromatin remodeling and was associated with other motifs such as the PHD, DDT, and bromo domains (Bozhenok et al. 2002; Poot et al. 2004). Similarly, the PINTOX domain was also found conserved in unicellular green algae, moss, and vascular plants. PINTOX proteins have been recently found to exert resistance against necrotrophic fungal pathogens (Coego et al. 2005). We have also characterized a novel zinc finger motif of Cys2/Cys2 type in the DDT class of proteins that we named D-TOX ZF. This type of zinc finger DNA-binding domain is common among glucocorticoid hormone receptors and also among GATA transcription factors, suggesting a possible common functional role in this group of DDT proteins. A typical characteristic of the Cys2/Cys2 type zinc finger domain is its ability to interact with DNA as well as with proteins, allowing it to play an essential role in chromatin rearrangement (Gronenborn 2004; Matsushita et al. 2007). Specifically, the DDT domain is known to bind DNA and to take part in chromosome remodeling (Doerks et al. 2001). It is also found to be associated with codomains, such as the PHD, bromo, MBD, and ring finger domains. Little experimental information on the function of the SAWADEE homeodomain proteins is available, and their roles can only be speculated from sequence features and expression data. Conservation of basic, cysteine, and histidine residues suggest that SAWADEE may be a DNA-binding domain. EST data indicate that SAWADEE is expressed in roots, leaves, developing seeds, embryos, flower buds, and endosperm.
Some of the newly identified domains, as well as previously known domains, are also found fused with other nonhomeodomain proteins (DDT, LUMI, PHD, and WSD) or as independent proteins (KNOX, SAWADEE, PLINC, START, and HD-SAD). It is possible that these proteins act as negative regulators of the corresponding HD proteins by competing for protein–protein interaction or DNA-binding sites. This has been recently shown in the case of the non-HD, KNOX domain–containing protein PTS of tomato, which competes for protein–protein interaction sites with the HD proteins KNOX I and BIP (Kimura et al. 2008).
Evolutionary History of Plant Homeobox Genes
Identification of the different classes and codomains of homeodomain proteins in various plants and other evolutionary groups allows us to coherently reconstruct the overall evolutionary history of this protein family in plants, summarized in fig. 11. The overall picture that emerges from our analysis is that the family of homeobox genes has expanded and differentiated together with the differentiation of plants into organisms of increasing complexity. The functional differentiation of homeodomain proteins appears to have first been achieved through the acquisition of different codomain architectures. This stage had already achieved its current state during the evolution of land plants before separation of moss and vascular plants, when all 14 different codomain architectures represented in modern homeodomain classes must have been already present. Further specialization was successively achieved by the proliferation of gene paralogs within each class, as observed in modern flowering plants. It must be noted that our interpretation of a progressive increase in the number of homeobox genes in the lineage leading to flowering plants could be equivalently interpreted as corresponding loss of genes in the respective lineages. Although this interpretation is in principle coherent, we find the alternative view of a progressive lineage-specific expansion of the homeobox gene family more plausible because 1) it is more parsimonious, requiring for each new gene one lineage-specific duplication event rather than one duplication in a common ancestor plus one or more lineage-specific gene loss(es), and 2) it correlates with a corresponding increase in organism developmental and functional complexity. Nevertheless, some of the results suggest gene loss events. For example, a representative of the WOX class is found in the genome of the Ostreococcus unicellular green algae, but not in the other green alga Chl. reinhardtii.
FIG. 11.—
Schematic representation of the proposed evolutionary history of plant homeobox gene classes and codomains. A class or motif represented in a parental branch indicates that the same class/motif is also present in all of its child-branches unless otherwise indicated. The HD-ZIP I to IV classes are represented by the single domain architecture HD-LZ-CPSC-START-HD-SAD-MEKHLA. In this representation, arrows separate motif groups whose addition defines, in the order, HD-ZIP classes I, II, IV, and III. Loss of the START domain in the genomes of unicellular green algae and red algae is represented by the domain crossed in red. Acquisition of the MEKHLA domain through the cyanobacteria/chloroplast is indicated by an arrow.
HD Classes and Domains Common to All Examined Plant Groups (Rhodophyta, Chlorophyta, Moss, and Vascular Plants)
The two classes of TALE HD proteins found in plants, KNOX and BEL, are characterized by a conserved codomain structure that is also recognized in two of the several classes of animal TALE HD proteins, MEIS and PBC. This suggests that these proteins derived from a common ancestor, and their codomain was named MEINOX (Bürglin 1997). The duplication of MEINOX genes observed in animals and plants has been previously described as two independent events in the plant and animal lineages (Bürglin 1998a). However, the presence of a common intron insertion position in the plant BEL class and in the animal PBC class but not in the KNOX (plant) and MEIS (animal) classes suggests that their common ancestor duplicated and that one intron was acquired by one of the two gene copies before separation of animals and plants. In either case, KNOX and BEL HD proteins must have been already present in the last common ancestor of the plant clade. Typical (non-TALE) HD proteins are also found in animals, plants, including unicellular green and red algae, and many other anciently diverged eukaryotic groups (Derelle et al. 2007), suggesting that TALE and non-TALE HD proteins were already present in the common ancestor of plants and animals. However, none of the animal non-TALE HD proteins can be reliably associated with a specific non-TALE plant class. The START, PHD, and DDT-WSD protein domains are found associated with HD in modern land plants and associated with other proteins (PHD and DDT-WSD) or absent (START) in unicellular green and red algae. Because these domains are also found in animals not associated with HD, we most parsimoniously predict that they were present but separated from HD in the common ancestor of plants and animals (fig. 11). Among these domains, START is not found in unicellular green algae and red algae, suggesting that it has been secondarily lost in these groups.
Appearance of Three New HD Classes in the Common Ancestor Lineage of Green Plants
In moss, vascular plants, and unicellular green algae, we found the DDT-WSD domain associated with HD, identifying the presence of the homeodomain protein DDT class in the last common ancestor of green plants (Chlorobionta). We also infer that at this time HD was also associated with the green plant–specific PINTOX and WUS motifs, giving rise to the corresponding PINTOX and WOX classes. The evolution of green plants can then be associated with the addition of three more HD classes to the KNOX and BEL classes present in all plants. Presumably each of these classes was originally represented by one gene, as observed in modern-day unicellular green algae. In the genome of Chl. reinhardtii, we found the bacterial PAS-like MEKHLA domain, so far found associated with HD in land plants, including the liverwort Marchantia polymorpha, and in the charophycean green alga Chara corallina (Floyd et al. 2006). We speculated that this domain was acquired from the cyanobacterial genome of the chloroplast at the onset of the green plant clade (Mukherjee and Bürglin 2006).
Multiple HD Genes and HD Classes in Embryophyta (Land Plants)
Embryophyta are characterized by the appearance, at least in the lineage leading to moss and vascular plants, of nine classes of HD proteins not present in Rhodophyta or Chlorophyta, namely HD-ZIP I to IV, LD, SAWADEE, NDX, PHD, and PLINC. These classes are identified by fusion of HD with codomains (PHD, START, MEKHLA, CPSC, HD-SAD, LD, SAWADEE, NDX, and PLINC). All HD classes not found in Rhodophyta and Chlorophyta were found both in moss and in flowering plants, suggesting that they originated early in land plant evolution (at least before separation of moss and vascular plants and possibly before transition to land) in relation to newly acquired developmental features. In fact, a full-length HD-ZIP III gene with all functional domains conserved has been recently detected also in Cha. corallina (Floyd et al. 2006), a freshwater green alga belonging to the charophycean group Charales, believed to be a sister group to land plants (Karol et al. 2001; McCourt et al. 2004). This suggests that at least HD-ZIP III homeobox genes (and presumably all HD-ZIP homeobox gene classes) must have been present in the last common ancestor of Charales algae and land plants. Interestingly, no new HD classes (defined by their codomain associations) developed in the evolution of vascular plants or moss after their separation. Instead, further complexity and functional differentiation have been achieved, at least in flowering plants and in moss, through a great proliferation of gene paralogs within each of the original 14 classes.
Rooting the Tree of Plant Homeodomain Proteins
The evolutionary trees based on homeodomain sequences (fig. 2 and supplementary figs. 2 and 3, Supplementary Material online) robustly identify the different classes of plant homeodomain proteins, which are also fully supported by their characteristic codomain architectures. However, the trees do not provide reliable information on the evolutionary relations among most of the classes, which are connected by poorly supported topologies. These are also contradicted by the partial information provided by intron insertion analysis, including two parsimony-informative intron insertion sites (fig. 4). One intron insertion site is conserved between the non-TALE classes LD, SAWADEE, and PHD. The same intron insertion site is also found in the TALE class BEL and in some animal and fungus TALE HD classes—PBC, TGIF, IRO (Bürglin 1997), and in MKX (Mukherjee K, unpublished data). However, this intron is not present in KNOX, the other plant TALE class. KNOX instead has an intron insertion position in common with the non-TALE class PINTOX, neither present in BEL nor in any animal or fungus HD gene. This arrangement would question the monophyletic origins of plant TALE as well as of plant non-TALE proteins. The evolutionary significance of these few common intron insertions is, however, dubious because we do not know the likelihood of parallel intron insertion or deletion events in different lineages. For example, we also find an intron insertion position conserved between the SAWADEE class and the Arabidopsis gene ATHB1, which clearly belongs to the HD-ZIP I class, suggesting homoplasy or multiple intron losses in different classes. In fig. 12 we show a summary representation of the results from the trees of fig. 2 and supplementary figs. 2 and 3 (Supplementary Material online), where each class is represented by a single branch, and the topology of branches connecting TALE or non-TALE classes with a bootstrap support of less than 50% is replaced by circles representing their uncertainty. Sequence representatives of animal TALE and non-TALE proteins cluster with plant TALE and non-TALE proteins, respectively (supplementary fig. 3 [Supplementary Material online] and Derelle et al. [2007]), consistent with previous observations that both types of homeodomain proteins are found in anciently diverged eukaryotic groups (Derelle et al. 2007) and must have been present early in eukaryote evolution. However, TALE proteins from plants, animals, or any other phylogenetic group cannot be used as an outgroup to root the subtree of plant non-TALE proteins unless we assume that all modern classes of plant non-TALE homeodomain proteins originated monophyletically after separation of TALE and non-TALE genes. With this assumption, the root of plant non-TALE proteins would lie in the branch connecting the two types (bold faced in fig. 12). If this is not the case (i.e., some non-TALE classes separated before TALE genes separated from other non-TALE classes), the root of all non-TALE proteins should be positioned within the circle of uncertain topology shown in fig. 12 connecting the different non-TALE plant classes. Similar arguments apply to the rooting of TALE proteins, although conservation of their signature insertion suggests that all TALE genes have a monophyletic origin, a notion that is challenged, however, by the pattern of intron conservation (see above). The two TALE gene classes found in plants resulted either from a duplication in the common ancestor of plants and animals or after separation of the two phyla (Bürglin 1997).
FIG. 12.—
Summary representation of the trees shown in fig. 2 and supplementary figs. 2 and 3 (Supplementary Material online), where sequences from each class are represented by one branch and the low–bootstrap support (<50%) branches connecting different TALE or non-TALE classes are hidden by circles representing uncertainty of their relations.
Throughout plant evolution, the homeobox gene family has proliferated and diversified in accordance with the growth in structural and developmental complexity of the organisms in which they were expressed. Some homeodomain proteins have been intensely studied, but little or nothing is known about the functionality of many other homeobox genes and classes. In this study of multiple plant genomes, we newly uncovered HD protein classes and a greater abundance of homeobox genes than previously known. We also identified many previously unnoticed conserved motifs whose specific role in protein–protein or protein–DNA interaction remains to be experimentally verified. Our findings provide a rich data set for future experimental analyses and characterizations.
Supplementary Material
Supplementary figures 1–9 and table 1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We thank Maria Thalén and Tobias Klingenfuss for the initial compilation of sequences, Shaina Wallach and Samir Saha for critical reading of the manuscript, and two anonymous reviewers for their constructive suggestions. T.R.B. was supported by the Swedish Foundation for Strategic Research and the Karolinska Institutet, Södertörns Högskola. L.B. was supported by the University of Florida Genetics Institute.
References
- Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Agalou A, Purwantomo S, Overnas E, et al. (14 co-authors) A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol Biol. 2008;66:87–103. doi: 10.1007/s11103-007-9255-7. [DOI] [PubMed] [Google Scholar]
- Aso K, Kato M, Banks JA, Hasebe M. Characterization of homeodomain-leucine zipper genes in the fern Ceratopteris richardii and the evolution of the homeodomain-leucine zipper gene family in vascular plants. Mol Biol Evol. 1999;16:544–552. doi: 10.1093/oxfordjournals.molbev.a026135. [DOI] [PubMed] [Google Scholar]
- Baima S, Possenti M, Matteucci A, Wisman E, Altamura MM, Ruberti I, Morelli G. The arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 2001;126:643–655. doi: 10.1104/pp.126.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A, Bey M, Bürglin TR, Saedler H, Theissen G. Ancestry and diversity of BEL1-like homeobox genes revealed by gymnosperm (Gnetum gnemon) homologs. Dev Genes Evol. 2002;212:452–457. doi: 10.1007/s00427-002-0259-7. [DOI] [PubMed] [Google Scholar]
- Bellaoui M, Pidkowich MS, Samach A, Kushalappa K, Kohalmi SE, Modrusan Z, Crosby WL, Haughn GW. The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell. 2001;13:2455–2470. doi: 10.1105/tpc.010161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino E, Reimund B, Wildt-Perinic D, Clerc RG. A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J Biol Chem. 1995;270:31178–31188. doi: 10.1074/jbc.270.52.31178. [DOI] [PubMed] [Google Scholar]
- Bharathan G, Janssen BJ, Kellogg EA, Sinha N. Did homeodomain proteins duplicate before the origin of angiosperms, fungi, and metazoa? Proc Natl Acad Sci USA. 1997;94:13749–13753. doi: 10.1073/pnas.94.25.13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozhenok L, Wade PA, Varga-Weisz P. WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. Embo J. 2002;21:2231–2241. doi: 10.1093/emboj/21.9.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürglin TR. A comprehensive classification of homeobox genes. In: Duboule D, editor. Guidebook to the homeobox genes. Oxford: Oxford University Press; 1994. pp. 25–71. [Google Scholar]
- Bürglin TR. The evolution of homeobox genes. In: Arai R, Kato M, Doi Y, editors. Biodiversity and evolution. Tokyo: The National Science Museum Foundation; 1995. pp. 291–336. [Google Scholar]
- Bürglin TR. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997;25:4173–4180. doi: 10.1093/nar/25.21.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürglin TR. The PBC domain contains a MEINOX domain: coevolution of Hox and TALE homeobox genes? Dev Genes Evol. 1998a;208:113–116. doi: 10.1007/s004270050161. [DOI] [PubMed] [Google Scholar]
- Bürglin TR. PPCMatrix: a PowerPC dotmatrix program to compare large genomic sequences against protein sequences. Bioinformatics. 1998b;14:751–752. doi: 10.1093/oxfordjournals.bioinformatics.a011055. [DOI] [PubMed] [Google Scholar]
- Bürglin TR. Homeodomain proteins. In: Meyers RA, editor. Encyclopedia of molecular cell biology and molecular medicine. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co; 2005. pp. 179–222. [Google Scholar]
- Bürglin TR, Cassata G. Loss and gain of domains during evolution of cut superclass homeobox genes. Int J Dev Biol. 2002;46:115–123. [PubMed] [Google Scholar]
- Chan RL, Gago GM, Palena CM, Gonzalez DH. Homeoboxes in plant development. Biochim Biophys Acta. 1998;1442:1–19. doi: 10.1016/s0167-4781(98)00119-5. [DOI] [PubMed] [Google Scholar]
- Chen H, Rosin FM, Prat S, Hannapel DJ. Interacting transcription factors from the three-amino acid loop extension superclass regulate tuber formation. Plant Physiol. 2003;132:1391–1404. doi: 10.1104/pp.103.022434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarbelli AR, Ciolfi A, Salvucci S, Ruzza V, Possenti M, Carabelli M, Fruscalzo A, Sessa G, Morelli G, Ruberti I. The Arabidopsis homeodomain-leucine zipper II gene family: diversity and redundancy. Plant Mol Biol. 2008;68:465–478. doi: 10.1007/s11103-008-9383-8. [DOI] [PubMed] [Google Scholar]
- Coego A, Ramirez V, Gil MJ, Flors V, Mauch-Mani B, Vera P. An Arabidopsis homeodomain transcription factor, Overexpressor of Cationic Peroxidase 3, mediates resistance to infection by necrotrophic pathogens. Plant Cell. 2005;17:2123–2137. doi: 10.1105/tpc.105.032375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper B, Clarke JD, Budworth P, et al. (12 co-authors) A network of rice genes associated with stress response and seed development. Proc Natl Acad Sci USA. 2003;100:4945–4950. doi: 10.1073/pnas.0737574100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- Cus R, Maurus D, Kuhl M. Cloning and developmental expression of WSTF during Xenopus laevis embryogenesis. Gene Expr Patterns. 2006;6:340–346. doi: 10.1016/j.modgep.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Derelle R, Lopez P, Le Guyader H, Manuel M. Homeodomain proteins belong to the ancestral molecular toolkit of eukaryotes. Evol Dev. 2007;9:212–219. doi: 10.1111/j.1525-142X.2007.00153.x. [DOI] [PubMed] [Google Scholar]
- Doerks T, Copley R, Bork P. DDT—a novel domain in different transcription and chromosome remodeling factors. Trends Biochem Sci. 2001;26:145–146. doi: 10.1016/s0968-0004(00)01769-2. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd SK, Zalewski CS, Bowman JL. Evolution of class III homeodomain-leucine zipper genes in streptophytes. Genetics. 2006;173:373–388. doi: 10.1534/genetics.105.054239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Affolter M, Bürglin TR. Homeodomain proteins. Annu Rev Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- Gertz EM, Yu YK, Agarwala R, Schaffer AA, Altschul SF. Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol. 2006;4:41. doi: 10.1186/1741-7007-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronenborn AM. The DNA-binding domain of GATA transcription factors-a prototypical type IV Cys2-Cys2 zinc finger. In: Shiro I, Kuldell N, editors. Zinc finger proteins: from atomic contact to cellular function. New York: Kluwer Academic/Plenum Publishers; 2004. pp. 26–30. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development. 2004;131:657–668. doi: 10.1242/dev.00963. [DOI] [PubMed] [Google Scholar]
- Henriksson E, Olsson AS, Johannesson H, Johansson H, Hanson J, Engstrom P, Soderman E. Homeodomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiol. 2005;139:509–518. doi: 10.1104/pp.105.063461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland PW, Booth HA, Bruford EA. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007;5:47. doi: 10.1186/1741-7007-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP. Genome-wide identification, classification, evolutionary expansion and expression analyses of homeobox genes in rice. Febs J. 2008;275:2845–2861. doi: 10.1111/j.1742-4658.2008.06424.x. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Jørgensen JE, Gronlund M, Pallisgaard N, Larsen K, Marcker KA, Jensen EO. A new class of plant homeobox genes is expressed in specific regions of determinate symbiotic root nodules. Plant Mol Biol. 1999;40:65–77. doi: 10.1023/a:1026463506376. [DOI] [PubMed] [Google Scholar]
- Kaplan W, Littlejohn TG. Swiss-PDB Viewer (Deep View) Brief Bioinform. 2001;2:195–197. doi: 10.1093/bib/2.2.195. [DOI] [PubMed] [Google Scholar]
- Karol KG, McCourt RM, Cimino MT, Delwiche CF. The closest living relatives of land plants. Science. 2001;294:2351–2353. doi: 10.1126/science.1065156. [DOI] [PubMed] [Google Scholar]
- Kerstetter R, Vollbrecht E, Lowe B, Veit B, Yamaguchi J, Hake S. Sequence analysis and expression patterns divide the maize knotted1-like homeobox genes into two classes. Plant Cell. 1994;6:1877–1887. doi: 10.1105/tpc.6.12.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Koenig D, Kang J, Yoong FY, Sinha N. Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr Biol. 2008;18:672–677. doi: 10.1016/j.cub.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. (13 co-authors) Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, DeWeese-Scott C, et al. (27 co-authors) CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 2003;31:383–387. doi: 10.1093/nar/gkg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita A, Sasaki S, Kashiwabara Y, Nagayama K, Ohba K, Iwaki H, Misawa H, Ishizuka K, Nakamura H. Essential role of GATA2 in the negative regulation of thyrotropin beta gene by thyroid hormone and its receptors. Mol Endocrinol. 2007;21:865–884. doi: 10.1210/me.2006-0208. [DOI] [PubMed] [Google Scholar]
- McCourt RM, Delwiche CF, Karol KG. Charophyte algae and land plant origins. Trends Ecol Evol. 2004;19:661–666. doi: 10.1016/j.tree.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Meijer AH, Scarpella E, van Dijk EL, Qin L, Taal AJ, Rueb S, Harrington SE, McCouch SR, Schilperoort RA, Hoge JH. Transcriptional repression by Oshox1, a novel homeodomain leucine zipper protein from rice. Plant J. 1997;11:263–276. doi: 10.1046/j.1365-313x.1997.11020263.x. [DOI] [PubMed] [Google Scholar]
- Mukherjee K, Bürglin TR. MEKHLA, a novel domain with similarity to PAS domains, is fused to plant homeodomain-leucine zipper III proteins. Plant Physiol. 2006;140:1142–1150. doi: 10.1104/pp.105.073833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Katsumata H, Abe M, Yabe N, Komeda Y, Yamamoto KT, Takahashi T. Characterization of the class IV homeodomain-leucine zipper gene family in Arabidopsis. Plant Physiol. 2006;141:1363–1375. doi: 10.1104/pp.106.077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesch G, Stormann K, Torres JT, Walden R, Somssich IE. Developmental and auxin-induced expression of the Arabidopsis prha homeobox gene. Plant J. 1997;12:635–647. doi: 10.1046/j.1365-313x.1997.00635.x. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Aravind L. START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem Sci. 1999;24:130–132. doi: 10.1016/s0968-0004(99)01362-6. [DOI] [PubMed] [Google Scholar]
- Poot RA, Bozhenok L, van den Berg DL, Steffensen S, Ferreira F, Grimaldi M, Gilbert N, Ferreira J, Varga-Weisz PD. The Williams syndrome transcription factor interacts with PCNA to target chromatin remodelling by ISWI to replication foci. Nat Cell Biol. 2004;6:1236–1244. doi: 10.1038/ncb1196. [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, et al. (70 co-authors) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- Ruberti I, Sessa G, Lucchetti S, Morelli G. A novel class of plant proteins containing a homeodomain with a closely linked leucine zipper motif. Embo J. 1991;10:1787–1791. doi: 10.1002/j.1460-2075.1991.tb07703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara K, Nishiyama T, Deguchi H, Hasebe M. Class 1 KNOX genes are not involved in shoot development in the moss Physcomitrella patens but do function in sporophyte development. Evol Dev. 2008;10:555–566. doi: 10.1111/j.1525-142X.2008.00271.x. [DOI] [PubMed] [Google Scholar]
- Sakakibara K, Nishiyama T, Kato M, Hasebe M. Isolation of homeodomain-leucine zipper genes from the moss Physcomitrella patens and the evolution of homeodomain-leucine zipper genes in land plants. Mol Biol Evol. 2001;18:491–502. doi: 10.1093/oxfordjournals.molbev.a003828. [DOI] [PubMed] [Google Scholar]
- Schindler U, Beckmann H, Cashmore AR. HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant J. 1993;4:137–150. doi: 10.1046/j.1365-313x.1993.04010137.x. [DOI] [PubMed] [Google Scholar]
- Schrick K, Nguyen D, Karlowski WM, Mayer KF. START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 2004;5:R41. doi: 10.1186/gb-2004-5-6-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G, Carabelli M, Ruberti I. Identification of distinct families of HD-Zip proteins in Arabidopsis thaliana. In: Coruzzi P, editor. Plant molecular biology. Berlin: Springer-Verlag; 1994. pp. 412–426. [Google Scholar]
- Shiu SH, Shih MC, Li WH. Transcription factor families have much higher expansion rates in plants than in animals. Plant Physiol. 2005;139:18–26. doi: 10.1104/pp.105.065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HM, Boschke I, Hake S. Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc Natl Acad Sci USA. 2002;99:9579–9584. doi: 10.1073/pnas.092271599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatori N, Butts T, Candiani S, Pestarino M, Ferrier DE, Saiga H, Holland PW. Comprehensive survey and classification of homeobox genes in the genome of amphioxus, Branchiostoma floridae. Dev Genes Evol. 2008;218:579–590. doi: 10.1007/s00427-008-0245-9. [DOI] [PubMed] [Google Scholar]
- Tan QK, Irish VF. The Arabidopsis zinc finger-homeodomain genes encode proteins with unique biochemical properties that are coordinately expressed during floral development. Plant Physiol. 2006;140:1095–1108. doi: 10.1104/pp.105.070565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocke S, Muszynski M, Briggs K, Amasino RM. Characterization of a gene from Zea mays related to the Arabidopsis flowering-time gene LUMINIDEPENDENS. Plant Mol Biol. 2000;44:107–122. doi: 10.1023/a:1006472929800. [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Veit B, Sinha N, Hake S. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature. 1991;350:241–243. doi: 10.1038/350241a0. [DOI] [PubMed] [Google Scholar]
- Windhovel A, Hein I, Dabrowa R, Stockhaus J. Characterization of a novel class of plant homeodomain proteins that bind to the C4 phosphoenolpyruvate carboxylase gene of Flaveria trinervia. Plant Mol Biol. 2001;45:201–214. doi: 10.1023/a:1006450005648. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.