Abstract
The PRC2 complex protein EZH2 is a histone methyltransferase that is known to bind and recruit DNMT1 to the DNA to modulate DNA methylation. Here, we determined that the pan-HDAC inhibitor panobinostat (LBH589) treatment depletes DNMT1 and EZH2 protein levels, disrupts the interaction of DNMT1 with EZH2, as well as de-represses JunB in human acute leukemia cells. Similar to treatment with the hsp90 inhibitor 17-DMAG, treatment with panobinostat also inhibited the chaperone association of heat shock protein 90 with DNMT1 and EZH2, which promoted the proteasomal degradation of DNMT1 and EZH2. Unlike treatment with the DNA methyltransferase inhibitor decitabine, which demethylates JunB promoter DNA, panobinostat treatment mediated chromatin alterations in the JunB promoter. Combined treatment with panobinostat and decitabine caused greater attenuation of DNMT1 and EZH2 levels than either agent alone, which was accompanied by more JunB de-repression and loss of clonogenic survival of K562 cells. Co-treatment with panobinostat and decitabine also caused more loss of viability of primary AML but not normal CD34+ bone marrow progenitor cells. Collectively, these findings indicate that co-treatment with panobinostat and decitabine targets multiple epigenetic mechanisms to de-repress JunB and exerts antileukemia activity against human acute myeloid leukemia cells.
INTRODUCTION
Gene silencing and loss of gene function mediated through epigenetic mechanisms collaborates with genetic mutations and alterations leading to cancer.1–3 As multiprotein complexes, Polycomb group proteins epigenetically silence gene expression, including tumor suppressor genes (TSGs).1–3 EZH2 is the catalytic subunit of the polycomb repressive complex 2 (PRC2), which also includes SUZ12, EED and YY1.1–3 EZH2 acts as a histone lysine methyltransferase (HKMT), which mediates methylation of lysine (K) 27 on the N-terminal tail of histone H3 to silence expression of PRC2 target genes involved in lineage differentiation.3–5 EZH2 has also been recognized as a senescence preventing gene in embryonic fibroblasts, as well as shown to be abundantly expressed in purified hematopoietic stem cell (HSCs), where it preserves HSC potential and prevents exhaustion.6 EZH2 regulates cell proliferation by promoting S phase entry and G2-M transition, and it is highly expressed in tumor versus normal tissue.7,8 EZH2 mediated cell cycle progression promoted by gene repression also involves histone deacetylation by HDAC1, with which EZH2 interacts through its PRC2 binding partner EED.9 EZH2 is overexpressed in a variety of malignancies, including prostate, breast and bladder cancers with poor prognosis.10–12 Knockdown of EZH2 by siRNA has been demonstrated to inhibit breast cancer cell proliferation, while pharmacologic inhibition of EZH2 resulted in apoptosis of breast cancer but not normal cells.10,13 Recently, EZH2 was shown to directly interact with and regulate the activity of the DNA methyltransferases DNMT1, DNMT3a and DNMT3b.14,15 DNMTs function to transfer a methyl group from S-adenosyl-methionine to the 5′ position of cytosine in the CpG dinucleotides in the promoters of genes, thereby maintaining a consistent pattern of epigenetic gene silencing of TSGs in cancer cells.16–19 The heritable pattern of cytosine methylation is maintained during DNA replication by DNMT1, which has a preference for hemi-methylated DNA and is regarded as the primary maintenance DNMT.17–19 DNA methylation by DNMTs also recruits HDAC activity to the promoters of silenced genes.20,21 Similar to the PRC2 complex, DNMT1 has a direct interaction with histone deacetylases HDAC1 and HDAC2.22,23 In acute myeloid leukemia (AML) and chronic myeloid leukemia (CML), all three DNMTs are overexpressed and may play a significant role in the pathogenesis of leukemia due to aberrant hypermethylation of TSGs or DNA repair related genes.24,25 Treatment with the antisense oligonucleotides against DNMT1 was demonstrated to cause cell cycle arrest in the S-phase of the cell cycle and inhibited DNA replication with loss of DNMT1 at the replication fork.26,27 Although genes methylated in cancer cells are packaged with nucleosomes containing the trimethylated K27 on the H3 mark, gene silenced in cancer by H3K27 trimethylation has been shown to be independent of promoter DNA methylation.17,28 Consistent with this, DNA methylation and transcriptional silencing of cancer genes has been shown to persist despite the depletion of EZH2.29
In myeloid leukemia, overexpression of DNMT1 has been associated with promoter hypermethylation of a number of genes, including JunB.30–33 In AML, promoter hypermethylation of E cadherin, ERα and p15 have also been reported.34 Inactivation of JunB expression in stem cells leads to myeloproliferative disorders, and JunB overexpression and amplification has been associated with failure of differentiation.32,35 JunB deficient hematopoietic stem cells show concomitant down regulation of p16INK4a and up regulation of Bcl-2 and Bcl-xL.32 Indeed, JunB methylation and impaired expression was observed in blast crisis of CML.31 The DNMT1 inhibitor decitabine inhibits hypermethylation of the promoter DNA of tumor suppressor genes and decreased global methylation, which supports the rationale for the use of decitabine as a therapeutic agent in the treatment of hematologic malignancies.36,37 Further, co-treatment with a DNMT1 and HDAC inhibitor has been shown to synergistically hypomethylate the promoters of TSGs and significantly inhibit tumor growth.38,39 Previously, while the pan-HDAC inhibitor trichostatin A (TSA) was shown to downregulate both the mRNA and protein levels of DNMT1 in tumor cells, treatment with panobinostat (LBH589) was shown to release DNMT1 and HDAC1 from the ERα gene promoter, suggesting an association between DNMT1 and the HDACs.40,41 In a previous report, we had demonstrated that panobinostat treatment inhibits HDAC6 and induces hyperacetylation of the molecular chaperones heat shock protein (hsp) 90, associated with depletion of EZH2 and other PRC2 components.42–44 This occurred with the concomitant depletion of the transcription factors HOXA9 and MEIS 1 levels, resulting in loss of clonogenic survival of human leukemia cells.44 Based on these observations we determined the effects of panobinostat on the interaction between the EZH2 and PRC2 complex with DNMT1, as well as on the levels and chaperone association of DNMT1 and EZH2 with hsp 90. Here, we report that treatment with panobinostat or a geldanamycin analogue hsp90 inhibitor (17-DMAG) attenuated the levels of DNMT1 with EZH2 by inhibiting their chaperone association with hsp90 in acute leukemia cells.45 We further determined that, as compared to treatment with either agent alone, combined treatment with decitabine (DAC) and panobinostat resulted in greater depletion of DNMT1 levels, more derepression of JunB, and induced greater loss of cell viability and clonogenic survival of cultured and primary acute leukemia cells.
MATERIALS AND METHODS
Reagents
Panobinostat (PS) and 17-DMAG were kindly provided by Novartis Pharmaceuticals Inc. (East Hanover, NJ) and Kosan Biosciences (Hayward, CA), respectively. Decitabine was acquired from Sigma-Aldrich (St. Louis, MO). Bortezomib was kindly provided by Millennium Pharmaceuticals (Cambridge, MA). Monoclonal anti-DNMT1 antibody was purchased from Abcam (Cambridge, MA). Anti-EZH2 monoclonal antibody was purchased from BD Transduction labs (San Jose, CA). Rat monoclonal anti-hsp90 and rabbit polyclonal anti-hsp70 were purchased from Stressgen (Ann Arbor, MI). Anti-β-actin was purchased from Sigma Aldrich (St. Louis, MO). Monoclonal anti-JunB was purchased from Santa Cruz Biotechnologies (Santa Cruz, CA). Monoclonal anti-ubiquitin was purchased from Covance Biologicals (Berkeley, CA). Suv39H1, Trimethyl-lysine 9 H3 and acetyl H3 were acquired from Upstate biotechnologies (Charlottesville, VA). Normal mouse IgG and normal rat IgG were acquired from Santa Cruz (Santa Cruz, CA).
Cell Culture
K562 and LAMA-84 cells were cultured in complete RPMI-1640 medium and incubated at 37°C with 5% CO2 as previously described.46 Growth media was changed every 2–3 days and cells were maintained at a density of 250,000 cells/mL. Exponentially growing cell cultures were used for all experiments described below.
Leukemia Blast and Normal CD34+ Hemopoietic Progenitor Cells
Primary acute myeloid leukemia (AML) cells were obtained with informed consent as part of a clinical protocol approved by the Institutional Review Board of the Medical College of Georgia. As previously described, peripheral blood samples were collected in heparinized tubes, or were obtained from leukophoresis units, and mononuclear cells were separated using Lymphoprep (Axis-Shield, Oslo, Norway), washed once with complete RPMI-1640 media, resuspended in complete RPMI-1640 and counted to determine the number of cells isolated prior to their use in the various experiments.47 The purity of the blast population was confirmed to be 80% or better by morphologic evaluation of cytospun cell preparations stained with Wright stain. Banked, de-linked, and de-identified donor peripheral blood CD34+ mononuclear cells procured for recipients who had since died were purified by immunomagnetic beads conjugated with anti-CD34 antibody prior to use in thecell viability assay (StemCell Technologies).47
Cell Lysis and Protein Quantitation
Untreated or drug treated cells were centrifuged and the cell pellets were re-suspended in 200 μL of lysis buffer, then centrifuged and protein concentrations determined, as previously described.46–48
Immunoprecipitation of hsp90, DNMT-1, and EZH2 and immunoblot analysis
Following designated treatments, cells were lysed with the lysis buffer as described above. Five hundred micrograms of cell lysate was mixed gently with 5 μg of rat monoclonal anti-Hsp90 or 5μg of mouse monoclonal anti-DNMT1 or 5μg of mouse monoclonal EZH2 and incubated at 4°C for 1–2 hours on a rotator. Pre-washed protein-G beads were added to the lysate-antibody mixture and incubated overnight at 4°C on a rotator. The immunoprecipitates were washed 4 times with lysis buffer and eluted from the agarose beads by boiling with 6X SDS sample buffer before SDS-PAGE and Western blot analysis, as previously described.42
Preparation of detergent soluble and insoluble lysates
Following drug treatment with bortezomib (BZ) and panobinostat, cells were separated into detergent soluble and insoluble fractions as previously described.42 Briefly, cells were lysed with TNSEV buffer (50 mmol/L Tris-HCl, pH 7.5, 2 mM EDTA, 100 mmol/L NaCl, 1 mmol/Lsodium orthovanadate, 1% Nonidet P-40 containing 20 μg/mlaprotinin, 20 μg/ml leupeptin, 1 mmol/L PMSF, 25 mmol/L NaF, and 5 mmol/L N-ethylmaleimide). The insoluble fractions (pellet) were solubilized with SDS buffer (80 mmol/L Tris, pH 6.8, 2% SDS, 100 mmol/L dithiothreitol, and 10% glycerol). Fifty μg of proteins from the Nonidet P-40-soluble and insoluble fractionswere separated on 7.5% SDS-polyacrylamide gel and analyzed byWestern blotting.
SDS-PAGE and Western Blotting
One hundred micrograms of total cell lysate was used for SDS-PAGE. Western blot analyses of EZH2, DNMT1, hsp70, hsp90, ubiquitin, and JunB were performed on total cell lysates or immunoprecipitates using specific antisera or monoclonal antibodies, as previously described.46–48 The expression level of β-actin or α-tubulin was used as the loading control for the Western blots.
RNA isolation and RT-PCR
RNA was extracted from the cultured cells using the Trizol method (Invitrogen, Carlsbad, CA) as previously described.44,48 Purified RNA was quantified and reverse-transcribed using Superscript II according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA). Resulting cDNAs were used in subsequent PCR reactions for JunB (sequences available upon request). PCR reactions for β-actin were used as an internal loading control. PCR reactions were carried out in a gradient Mastercycler (Eppendorf, Westbury, NY) and consisted of a 3-minute denaturation at 95°C followed by 35 cycles of 95° C (30 sec), 52°C (30 sec) and 72°C (30 sec) with a final extension at 72°C (10 min). Amplified products were resolved on a 2% agarose gel and recorded with a UV transilluminator. Horizontal scanning densitometry was performed with ImageQuant 5.2 and the band intensity of each PCR product was compared to that of β-actin. For quantitative assessment of DNMT1 and JunB, qPCR was performed with TaqMan probes against the exon 38–39 boundary for DNMT1 and the exon 1 boundary for JunB. Expression levels were normalized to a TaqMan probe against GAPDH.
Colony culture assay
Following the designated treatments, cells were harvested and washed twice with 1X PBS and approximately 300 cells were plated in complete Methocult (StemCell Technologies, Vancouver, British Columbia) and cultured for 7–10 days at 37°C in a 5% CO2 environment. Colony growth was measured as a percentage of the control cell colony growth, as previously described.44
Bisulfite modification and methylation specific PCR
Following treatment with either decitabine or panobinostat, the genomic DNA was isolated from K562 cells using a DNeasy kit (Qiagen, Valencia, CA). One microgram of genomic DNA was subjected to bisulfite modification with a kit from Chemicon (Temecula, CA) according to the manufacturer’s protocol. DNA was eluted in 25 μL of TE Buffer. Methylation-specific PCR reactions were set up using a Hotmaster mix (Eppendorf, Westbury, NY). Primers were designed according to within a CpG island of the JunB promoter and consisted of the following sequences, JunB MF 5′-GATAGGGTTTTTGCGTATAGTTGTC - 3′, JunBMR 5′-TATTCCATTTTAATACACATCCGAA - 3′ and JunBUF 5′-TAGGGTTTTTGTGTATAGTTGTTGG-3′, JunBUR 5′-CTATTCCATTTTAATACACATCCAAA -3′.49 Primers were also designed against the E-cadherin and ER alpha promoters and consisted of sequence EcadMF 5′-TAATTTTAGGTTAGAGGGTTATCGC-3′, EcadMR 5′-CTCACAAATACTTTACAATTCCGAC-3′, EcadUF 5′-ATTTTAGGTTAGAGGGTTATTGTGT-3′, EcadUR 5′-CAAACTCACAAATACTTTACAATTCCA-3′, ERαMF 5′-TAAATAGAGATATATCGGAGTTTGGTACG-3′, ERαMR 5′-AACTTAAAATAAACGCGAAAAACGA-3′, ERαUF 5′-TAAATAGAGATATATTGGAGTTTGGTATGG-3′, and ERαUR 5′-AACTTAAAATAAACACAAAAAACAAA-3′. Internal loading control primers were designed to recognize the JunB promoter regardless of methylation status. These primers consisted of sequence JunBBSF or 5′-GAGTTAGTAGGGAGTTGGGAGTTG-3′ and JunBBSRev 5′-AACTATTCCATTTTAATACACATCC-3′. PCR reactions were carried out in an Eppendorf Gradient Mastercycler (Westbury, NY) and consisted of a 95°C hold followed by a 2-minute denaturation at 95°C and 30 cycles of 95° C (30 sec), 55°C (30 sec) and 72°C (30 sec) with a final extension at 72°C (10 min). PCR products were resolved on a 2% agarose gel and visualized with a UV-transilluminator. Horizontal scanning densitometry was performed with ImageQuant 5.2 and the band intensity of each PCR product was compared to that of the internal loading control.
Chromatin immunoprecipitation and PCR
K562 cells were treated with panobinostat for 16 hours. Following drug exposure, the chromatin in the cells was crosslinked with formaldehyde for 10 minutes at 37°C. The crosslinking reaction was quenched with 1/20 volume of 2.5 M glycine for 5 minutes at room temperature, then the cells were washed twice for 5 minutes in ice-cold 1X PBS. Cell lysis, sonication and chromatin immunoprecipitation was performed according to the manufacturer’s protocol (Upstate Biotechnologies, Charlottesville, VA), and as previously described.48 Primers for the amplification of the JunB promoter were synthesized by IDT (Coralville, IA). Sequences are available upon request. For quantitative assessment of JunB in the chromatin immunoprecipitates, a SYBR Green mastermix from Applied Biosystems was used (Foster City, CA). Relative enrichment was normalized against GAPDH in the input samples.
Statistical analysis
Significant differences between valuesobtained in a population of leukemia cells treated with differentexperimental conditions were determined using the Student’s t-test.
RESULTS
Panobinostat treatment disrupts the association of DNMT1 and EZH2 with hsp90, leading to proteasomal degradation of DNMT1 and EZH2
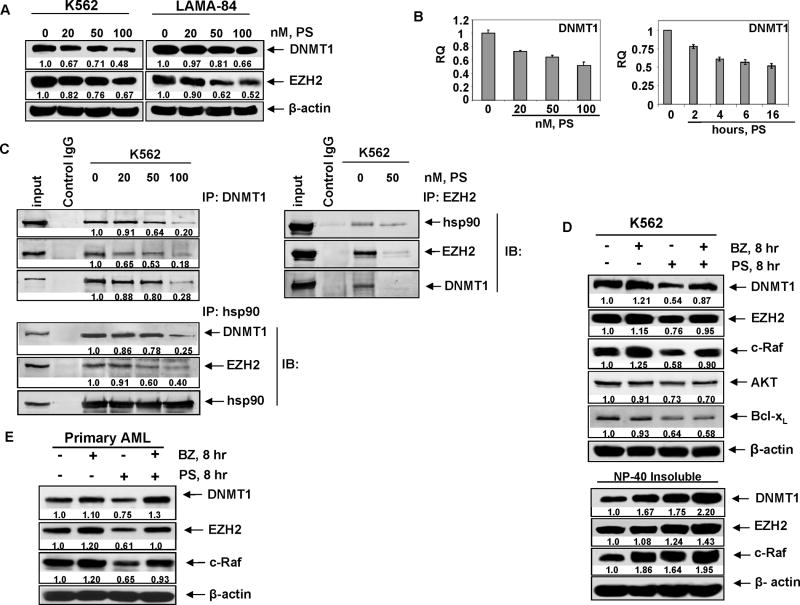
In a previous report we had shown that panobinostat depletes the levels and activity of EZH2.44 Based on this, and on the reports that EZH2 directly interacts with and regulates the activity of the DNMT1, we first determined the effects of panobinostat treatment on DNMT1 levels, as well as on the binding of EZH2 to DNMT1. Treatment with panobinostat dose-dependently reduced the protein levels of DNMT1 and EZH2 in the CML blast crisis (CML-BC) K562 and LAMA-84 cells (Figure 1A).14 This was accompanied by panobinostat-mediated decrease in the mRNA levels of DNMT1 in a time and dose dependent manner in K562 cells (Figure 1B). Similar effects were seen in LAMA84 cells (Supplemental Figure 1A). In contrast, panobinostat treatment did not lower EZH2 mRNA levels (Supplemental Figure 1B and 1C). We have previously demonstrated that, by inducing hyperacetylation of hsp90, pan-HDAC inhibitors can inhibit the chaperone function and association of hsp90 with its client proteins.42,43 Based on this, we next determined the effects of panobinostat treatment on the interaction of DNMT1 and EZH2, and whether panobinostat-mediated depletion of DNMT1 and EZH2 was due to inhibition of their chaperone association with hsp90. Figure 1C shows that exposure to panobinostat disrupted the binding of DNMT1 to EZH2, as well as decreased the binding of hsp90 to both EZH2 and DNMT1. Similar findings were observed in LAMA84 cells (Supplemental Figure 1D). Exposure intervals as short as 2 to 4 hours to panobinostat reduced the binding of hsp90 to EZH2 and DNMT1 (Supplemental Figure 1E). This was associated with increased accumulation of both DNMT1 and EZH2 in the detergent (NP40) insoluble fraction of the cytosol, suggesting increase in the levels of misfolded DNMT1 and EZH2 following treatment with panobinostat (Figure 1D). Additionally, co-treatment with the proteasome inhibitor bortezomib significantly increased the levels of DNMT1 and EZH2 in the detergent insoluble fraction, as well as restored the levels in the total cell lysates (Figure 1D). Co-treatment with bortezomib did not alter or rescue the levels of AKT or Bcl-xL in panobinostat treated cells (Figure 1D). Similar findings were observed in panobinostat-treated LAMA84 leukemia cells (Supplemental Figure 2A). Panobinostat-mediated decline in EZH2 levels was not due to the activity of caspases, since co-treatment with the caspase inhibitor ZVAD did not restore the levels of EZH2 (Supplemental Figure 2B). Panobinostat treatment also depleted DNMT1 and EZH2 levels in primary AML cells, which were also restored by co-treatment with bortezomib (Figure 1E). In these studies, the levels of the hsp90 client protein c-Raf served as the control protein that is a well recognized hsp90 client protein.42,43
Figure 1. Panobinostat depletes DNMT1 and disrupts its binding to EZH2 and hsp90.
A. Western blot of DNMT1 and EZH2 in K562 and LAMA84 following 24 hours treatment with the indicated doses of panobinostat (PS). The levels of β-actin served as the loading control. B. K562 cells were treated with the indicated concentrations of panobinostat for 16 hours. Alternatively, cells were treated with 100 nmol/L of panobinostat for the indicated times. Total RNA was isolated and qPCR was performed for DNMT1. The relative quantity (RQ) of DNMT1 mRNA expression was normalized against expression of GAPDH. C. K562 cells were treated with the indicated doses of panobinostat for 8 hours and DNMT1 (left panel), EZH2 (right panel), and hsp90 (bottom panel) were immunoprecipitated from the cell lysates. The immunoprecipitates were immunoblotted for hsp90, EZH2, and DNMT1. D. K562 cells were treated with 100 nmol/L of panobinostat and/or 100 nmol/L of bortezomib (BZ) for 8 hours. Following treatment, total cell lysates or NP-40 detergent insoluble fractions were immunoblotted for DNMT1, EZH2 and c-Raf, AKT and Bcl-xL. The levels of β-actin served as the loading control. E. Primary AML cells were treated with 100 nmol/L of panobinostat and/or 100 nmol/L of bortezomib for 8 hours. Following treatment, total cell lysates were immunoblotted for DNMT1, EZH2, and c-Raf. The levels of β-actin in the lysates served as the loading control.
Co-treatment with panobinostat enhances 17-DMAG mediated depletion of DNMT1 and EZH2 in acute leukemia cells
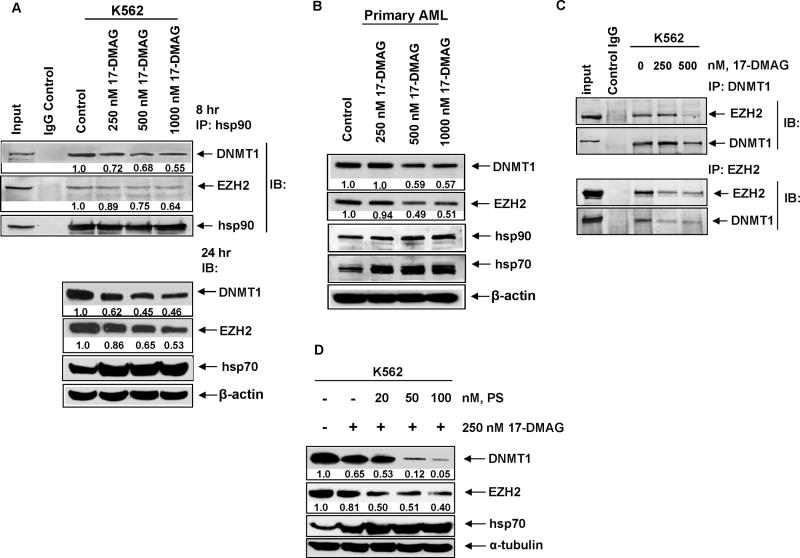
First, to confirm that DNMT1 and EZH2 are hsp90 client proteins, we determined the effects of the geldanamycin analogue hsp90 inhibitor 17-DMAG on the expression of DNMT1 in the K562 cells. Treatment with 17-DMAG depleted DNMT1 protein levels, while it concomitantly induced hsp70 levels (Figure 2A).50 Similar findings were noted, following treatment of primary leukemia cells with 17-DMAG (Figure 2B). We next determined the effects of 17-DMAG treatment on the binding of DNMT1 and EZH2 to hsp90. Similar to the results with LBH589, exposure to 17-DMAG reduced the binding of DNMT1 and EZH2 to hsp90 (Figure 2A), increased the binding to hsp70, as well as attenuated the binding of DNMT1 with EZH2 (Figure 2C). Additionally, co-treatment with bortezomib restored 17-DMAG induced depletion of DNMT1 and EZH2 (data not shown). We next determined the combined effects of panobinostat and 17-DMAG on the levels of DNMT1 and EZH2. Notably, co-treatment with panobinostat significantly increased 17-DMAG mediated depletion of DNMT1 and EZH2 in K562 cells (Figure 2D).
Figure 2. 17-DMAG depletes the levels of DNMT1 and EZH2 by inhibiting their chaperone association hsp90 in acute leukemia cells.
A. K562 cells were treated with the indicated does of 17-DMAG for 8 hours and hsp90 was immunoprecipitated. Immunoblot analysis was performed for DNMT1, EZH2 and hsp90 on the immunoprecipitates. Alternatively, K562 cells were treated with the indicated concentrations of 17-DMAG for 24 hours and immunoblot analysis was done for DNMT1, EZH2 and hsp70. The expression levels of β-actin in the lysates served as the loading control. B. Primary acute leukemia cells were treated with the indicated concentrations of 17-DMAG for 24 hours. Total cell lysates were harvested and immunoblot analysis was performed for DNMT1, EZH2 hsp90 and hsp70. The levels of β-actin in the lysates served as the loading control. C. K562 cells were treated with the indicated doses of 17-DMAG for 8 hours and DNMT1 (top panel) and EZH2 (bottom panel) were immunoprecipitated. Immunoblot analysis was performed for EZH2 and DNMT1 in the immunoprecipitates. D. K562 cells were treated with the indicated concentrations of panobinostat and 17-DMAG for 24 hours. Immunoblot analysis was performed for DNMT1, EZH2 and hsp70. The expression levels of α-tubulin in the lysates served as the loading control.
Panobinostat treatment induces JunB and enhances decitabine (DAC)-mediated induction of JunB
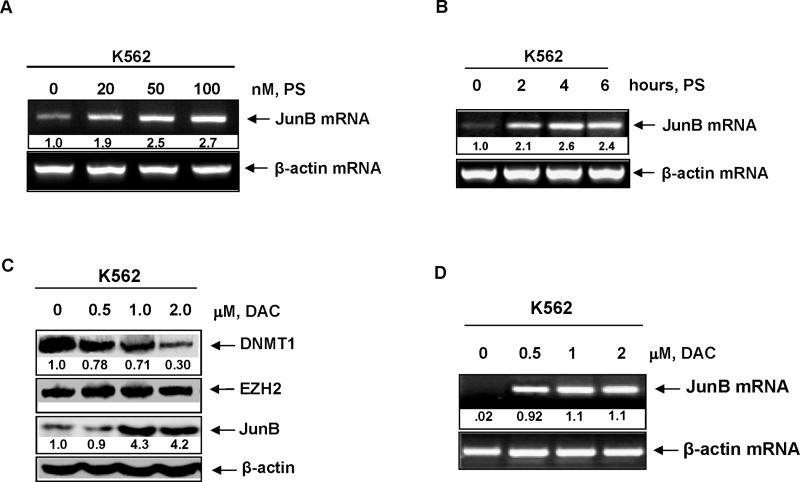
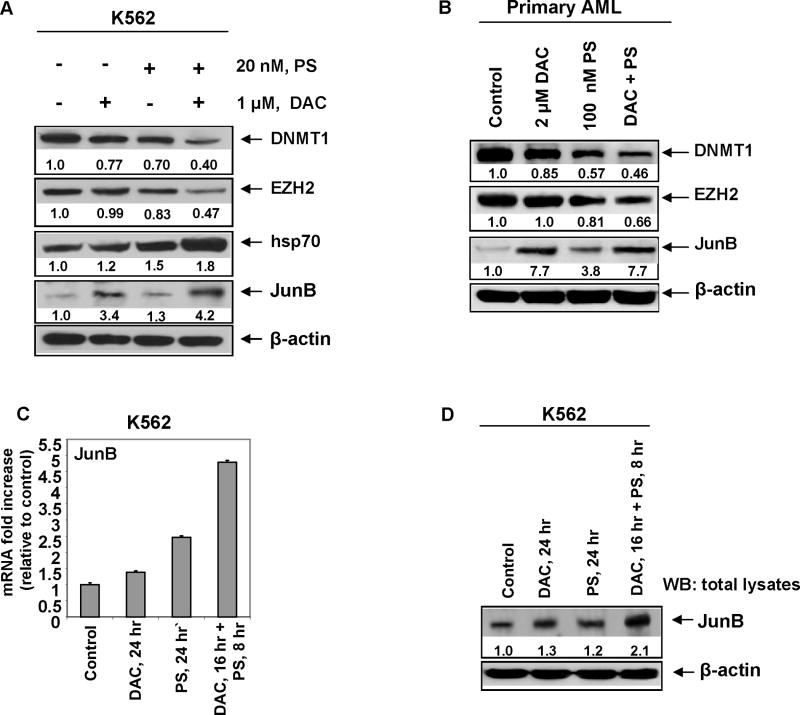
We next determined the effect of panobinostat on the mRNA expression JunB, a gene that is known to be silenced by promoter hypermethylation in CML cells.31,32 Treatment with panobinostat dose-dependently up-regulated the mRNA levels of JunB, with 2.67 fold increase seen following treatment with 100 nmol/L of panobinostat (Figure 3A). JunB mRNA induction was observed following exposure interval to panobinostat as short as two hours (Figure 3B). As noted above for panobinostat, treatment with decitabine (DAC) dose-dependently attenuated DNMT1 protein levels and up-regulated JunB mRNA and protein levels (Figures 3C and 3D). However, exposure to DAC did not lower the levels of EZH2 (Figure 3C). We next determined the effect of co-treatment with panobinostat and DAC on EZH2, DNMT1 and JunB levels. Figure 4A demonstrates that as compared to treatment with either agent alone, co-treatment with DAC and panobinostat caused greater attenuation of the levels of DNMT1 and EZH2 levels, while concomitantly inducing more JunB levels. Combined treatment with DAC and panobinostat also had a similar effect on EZH2, DNMT1 and JunB levels in primary AML blasts (Figure 4B). Consistent with previous reports, sequential treatment with DAC followed by panobinostat induced more JunB mRNA and protein levels than treatment with the reverse schedule of administration of panobinostat followed by decitabine (Figure 4C).
Figure 3. Panobinostat induces JunB mRNA in a dose and time dependent manner.
A. K562 cells were treated with the indicated concentrations of panobinostat for 16 hours. Total RNA was isolated and RT-PCR was performed for JunB mRNA expression. A β-actin specific PCR reaction and expression levels served as the loading control. B. K562 cells were treated with 100 nmol/L of panobinostat for the indicated times. Following this, RT-PCR analysis was performed for JunB mRNA expression. A β-actin specific PCR reaction and expression levels served as the loading control. C. K562 cells were treated with the indicated concentrations of decitabine (DAC) for 24 hours. Following this, total cell lysates were harvested and immunoblot analysis was performed for DNMT1, EZH2, and JunB. The levels of β-actin in the lysates served as the loading control. D. K562 cells were treated with the indicated concentrations of decitabine (DAC) for 16 hours. Following this, total RNA were isolated and RT-PCR analysis was performed for JunB mRNA expression. A β-actin specific PCR reaction and expression served as the loading control.
Figure 4. Co-treatment with decitabine and panobinostat enhances DNMT1 depletion and JunB induction.
A. K562 cells were treated with the indicated concentrations of DAC and panobinostat for 24 hours. Immunoblot analysis was performed for DNMT1, EZH2, hsp70, and JunB on the total cell lysates. The expression levels of β-actin in the cell lysates served as the loading control. B. Primary acute leukemia cells were treated with the indicated concentrations of DAC and panobinostat for 24 hours. Immunoblot analysis was performed for DNMT1, EZH2 and JunB on the total cell lysates. The levels of β-actin in the cell lysates served as the loading control. C. K562 cells were treated with 20 nmol/L of panobinostat and 1 μmol/L of decitabine in the manner indicated for 24 hours. Following this, total RNA were isolated and qPCR analysis was performed for exon 1 of JunB mRNA. Expression of JunB mRNA was normalized against GAPDH. Alternatively, Western blot analysis was performed for JunB on the total cell lysates. The expression levels of β-actin in the lysates served as the loading control.
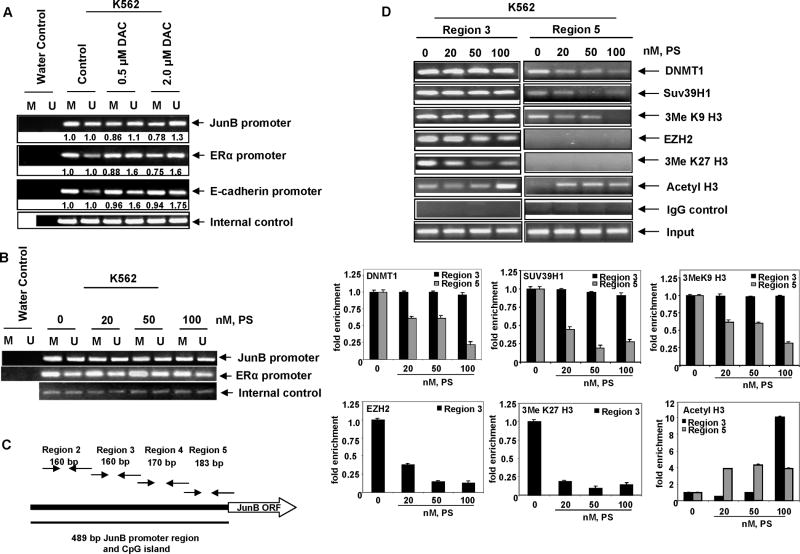
Decitabine and panobinostat induce JunB expression through different mechanisms
We next determined the mechanism by which DAC and panobinostat de-repress JunB in K562 cells. As shown in Figure 5A, treatment with as little as 0.5 μmol/L DAC for 16 hours caused demethylation of the JunB promoter as demonstrated by methylation specific PCR performed on bisulfite-treated DNA. DAC also demethylated the promoter of the E-cadherin and ERα genes, which are known to be silenced by methylation in hematologic malignancies (Figure 5A).41 In contrast, although it de-repressed JunB similar to DAC, panobinostat treatment did not alter the methylation status of JunB in K562 cells (Figure 5B). Panobinostat also did not affect the methylation status of ERα (Figure 5B) and E-cadherin (not shown). We next determined whether the panobinostat mediated derepression of JunB was due to chromatin alterations in the JunB promoter. Figure 5C shows a schematic representation of four regions in the CpG island of the JunB promoter. To perform ChIP analyses involving these regions in the JunB promoter, primers sets were tiled: four across the CpG island and one pair 2 kb upstream of the transcription start site (TSS) in another 80% GC rich region. As demonstrated in Figure 5D, ChIP analyses of untreated K562 cells confirmed that EZH2, DNMT1 and Suv39H1 were bound to the JunB promoter in region 3 (−378 to −218 relative to TSS) and DNMT1 and the histone methyltransferase Suv39H1 were bound to region 5 (−174 to +9 relative to TSS) nearest the transcriptional start site. EZH2 did not bind to region 5 of the JunB promoter. The repressive histone marks, tri-methlyated lysine 27 and tri-methylated lysine 9 were present in the untreated chromatin of region 3, corresponding to the localization of EZH2 and Suv39H1 (Figure 5D). In contrast, only the tri-methylated lysine 9 mark was detected in region 5 corresponding to the localization of Suv39H1 to this region (Figure 5D). Treatment with panobinostat dose-dependently depleted EZH2 and the tri-methlyated lysine 27 mark from region 3, and DNMT1 and Suv39H1 from region 5 of the JunB promoter (Figure 5D). Panobinostat-mediated depletion of the trimethylated lysine 9 and lysine 27 marks were also accompanied by increase in acetylation of histone H3 associated with the JunB promoter (Figure 5D). These findings clearly show that panobinostat mediated de-repression of JunB is accompanied by depletion of the repressive chromatin marks but not by demethylation of the CpG dinucleotides in the promoter DNA of JunB.
Figure 5. Decitabine and panobinostat alter the JunB promoter through different mechanisms.
A–B. K562 cells were treated with the indicated doses of DAC or panobinostat for 16 hours. Genomic DNA was isolated and bisulfite modified. PCR analysis was performed for the JunB, ERα, and E-cadherin promoters using primer sets for methylated and unmethylated DNA. Internal control primers for the amplification of bisulfite modified DNA regardless of methylation status were used to ensure equal loading. C. Schematic representation of the JunB promoter and CpG island with relative locations of primer sets used for ChIP PCR. D. K562 cells were treated with the indicated concentrations of panobinostat for 16 hours. Chromatin immunoprecipitations were performed with antibodies to DNMT1, SUV39H1, EZH2, 3MeK27 Histone H3, 3MeK9 Histone H3, and Acetyl H3. Normal mouse IgG was used as a control. PCR products were obtained for region 3 and region 5 on the chromatin immunoprecipitates. Quantitative assessment of the chromatin immunoprecipitates was also determined by qPCR. The bar graphs represent fold enrichment of the chromatin in each region of the promoter relative to the input chromatin.
Co-treatment with decitabine enhances panobinostat and 17-DMAG-mediated anti-leukemia activity in cultured and primary leukemia cells
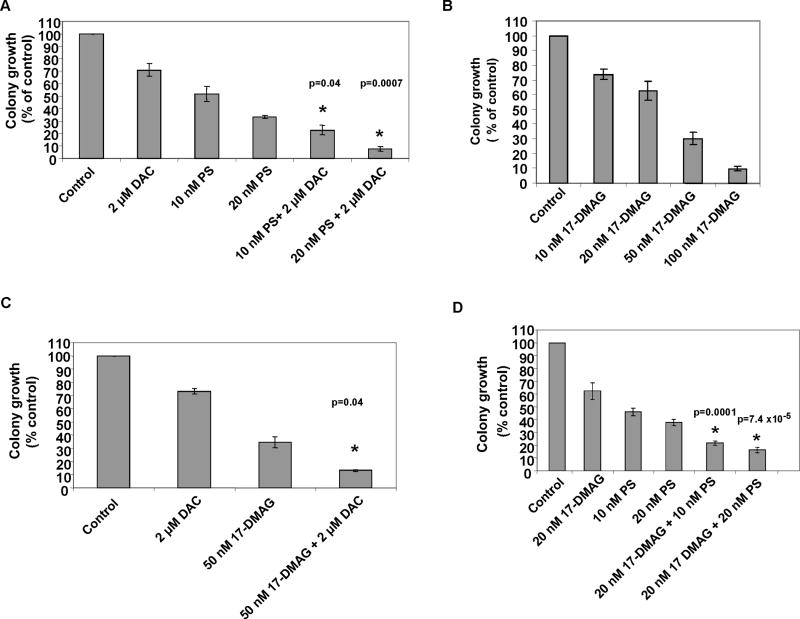
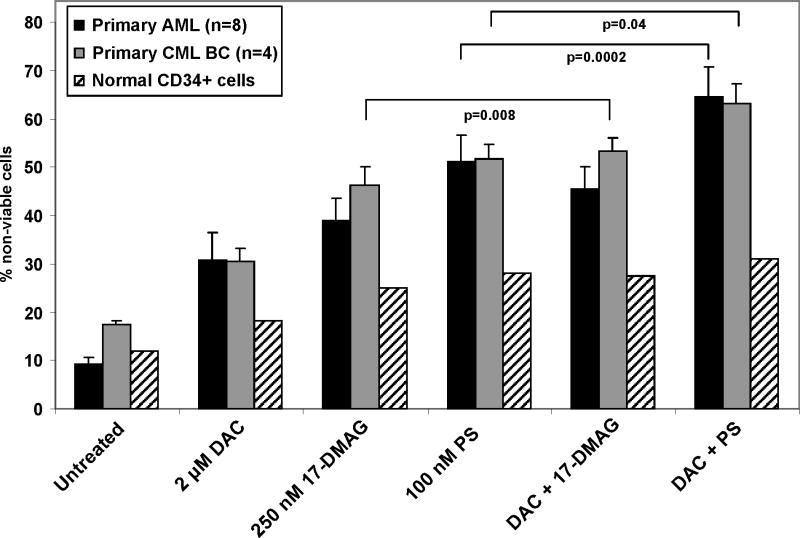
We next determined the effects of DAC alone and in combination with panobinostat or 17-DMAG on the clonogenic survival of leukemia cells. Treatment with DAC alone for 48 hours only modestly decreased the colony growth of K562 cells (Figure 6A). Treatment with panobinostat alone caused more inhibition of colony growth than DAC, whereas co-treatment with DAC and panobinostat mediated significantly greater loss of clonogenic survival than either agent alone (p=0.04 and p=0.007, respectively) (Figure 6A). The combined treatment also induced more apoptosis of K562 cells (data not shown). Treatment with 17-DMAG alone dose dependently inhibited colony growth of K562 cells (Figure 6B). Co-treatment with DAC and 17-DMAG also induced greater loss of clonogenic survival of K562 cells than treatment with either agent alone (p=0.04) (Figure 6C). Additionally, co-treatment with panobinostat and 17-DMAG significantly inhibited more colony growth of K562 cells than either agent alone (p=0.0001) (Figure 6D). We next determined the anti-leukemia activity of DAC and/or panobinostat and/or 17-DMAG against primary AML cells from 8 patients and primary CML-BC cells from 4 patients. Figure 7 demonstrates that treatment with clinically achievable concentrations of panobinostat > 17-DMAG > DAC mediated significant loss of viability in all samples tested. Notably, co-treatment with DAC significantly enhanced panobinostat or 17-DMAG-mediated loss of viability in the primary AML and CML cells (p=0.008 and p=0.0002; p=0.04 for panobinostat and DAC in CML). Co-treatment with DAC and panobinostat or 17-DMAG caused significantly lower loss of viability of normal bone marrow progenitor cells (Figure 7).
Figure 6. Co-treatment with decitabine and panobinostat, or 17-DMAG inhibits clonogenic survival greater than treatment with the single agents alone.
A. K562 cells were treated with the indicated doses of DAC and/or panobinostat for 48 hours. Following treatment the cells were washed and plated in methocult media for 7 days. (*) represents values significantly less than those following treatment with either agent alone at the indicated concentrations (p=0.04 and p=0.007, respectively). B–C. K562 cells were treated with the indicated doses of DAC and/or 17-DMAG for 48 hours. The cells were washed and plated as before. (*) represents values significantly less than those following treatment with either agent alone at the indicated concentrations (p=0.04). D. K562 cells were treated with the indicated concentrations of panobinostat and/or 17-DMAG for 48 hours. Following treatment the cells were washed and plated in methocult media for 7 days.
Figure 7. Co-treatment with panobinostat and decitabine exerts superior anti-leukemic activity against primary AML and CML cells than treatment with either agent alone.
Peripheral blood or bone marrow from 8 AML, 4 CML-BC and 2 normal CD34+ patients were treated with the indicated doses of 17-DMAG or panobinostat and DAC for 48 hours. Then, the percentages of non-viable cells for each drug alone or drug combination were determined by trypan blue uptake in a hemocytometer. Values represent the percentage of non-viable cells from each condition as compared to the untreated cells.
Discussion
Findings presented here demonstrate for the first time that both DNMT1 and EZH2 are hsp90 client proteins, and that treatment with pan-HDAC inhibitor panobinostat and the hsp90 inhibitor 17-DMAG disrupt the chaperone association between hsp90 and DNMT1 and EZH2, promoting their misfolding and depletion by the proteasome. Parenthetically, panobinostat-induced activities of caspases were not involved in degrading DNMT1 and EZH2. Although hsp90 is predominantly cytosolic, DNMT1 and EZH2 should now be included in the growing list of nuclear client proteins of hsp90.51 This highlights the involvement of molecular chaperones such as hsp90 not only in the DNA damage repair pathway but also in the epigenetic regulation of gene expression.52 Specifically, by regulating DNMT1 and EZH2, and also histone deacetylases, panobinostat regulates dual epigenetic mechanisms of DNA methylation and chromatin modifications through modulating hsp90 chaperone function.43,44 Previous reports have implicated hsp90 chaperone function and its regulation by panobinostat in the binding and action of nuclear hormone receptors with their co-activators and co-repressor in modulating the expression of hormone responsive genes.53 Collectively, these findings suggest that panobinostat treatment could influence multiple epigenetic mechanisms through its effects on HDAC and hsp90 functions.
Although the mechanism is not clear, panobinostat treatment also depleted the mRNA levels of DNMT1. This may further contribute toward panobinostat-mediated overall decline in the levels of DNMT1 in human leukemia cells. Despite the effect on DNMT1 levels, treatment with panobinostat had no effect on DNA methylation of JunB promoter. Panobinostat treatment also did not alter the mRNA levels of EZH2, but significantly depleted the protein levels of EZH2. We had previously reported that this was accompanied by disruption of the PRC2 complex and decline in the levels of SUZ12 and EED.44 Consistent with this, treatment with panobinostat markedly reduced the recruitment of EZH2 and DNMT1 to the promoter of JunB, as well as modified the epigenetic marks on the promoter chromatin of JunB. Specifically, panobinostat reduced the trimethylated K27 and K9 marks on histone H3. This was associated with significant de-repression of JunB. Therefore, collectively, panobinostat treatment appears to mediate its epigenetic effects by modifying lysine acetylation and methylation on histone proteins and by inhibiting DNMT1 recruitment on gene promoters without modifying promoter gene methylation. These data are in agreement with a previous report in which panobinostat was shown to induce expression of epigenetically silenced ERα through alteration of a multi-protein complex at the ERα promoter rather than through demethylation of the ERα promoter.41 In contrast, decitabine solely depletes DNMT1 and induces demethylation and de-repression of genes, e.g., JunB, ERα and E-cadherin. Hence, it is not surprising that the combination of DAC and panobinostat, by engaging separate and multiple epigenetic mechanisms, de-represses more JunB than treatment with either agent alone. Based on previous reports which had documented that JunB inactivation promotes leukemogenesis, accentuated derepression of JunB due to co-treatment with DAC and panobinostat could explain the loss of clonogenic survival and cell death of acute leukemia cells.31,32
Previous reports have shown that methylated genes in cancer cells are often packaged in nucleosomes containing H3 with trimethylated K27, which results from the activity of the EZH2 containing PRC2 complex recruiting DNMTs to the gene promoters.15 For example, PRC2 is required for the repression of E-cadherin by the Snail transcription factor.54 PRC2 is known to silence Hox and other differentiation program genes, thereby promoting self renewal of embryonic stem (ES) cells or of the cancer stem cells.3,43 Permanent silencing of tumor suppressors by acquisition of DNA methylation through DNMTs could be a specific feature of leukemogenesis.2,5 It appears that in cancer cells, the DNA methylome undergoes characteristic changes including genome wide loss of methylation and aberrant local gain of methylation, resulting in loss of TSG function.19,21 However, in addition to silencing due to DNA methylation, PRC2 may also be involved in down regulation of INK4A-ARF locus.7,55 Taken together, a picture of the epigenetic landscape in cancer is emerging that contains silenced genes that have a repressive trimethylated histone H3K27 mark and/or DNA methylation of TSGs.28,29 This is consistent with the observations, that selective down regulation of EZH2 restored the expression of trimethylated H3K27 marked genes, without affecting promoter DNA methylation or de-repressing genes silenced by DNA hypermethylation.28 The repressive trimethylated histone H3K27 along with trimethylated H3K4 mark represents the ‘bivalent mark’, which is a feature of the ‘bivalent domains’ in the silenced differentiation genes in ES and cancer stem cells.2,5 Thus, panobinostat treatment could erase the bivalent mark and de-repress the silenced differentiation genes in cancer stem cells. By depleting EZH2 levels and disrupting the PRC2 complex, as well as attenuating the DNMT1 levels, panobinostat treatment could also subsequently disrupt the localization of the PRC1 complex containing BMI1 and RING1 which contribute to the epigenetic silencing of differentiation genes in ES and cancer stem cells.3,5,56 Recently, activation of the ES cell transcriptional program by c-Myc has also been demonstrated to pathologic self-renewal characteristic in cancer stem cells.57 Whether panobinostat treatment alone or in combination with DAC also affects this program, remains to be elucidated.
Our data demonstrating greater de-repression of JunB and superior antileukemia efficacy of the combination of DAC and panobinostat are in agreement with previous reports showing robust re-expression of silenced genes when their promoters are de-methylated prior to HDAC inhibition.37,38 Increased anti-leukemia activity has also been demonstrated following treatment with the combination of DNMT and HDAC inhibitors.58 Recently, the combination of decitabine and valproic acid was reported to induce clinical responses in patients with advanced leukemia.59 In our studies, although co-treatment with DAC and panobinostat mediated greater loss of survival than either agent alone, how much of this is mediated by more pronounced attenuation of EZH2 and de-repression of JunB is not clear and remains to be established. It is noteworthy that induction of JunB in patients has been used as a prognostic marker for predicting clinical response to imatinib treatment in CML, and patients with increased JunB expression following treatment have been shown as more likely to achieve a clinical response than those with lower junB expression.60 Genetic or pharmacologic inhibition of EZH2 has also been shown to inhibit growth of transformed cells.10,13,61 Additionally, co-treatment with the HDAC inhibitor trichostatin A with pharmacologic inhibition of EZH2 has been shown to result in massive apoptosis of transformed cells.62 It should be noted that pan-HDAC inhibitors, such as panobinostat, induce apoptosis of acute leukemia cells through epigenetic and a variety of other mechanisms.63–65 Therefore, epigenetic targeting and de-repression of silenced genes is likely to be mechanistically only partially responsible for the enhanced antileukemia efficacy of the combination of DAC and panobinostat. Nevertheless, because of the relevance of the activity of EZH2 and PRC2 for the survival and self-renewal of leukemia stem cells, the combined epigenetic therapy with DAC and panobinostat represents an attractive strategy for the treatment of acute leukemia, especially in the minimal residual disease state.
Supplementary Material
Acknowledgments
Co-author PA is an employee of Novartis Institute for Biomedical Research Inc. and the corresponding author, KNB, has received clinical and laboratory research support from Novartis Institute for Biomedical Research Inc. All other authors have no competing financial interests.
Footnotes
Authorship
WF, KB, RR, AM, YY, RJ, YW, and performed the in vitro biochemical and molecular studies with the cultured leukemia cells. RB, JC, SK, AJ and SU performed the in vitro biologic studies with primary leukemia and normal CD34+ normal bone marrow progenitor cells. PA provided reagents for the study. KNB planned and supervised the in vitro and in vivo studies and prepared the report.
References
- 1.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–45. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein B, Meissner A, Lander E. The Mammalian Epigenome. Cell. 2007;128:669–81. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 3.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nature Rev Cancer. 2006;6:846–56. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 4.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–64. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008 Aug 3; doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 6.Kamminga LM, Bystrykh LV, de Boer A, Houwer S, Douma J, Weersing E, et al. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood. 2006;107:2170–9. doi: 10.1182/blood-2005-09-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–35. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otte AP, Kwaks TH. Gene repression by Polycomb group protein complexes: a distinct complex for every occasion? Curr Opin Genet Dev. 2003;13:448–54. doi: 10.1016/s0959-437x(03)00108-4. [DOI] [PubMed] [Google Scholar]
- 9.van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999;23:474–8. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- 10.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 11.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bachmann IM, Halvorsen OJ, Collett K, Stefansson IM, Straume O, Haukaas SA, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol. 2006;24:268–73. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 13.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–63. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–4. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 15.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–6. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 16.Ting AH, Jair K, Suzuki H, Yen R-WC, Baylin SB, Schuebel KE. Mammalian DNA methyltransferase 1: inspiration for new directions. Cell Cycle. 2004;3:1024–6. [PubMed] [Google Scholar]
- 17.Jeltsch A. On the enzymatic properties of Dnmt1: specificity, processivity, mechanism of linear diffusion and allosteric regulation of the enzyme. Epigenetics. 2006;1:63–6. doi: 10.4161/epi.1.2.2767. [DOI] [PubMed] [Google Scholar]
- 18.Svedruzic ZM. Mammalian cytosine DNA methyltransferase Dnmt1: enzymatic mechanism, novel mechanism-based inhibitors, and RNA-directed DNA methylation. Curr Med Chem. 2008;15:92–106. doi: 10.2174/092986708783330700. [DOI] [PubMed] [Google Scholar]
- 19.Teodoridis JM, Hardie C, Brown R. CpG island methylator phenotype (CIMP) in cancer: causes and implications. Cancer Lett. 2008;268:177–86. doi: 10.1016/j.canlet.2008.03.022. [DOI] [PubMed] [Google Scholar]
- 20.Robertson K. DNA methylation and chromatin - unraveling the tangled web. Oncogene. 2002;21:5361–79. doi: 10.1038/sj.onc.1205609. [DOI] [PubMed] [Google Scholar]
- 21.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 assoicates with histone deacetylase activity. Nat Genetics. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 23.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-reporessor, DMAP1, to form a complex at replication foci. Nat Genetics. 2000;25:269–77. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 24.Mizuno S, Chijiwa T, Okamura T, Akashi K, Fukumaki Y, Niho Y, Sasaki H. Expression of DNA methyltransferases DNMT-1, 3A and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood. 2001;97:1172–9. doi: 10.1182/blood.v97.5.1172. [DOI] [PubMed] [Google Scholar]
- 25.Melnick AM, Adelson K, Licht JD. The theoretical basis of transcriptional therapy of cancer: Can it be put into practice? J Clin Oncol. 2005;23:3957–70. doi: 10.1200/JCO.2005.14.498. [DOI] [PubMed] [Google Scholar]
- 26.Unterberger A, Andrews SD, Weaver ICG, Szyf M. DNA methyltransferase 1 knockdown activates a replication stress checkpoint. Mol Cell Biol. 2006;26:7575–86. doi: 10.1128/MCB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milutinovic S, Zhuang Q, Niveleau A, Szyf M. Epigenomic stress response. Knockdown of DNA methyltransferase 1 triggers an intra-S-phase arrest of DNA replication and induction of stress response genes. J Biol Chem. 2003;278:14985–95. doi: 10.1074/jbc.M213219200. [DOI] [PubMed] [Google Scholar]
- 28.Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–50. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 29.McGarvey KM, Greene E, Fahrner JA, Jenuwein T, Baylin SB. DNA methylation and complete transcriptional silencing of cancer genes persist after depletion of EZH2. Cancer Res. 2007;67:5097–102. doi: 10.1158/0008-5472.CAN-06-2029. [DOI] [PubMed] [Google Scholar]
- 30.Melki JR, Vincent PC, Clark SJ. Concurrent DNA hypermethylation of multiple genes in acute myeloid leukemia. Cancer Res. 1999;59:3730–3740. [PubMed] [Google Scholar]
- 31.Yang MY, Liu TC, Chang JG, Lin PM, Lin SF. JunB gene expression is inactivated by methylation in chronic myeloid leukemia. Blood. 2003;101:3205–11. doi: 10.1182/blood-2002-05-1598. [DOI] [PubMed] [Google Scholar]
- 32.Passegué E, Wagner EF, Weissman IL. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell. 2004;119:431–43. doi: 10.1016/j.cell.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 33.Furukawa Y, Sutheesophon K, Wada T, Nishimura M, Saito Y, Ishii H, et al. Methylation silencing of the Apaf-1 gene in acute leukemia. Mol Cancer Res. 2005;3:325–34. doi: 10.1158/1541-7786.MCR-04-0105. [DOI] [PubMed] [Google Scholar]
- 34.Shimamoto T, Ohyashiki JH, Ohyashiki K. Methylation of p15INK4b and E-cadherin genes is independently correlated with poor prognosis in acute myeloid leukemia. Leukemia Res. 2005;29:653–9. doi: 10.1016/j.leukres.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 35.Mariani O, Brennetot C, Coindre JM, Gruel N, Ganem C, Delattre O, et al. JUN oncogene amplification and overexpression block adipocytic differentiation in highly aggressive sarcomas. Cancer Cell. 2007;11:361–74. doi: 10.1016/j.ccr.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 36.Issa JP. Decitabine. Curr Opin Oncol. 2003;15:446–51. doi: 10.1097/00001622-200311000-00007. [DOI] [PubMed] [Google Scholar]
- 37.Issa JP, Gharibyan V, Cortes J, Jelinek J, Morris G, Verstovsek S, et al. Phase II Study of Low-Dose Decitabine in Patients With Chronic Myelogenous Leukemia Resistant to Imatinib Mesylate. J Clin Oncol. 2005;23:3948–56. doi: 10.1200/JCO.2005.11.981. [DOI] [PubMed] [Google Scholar]
- 38.Cameron EE, Bachman KE, Myöhänen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nature Gen. 1999;21:103–7. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 39.Belinsky SA, Klinge DM, Stidley CA, Issa JP, Herman JG, March TH, et al. Inhibition of DNA methylation and histone deacetylation prevents murine lung cancer. Cancer Res. 2003;63:7089–93. [PubMed] [Google Scholar]
- 40.Januchowski R, Dabrowski M, Ofori H, Jagodzinski PP. Trichostatin A down regulate DNA methyltransferase 1 in Jurkat T cells. Cancer Lett. 2007;246:313–17. doi: 10.1016/j.canlet.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Q, Atadja P, Davidson NE. Histone deacetylase inhibitor LBH589 reactivates silenced estrogen receptor alpha (ER) expression without loss of DNA hypermethylation. Cancer Biol Ther. 2007;6:64–9. doi: 10.4161/cbt.6.1.3549. [DOI] [PubMed] [Google Scholar]
- 42.Bali P, Pranpat M, Bradner J, Balasis M, Fiskus W, Guo F, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280:26729–34. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 43.Yang Y, Rao R, Shen J, Tang Y, Fiskus W, Nechtman J, Atadja P, Bhalla K. Role of acetylation and extracellular location of heat shock protein 90alpha in tumor cell invasion. Cancer Res. 2008;68:4833–42. doi: 10.1158/0008-5472.CAN-08-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fiskus W, Pranpat M, Balasis M, Bali P, Estrella V, Kumaraswamy S, et al. Histone deacetylase inhibitors deplete EZH2 and associated Polycomb Repressive Complex 2 proteins with attenuation of HOXA9 and MEIS1 and loss of survival of human acute leukemia cells. Mol Cancer Ther. 2006;5:3096–104. doi: 10.1158/1535-7163.MCT-06-0418. [DOI] [PubMed] [Google Scholar]
- 45.Drysdale MJ, Brough PA, Massey A, Jensen MR, Schoepfer J. Targeting Hsp90 for the treatment of cancer. Curr Opin Drug Discov Devel. 2006;9:483–95. [PubMed] [Google Scholar]
- 46.Nimmanapalli R, O’Bryan E, Huang M, Bali P, Burnette PK, Loughran T, et al. Molecular characterization and sensitivity of STI-571 (Imatinib Mesylate, Gleevec)-resistant, Bcr-Abl positive, human acute leukemia cells retain sensitivity to SRC kinase inhibitor PD180970 and 17-allylamino-17-demethoxygeldanomycin (17AAG) Cancer Res. 2002;62:5761–9. [PubMed] [Google Scholar]
- 47.Fiskus W, Wang Y, Joshi R, Rao R, Yang Y, Chen J, et al. Cotreatment with vorinostat enhances activity of MK-0457 (VX-680) against acute and chronic myelogenous leukemia cells. Clin Cancer Res. 2008;14:6106–15. doi: 10.1158/1078-0432.CCR-08-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo F, Sigua C, Tao J, Bali P, George P, Li Y, et al. Cotreatment with histone deacetylase inhibitor LAQ824 enhances Apo-2L/tumor necrosis factor-related apoptosis inducing ligand-induced death inducing signaling complex activity and apoptosis of human acute leukemia cells. Cancer Res. 2004;64:2580–9. doi: 10.1158/0008-5472.can-03-2629. [DOI] [PubMed] [Google Scholar]
- 49.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo F, Rocha K, Bali P, Pranpat M, Fiskus W, Boyapalle S, et al. Abrogation of heat shock protein 70 induction as a strategy to increase antileukemia activity of heat shock protein 90 inhibitor 17-allylamino-demethoxy geldanamycin. Cancer Res. 2005;65:10536–44. doi: 10.1158/0008-5472.CAN-05-1799. [DOI] [PubMed] [Google Scholar]
- 51.Dezwaan DC, Freeman BC. HSP90: the Rosetta stone for cellular protein dynamics? Cell Cycle. 2008;7:1006–12. doi: 10.4161/cc.7.8.5723. [DOI] [PubMed] [Google Scholar]
- 52.Camphausen K, Tofilon PJ. Inhibition of Hsp90: a multitarget approach to radiosensitization. Clin Cancer Res. 2007;13:4326–30. doi: 10.1158/1078-0432.CCR-07-0632. [DOI] [PubMed] [Google Scholar]
- 53.Fiskus W, Ren Y, Mohapatra A, Bali P, Mandawat A, Rao R, et al. Hydroxamic acid analogue histone deacetylase inhibitors attenuate estrogen receptor alpha levels and transcriptional activity: a result of hyperacetylation and inhibition of chaperone function of heat shock protein 90. Clin Cancer Res. 2007;13:4882–90. doi: 10.1158/1078-0432.CCR-06-3093. [DOI] [PubMed] [Google Scholar]
- 54.Herranz N, Pasini D, Díaz VM, Francí C, Gutierrez A, Dave N, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol. 2008;28:4772–81. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kotake Y, Cao R, Viatour P, Sage J, Zhang Y, Xiong Y. pRB family proteins are required for H3K27 trimethylation and polycomb repression complexes binding to and silencing p16INK4a tumor suppressor gene. Genes Dev. 2007;21:49–54. doi: 10.1101/gad.1499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 57.Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–44. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gore SD, Baylin S, Sugar E, Carraway H, Miller CB, Carducci M, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 2006;66:6361–9. doi: 10.1158/0008-5472.CAN-06-0080. [DOI] [PubMed] [Google Scholar]
- 59.Garcia-Manero G, Kantarjian HM, Sanchez-Gonzalez B, Yang H, Rosner G, Verstovsek S, et al. Phase 1/2 study of the combination of 5-aza-2′-deoxycytidine with valproic acid in patients with leukemia. Blood. 2006;108:3271–9. doi: 10.1182/blood-2006-03-009142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu YC, Hsiao HH, Chang JG, et al. Usefulness of quantitative assessment of JunB gene expression as a marker for monitoring chronic myeloid leukemia patients undergoing imatinib therapy. Int J Hematol. 2006;84:425–31. doi: 10.1532/IJH97.A10514. [DOI] [PubMed] [Google Scholar]
- 61.Chen Y, Lin MC, Yao H, Wang H, Zhang AQ, Yu J, et al. Lentivirus-mediated RNA interference targeting enhancer of zeste homolog 2 inhibits hepatocellular carcinoma growth through down-regulation of stathmin. Hepatology. 2007;46:200–8. doi: 10.1002/hep.21668. [DOI] [PubMed] [Google Scholar]
- 62.Jiang X, Tan J, Li J, Kivimäe S, Yang X, Zhuang L, et al. DACT3 is an epigenetic regulator of Wnt/beta-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell. 2008;13:529–41. doi: 10.1016/j.ccr.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.George P, Bali P, Annavarapu S, Scuto A, Fiskus W, Guo F, et al. Combination of the histone deacetylase inhibitor LBH589 and the hsp90 inhibitor 17-AAG is highly active against human CML-BC cells and AML cells with activating mutation of FLT-3. Blood. 2005;105:1768–76. doi: 10.1182/blood-2004-09-3413. [DOI] [PubMed] [Google Scholar]
- 64.Fiskus W, Pranpat M, Bali P, Balasis M, Kumaraswamy S, Boyapalle S, et al. Combined effects of novel tyrosine kinase inhibitor AMN107 and histone deacetylase inhibitor LBH589 against Bcr-Abl expressing human leukemia cells. Blood. 2006;108:645–52. doi: 10.1182/blood-2005-11-4639. [DOI] [PubMed] [Google Scholar]
- 65.Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res. 2007;5:981–9. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.