Abstract
Aberrant messenger RNAs containing a premature termination codon (PTC) are eliminated by the nonsense-mediated mRNA decay (NMD) pathway. Here, we show that a crucial NMD factor, up frameshift 1 protein (Upf1), is required for rapid proteasome-mediated degradation of an aberrant protein (PTC product) derived from a PTC-containing mRNA. Western blot and pulse–chase analyses revealed that Upf1 stimulates the degradation of specific PTC products by the proteasome. Moreover, the Upf1-dependent, proteasome-mediated degradation of the PTC product was also stimulated by mRNAs harbouring a faux 3′ untranslated region (3′-UTR). These results indicate that protein stability might be regulated by an aberrant mRNA 3′-UTR.
Keywords: Upf1, quality control, protein degradation, proteasome, PTC product
Introduction
Gene expression is highly accurate because of quality control systems that prevent the expression of potentially harmful protein products. One such system, the nonsense-mediated messenger RNA decay pathway (NMD), recognizes and eliminates aberrant mRNAs containing a premature termination codon (PTC). Three NMD factors, up frameshift 1 protein (Upf1), Upf2 and Upf3, are highly conserved, and aberrant termination at a premature nonsense codon is thought to allow the binding of NMD factors to the ribosome, triggering mRNA degradation. In mammalian cells, translation termination codons and exon–exon junctions are cis-acting elements that allow the recognition of PTCs (Holbrook et al, 2004; Maquat, 2004). An abnormally long 3′ untranslated region (3′-UTR) can also trigger NMD in yeast, mammalian, Drosophila melanogaster, Caenorhabditis elegans and plant cells (Kertesz et al, 2006; Schwartz et al, 2006; Behm-Ansmant et al, 2007; Longman et al, 2007), although not all mRNAs with an extended 3′-UTR are subjected to NMD. The poly(A)-binding protein 1 (Pab1) interacts with the translation termination factor, eukaryotic release factor 3, to enhance normal termination efficiency (Cosson et al, 2002; Uchida et al, 2002). Premature termination, however, is thought to be characterized by the absence of proximal Pab1, leading to a reduction in translation termination efficiency and in the association of NMD factors with the termination complex (Amrani et al, 2004). This faux 3′-UTR model is supported by the observation that NMD is suppressed by the tethering of Pab1 to the proximal position of a PTC in yeast (Amrani et al, 2004), Drosophila (Behm-Ansmant et al, 2007) and mammalian cells (Ivanov et al, 2008), indicating that the role of Pabs in the identification of PTCs is conserved across eukaryotes. By contrast, it has been reported that PTC is recognized even in a pab1 deletion mutant (Meaux et al, 2008), suggesting that PTC could be recognized independently of Pab1 in yeast.
Furthermore, it has been shown that post-translational regulation could have a crucial function in quality control (Inada & Aiba, 2005; Ito-Harashima et al, 2007). These findings indicate that translation arrest and protein degradation are crucial for preventing potentially harmful products of aberrant mRNAs working together with mRNA degradation. Translation is essential for discriminating PTC-containing mRNAs, and therefore the truncated products might be synthesized from PTC-containing mRNAs until NMD factors are able to recognize and identify these aberrant mRNAs. It should be emphasized that NMD generally reduces the abundance of nonsense-containing mRNAs to approximately 5–25% of the nonsense-free level, indicating that aberrant products might be produced to some extent. Aberrant proteins can acquire dominant-negative activities; therefore it is crucial that cells repress the production of aberrant proteins derived from PTC-containing mRNAs. It has been shown that translation of PTC-containing aberrant mRNAs is repressed at the translation initiation step (Isken et al, 2008). However, it is unknown whether other steps in gene expression have crucial functions in repressing the aberrant proteins of PTC-containing mRNAs.
Here, we examined the levels of protein produced by aberrant PTC-containing mRNAs to investigate the potential role of other regulatory processes in the quality control system that represses PTC products. By stimulating proteasome-dependent protein degradation and mRNA degradation, we found that Upf1 downregulates the level of aberrant imidazoleglycerol-phosphate dehydratase (His3) or 3-phosphoglycerate kinase 1 (Pgk1) proteins derived from mRNAs containing a PTC at specific positions. The downregulation of specific PTC products by Upf1p requires aberrant 3′-UTRs and proteasome activity, indicating that Upf1 stimulates the proteasome-mediated degradation of specific PTC products, wherein the stability of these proteins is regulated by the length of the 3′-UTR. These results lead us to propose that, working together with rapid mRNA degradation, protein degradation by the proteasome has a function in preventing the expression of abnormal proteins derived from aberrant mRNAs.
Results And Discussion
Downregulation of aberrant proteins by Upf1
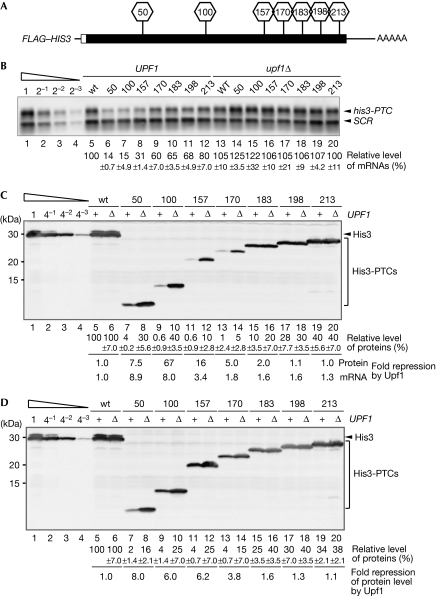
It has been shown that PTC-containing aberrant mRNAs are recognized by NMD factors and are rapidly degraded by general mRNA decay pathways. To examine whether other steps in gene expression have crucial functions in repressing proteins derived from aberrant mRNAs, we constructed FLAG–HIS3 reporter genes containing a PTC at different positions (Fig 1A), and determined the levels of aberrant mRNAs and proteins. As shown in Fig 1B, the decrease in the level of PTC-containing mRNAs was successively less marked as the PTC was positioned further towards the 3′ end within the HIS3 open reading frame (ORF), as previously reported (Peltz et al, 1993; Cao & Parker, 2003; Meaux et al, 2008). We also determined the levels of protein derived from FLAG–HIS3 reporter genes containing a PTC at various codon positions. Western blot analysis using secondary antibodies conjugated to quantum dots (Bakalova et al, 2005), which allows the detection of proteins at low concentrations, revealed that Upf1 reduced protein levels more efficiently than it did mRNA levels, especially for constructs with PTCs at positions 100 and 157 (Fig 1B, lanes 7–8 and 15–16; Fig 1C, lanes 9–12). For example, the level of FLAG–his3-100 mRNA relative to FLAG–HIS3 mRNA was 15% in wild-type cells, whereas the protein level of FLAG–his3-100 relative to FLAG–HIS3 was less than 1% (Fig 1B, lane 7; Fig 1C, lane 9). However, in the absence of Upf1, the relative FLAG–his3-100 mRNA level was 122%, whereas the relative protein level was 40% (Fig 1B, lane 15; Fig 1C, lane 10). The decrease to 40% protein in comparison with 122% mRNA indicates that protein degradation or reduced translation also occurs in the absence of Upf1. These results show that truncated protein levels are significantly lower than mRNA levels, and indicate that Upf1 contributes to the downregulation of severely truncated PTC products at the translational or post-translational level, as well as at the mRNA level.
Figure 1.
Upf1 strongly downregulates the level of proteins produced from HIS3 genes containing a PTC at specific positions. (A) A schematic of the reporter FLAG–HIS3 gene that expresses mRNAs containing a PTC at the indicated codon number. The filled box indicates the open reading frame of HIS3 and the open box indicates the FLAG tag; lines represent untranslated regions, and AAAAA denotes the poly(A) tail. (B) The ability of Upf1 to downregulate aberrant mRNAs is increased, depending on the length of the 3′-UTR. The reporter mRNA levels were determined by northern blot analysis with a DIG-labelled HIS3 probe. The relative levels for each mRNA were normalized to the FLAG–HIS3 mRNA level in wild-type cells, which was assigned a value of 100, and SCR RNA levels were used as a loading control for RNA samples. The mean values of three independent experiments are shown. (C) Upf1 downregulates PTC product levels in a PTC position-specific manner. Protein levels were determined by quantitative western blot analysis using secondary antibodies conjugated to quantum dots (Invitrogen, Carlsbad, CA, USA). The relative levels of each product were normalized to FLAG–His3 protein levels in wild-type cells (assigned a value of 100). The mean values of three independent experiments are shown. (D) The strong downregulation of FLAG–His3-100 or FLAG–His3-157 protein levels by Upf1 is partly suppressed in the presence of MG132. W303 (+) or W303upf1Δ (Δ) cells harbouring the plasmids shown in (A) were grown in SC-Ura (BD Difco, Detroit, MI, USA) in the presence of 0.2 mM MG132. Protein levels were determined as described in (C). DIG, digoxigenin; mRNA, messenger RNA; PTC, premature termination codon; SCR, RNA subunit of signal recognition particle; SC-Ura, synthetic complete medium lacking uracil; Upf1, up frameshift protein 1; 3′-UTR, 3′ untranslated region; wt, wild type.
To determine whether the proteasome is involved in this process, we measured the levels of protein derived from various reporter genes in UPF1 and upf1Δ cells after the addition of MG132. The FLAG-His3-100 and FLAG-His3-157 protein levels in wild-type cells were low (0.6%) in comparison with the levels of FLAG-His3 proteins in wild type (100%), but were increased significantly in the presence of 0.2 mM MG132, up to 4% of His3 (Fig 1D, lanes 9–12). By contrast, the levels of longer PTC proteins were minimally affected by the addition of MG132, whereas the levels of FLAG-His3-170 and FLAG-His3-183 protein slightly increased in the presence of MG132 (Fig 1D, lanes 13–20). Unexpectedly, the levels of FLAG-His3-50 protein in both UPF1 and upf1Δ cells were decreased in the presence of MG132 by an unknown mechanism (Fig 1D, lanes 7–8). Upf1 downregulated the FLAG–His3-100 protein sixfold in the presence of MG132 (Fig 1D, lanes 9–10) to nearly the same degree as the downregulation of FLAG–his3-100 mRNA (Fig 1B, lanes 7 and 15). It has been reported that proteasome inhibitors also inhibit translation through phosphorylation of eukaryotic initiation factor 2α, which interferes with NMD (Mazroui et al, 2007). Pulse-label experiments indicate that the rate of protein synthesis was only slightly affects by MG132 treatment (supplementary Fig S1 online). Consistently, MG132 treatment did not affect either the steady-state levels (supplementary Fig S2 online) or the stability of FLAG–his3-100 mRNA in wild-type and upf mutant yeast cells (Fig 2A). Therefore, we concluded that MG132 treatment minimally affects NMD under the conditions used in our study. In wild-type cells, the level of FLAG–his3-100 mRNA relative to FLAG–HIS3 mRNA was 15%, and that of the protein was only 4%, even though the proteasome was inhibited by the addition of MG132. This is consistent with previous observations showing that the translation of PTC-containing mRNAs is repressed (Amrani et al, 2004; Sheth & Parker, 2006; Isken et al, 2008). However, we could not exclude the possibility that another type of proteolysis, in addition to proteasomal degradation, could be involved in the downregulation of the FLAG–His3-100 protein.
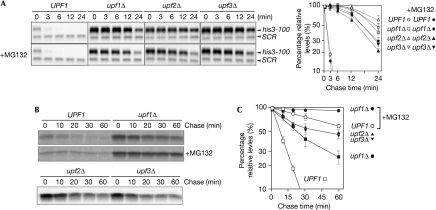
Figure 2.
The stability of the FLAG–His3-100 protein is decreased in cells expressing Upf1. (A) MG132 treatment does not affect the stability of FLAG–his3-100 mRNA in wild-type and upf mutants. W303 or W303upf1Δ, W303upf2Δ or W303upf3Δ cells harbouring pGAL1p-his3-100 were grown in SC-Ura (BD Difco, Detroit, MI, USA) containing 2% galactose. Cells were collected at the indicated times, after the addition of glucose to inhibit transcription from the GAL1 promoter. RNA samples were analysed by northern blot, as shown in Fig 1B. Relative quantities are shown as the mean values of three independent experiments with s.d. values. (B) FLAG–His3-100 is more stable in upf mutants. Pulse–chase experiments were carried out using W303 or W303upf1Δ, W303upf2Δ or W303upf3Δ cells harbouring pGPD-FLAG–his3-100, as described in the Methods section. When indicated, cells were grown in the presence of 0.2 mM MG132. (C) The relative levels of protein were shown as a function of the chase time. Relative amounts are shown as the mean values of three independent experiments with s.d. values. GAL1, galactokinase; GPD, glycerol-3-phosphate dehydrogenase; mRNA, messenger RNA; SC-Ura, synthetic complete medium lacking uracil; SCR, RNA subunit of signal recognition particle; Upf1, up frameshift protein 1.
FLAG–His3-100 is more stable in upf mutants
To address the possibility that Upf1 might stimulate proteasome-dependent protein degradation of truncated PTC products, the stability of these proteins was examined directly by pulse–chase analysis. We found that the FLAG–His3-100 protein was more labile in W303 cells (t1/2=7 min) than in upf1Δ (t1/2=30 min), upf2Δ or upf3Δ cells (Fig 2B,C). In addition, FLAG–His3-100 proteins were significantly stabilized in wild-type (t1/2 ≈90 min) and upf1Δ cells (t1/2>120 min) in the presence of MG132 (Fig 2B,C). These results indicate that Upf1 destabilizes FLAG–His3-100 proteins by stimulating their degradation, which is at least partly proteasome dependent. There is a possibility that protease(s) other than the proteasome might be involved in destabilizing truncated proteins, because MG132 does not completely inhibit proteasome function. We propose that Upf1 stimulates proteasome-dependent degradation of the FLAG–His3-100 protein, at least partly, to reduce PTC product levels together with the degradation of mRNA.
One possible explanation for the Upf1-mediated destabilization of FLAG–His3-100 protein (partly through a proteasome-dependent pathway) is that proteasome activity is generally impaired in the upf1Δ mutant. To investigate this possibility, we first measured the levels of ubiquitinated proteins by western blot analysis using an FK2 antibody (anti-ubiquitinylated proteins, clone FK2) that recognizes both mono-ubiquitin and poly-ubiquitin forms, but not free ubiquitin (Fujimuro et al, 1994). Rpn6 is a component of the proteasome lid, and the activity of the proteasome is impaired in an rpn6-2 temperature-sensitive mutant (Saeki et al, 2005). We found that levels of ubiquitinated proteins increased markedly in the rpn6-2 mutant but not in the upf1Δ mutant (supplementary Fig S3 online). Furthermore, the half-life of the well-known substrate of the proteasome, Sic1 (Petroski & Deshaies, 2005), was almost the same in wild type and upf1Δ mutants (supplementary Fig S4 online). These results also indicate that proteasome activity is not impaired in upf1Δ mutant strains.
Long 3′-UTR is required for downregulation of His3-100
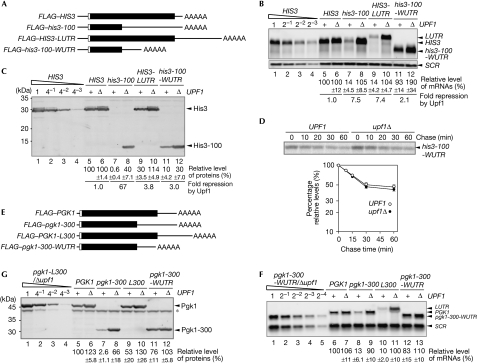
To investigate the mechanism by which Upf1 mediates the efficient downregulation of the FLAG–His3-100 protein, we constructed reporter genes as shown in Fig 3A. Upf1 downregulated FLAG–his3-100 mRNA levels by 7.5-fold (Fig 3B, lanes 7–8), and this reduction was significantly suppressed when the long 3′-UTR was replaced by wild-type 3′-UTR (FLAG–his3-100-WUTR; Fig 3B, lanes 11–12). Upf1 also reduced the levels of a FLAG–HIS3-LUTR mRNA that contained an intact ORF flanked by a long 3′-UTR (Fig 3B, lanes 9–10). These results are consistent with the faux 3′-UTR model that states that NMD requires improperly long 3′-UTRs (Amrani et al, 2004). We found that Upf1 reduced both FLAG–HIS3-LUTR mRNA and protein levels by almost the same extent (Fig 3B,C, lanes 9–10). These results indicate that Upf1 mainly downregulates the levels of the protein derived from FLAG–HIS3-LUTR by reducing mRNA levels, rather than by proteolysis. On the basis of these results, we propose that post-translational downregulation by proteolysis occurs only when the protein is truncated. In the case of FLAG–HIS3-LUTR, Upf1 regulated the levels of mRNA but not of the properly folded wild-type protein. We also found that Upf1 moderately downregulated the mRNA levels of a short ORF flanked by wild-type 3′-UTR (Fig 3B, lanes 11–12), but minimally affected the mRNA and protein levels derived from FLAG–HIS3 (Fig 3B,C, lanes 5–6). Therefore, we suspect that the shorter ORF might stimulate the effects of Upf1 on mRNA levels, and the wild-type HIS3 3′-UTR may not be long enough to repress NMD when the ORF is shorter than normal. To our surprise, the downregulation of the FLAG–His3-100 protein was significantly suppressed by replacing the long 3′-UTR with a wild-type 3′-UTR (FLAG–his3-100-WUTR; Fig 3C, lanes 11–12). Pulse–chase experiments also revealed that Upf1 minimally destabilized the FLAG–His3-100 protein expressed from FLAG–his3-100-WUTR (Fig 3D). These results indicate that the Upf1 downregulation of the levels of protein derived from FLAG–his3-100 depends more strongly on a faux 3′-UTR than does the Upf1 downregulation of mRNA levels. We also found that Upf1 downregulates both protein and mRNA levels derived from FLAG–pgk1-300, a construct with a PTC at codon 300 of PGK1, depending on the length of the 3′-UTR (Fig 3E–G). In addition, Upf1 stimulates proteasome-dependent degradation of the FLAG–Pgk1-300 protein, depending on the faux 3′-UTR (Fig 4). Considering these results, we propose that protein stability could be regulated by the length of the 3′-UTR of an mRNA.
Figure 3.
Upf1 downregulates His3-100 levels depending on faux 3′-UTR. (A) Schematic drawing of the mRNA derived from FLAG–HIS3 reporter genes. Filled boxes indicate the open reading frame, the open box indicates FLAG tag, lines represent UTRs and AAAAA denotes the poly(A) tail. (B) Upf1 downregulates aberrant mRNAs with long 3′-UTRs. W303 (+) or W303upf1Δ (Δ) cells harbouring the plasmids shown in (A) were grown and RNA samples were prepared. Relative mRNA levels were determined as described in Fig 1B. (C) Upf1 strongly reduces the levels of the FLAG–His3-100 protein in the presence of a long 3′-UTR. Relative protein levels were determined as described in Fig 1C. (D) The stability of the FLAG–His3-100 protein derived from pGPD-FLAG–his3-100-WUTR is unaffected by Upf1. Top: pulse–chase experiments were carried out using W303 or W303upf1Δ cells harbouring pGPD-FLAG–his3-100-WUTR. Bottom: relative levels of proteins were shown as a function of chase time. (E) Schematic for the mRNAs expressed from FLAG–PGK1 reporter genes constructed as described in the supplementary information online. (F) Upf1 downregulates aberrant PGK1 mRNAs with a long 3′-UTR. W303 or W303upf1Δ cells were transformed with the indicated plasmids. The levels of reporter mRNAs were analysed by northern blotting with a DIG-labelled anti-FLAG LNA oligonucleotide probe as described in the supplementary information online. The relative levels of each RNA product normalized to the FLAG–PGK1 mRNA level in wild-type cells (assigned a value of 100) are shown as mean values from three independent experiments. (G) Upf1 strongly reduces the level of the Pgk1-300 protein in a long 3′-UTR-dependent manner. W303 or W303upf1Δ cells containing indicated plasmids were grown in SC-Ura medium (BD Difco, Detroit, MI, USA). Samples were prepared, and protein levels were analysed by western blotting using a FLAG antibody. The relative levels of each protein product, normalized to the FLAG–Pgk1 protein level in wild-type cells (assigned a value of 100), are shown as mean values from three independent experiments. The asterisk indicates the endogenous products that are recognized by quantum dot streptavidin conjugates. DIG, digoxigenin; LNA, locked nucleic acid; mRNA, messenger RNA; PGK1, 3-phosphoglycerate kinase 1; SC-Ura, synthetic complete medium lacking uracil; SCR, RNA subunit of signal recognition particle; Upf1, up frameshift protein 1; 3′-UTR, 3′ untranslated region.
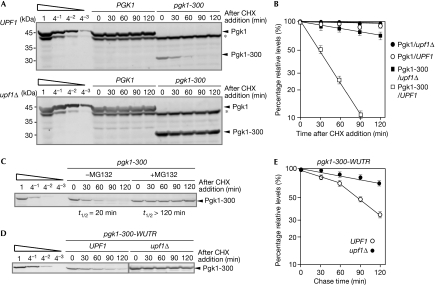
Figure 4.
Upf1 destabilizes the Pgk1-300 protein depending on proteasome activity. (A) Upf1 destabilizes the FLAG–Pgk1-300 protein. Samples of W303 (top panel) or W303upf1Δ (bottom panel) cells containing a pGPD-FLAG-pgk1-300 or pGPD-FLAG-PGK1 plasmid were prepared at the indicated times after the addition of cycloheximide (0.5 mg/ml). The levels of the remaining proteins were determined by quantum dot western blotting using a FLAG antibody. The asterisk indicates the endogenous products that are recognized by quantum dot streptavidin conjugates. (B) The half-life of reporter proteins. The levels of remaining FLAG–Pgk1 or FLAG–Pgk1-300 protein after inhibition of translation elongation were determined. The mean±s.d. values obtained from at least three independent experiments are shown. (C) The Pgk1-300 protein is stabilized by the addition of MG132. W303 cells containing a pGPD-FLAG-pgk1-300 plasmid were grown in the presence of 0.2 mM MG132, and samples were prepared and analysed by western blot analysis. (D) Upf1 destabilizes the FLAG–Pgk1-300 protein depending on a faux 3′-UTR. The stability of the FLAG–Pgk1-300 protein derived from FLAG-pgk1-300-WUTR mRNA in W303 or W303upf1Δ cells was measured, as shown in (A). (E) The half-life of Pgk1-300 reporter proteins derived from pgk1-300-WUTR reporter gene. The levels of the remaining FLAG–Pgk1-300 protein after inhibition of translation elongation were determined, as shown in (A). GPD, glycerol-3-phosphate dehydrogenase; PGK1, 3-phosphoglycerate kinase 1; Upf1, up frameshift protein 1; 3′-UTR, 3′ untranslated region.
Recent results, combined with the results reported here, indicate a possible mechanism for the stimulation of protein degradation by Upf1. Recently, it was reported that Upf1 potentially acts as an E3 ubiquitin ligase by its association with Upf3 in yeast (Takahashi et al, 2008). Therefore, Upf1 might act as an E3 ligase of the truncated protein, leading to the degradation of ubiquitinated protein by the proteasome. As Upf1 is recruited to aberrant mRNAs by an interaction with eukaryotic release factor 3 (Amrani et al, 2004; Kashima et al, 2006; Behm-Ansmant et al, 2007; Ivanov et al, 2008; Singh et al, 2008), the ubiquitination of the aberrant protein might occur after the binding of eukaryotic release factors to the ribosome. It is unknown how quickly the peptide is released from the ribosome after hydrolysis of the peptidyl-tRNA bond. To understand the mechanism by which Upf1 stimulates protein degradation, the kinetics of translation termination must be analysed more precisely in vivo. Mutational analysis of Upf1 is required for clarifying the mechanism of stimulation of protein degradation by Upf1.
We previously reported that translation of the poly(A) tail has a crucial function in repressing the production of aberrant proteins from nonstop mRNAs by both translation repression and proteasome-dependent nascent protein destabilization in yeast (Inada & Aiba, 2005; Ito-Harashima et al, 2007). This clearly indicates that translation arrest and protein degradation, in addition to mRNA degradation, are crucial for repressing the expression of nonstop mRNAs. We also recently reported that translation arrest caused by the presence of consecutive basic amino acids in a nascent protein induces Not4-dependent co-translational protein degradation by the proteasome (Dimitrova et al, 2009). In this study, we found that Upf1 prevents the accumulation of aberrant proteins by stimulating the degradation of specific PTC products. Taking these results together, we propose that protein degradation by the proteasome, working together with rapid mRNA degradation, has an important role in preventing the expression of abnormal proteins derived from aberrant mRNAs.
Methods
Yeast strains and plasmids. Details of the yeast strains and plasmids used in this study are described in supplementary Table S1 online, and information about the oligonucleotides is in supplementary Table S2 online. The construction of plasmids and other methods are described in the supplementary information online.
Determining relative protein levels by western blot with quantum dots. To quantify the protein levels, protein samples equivalent to 0.1 OD600 (optical density at 600 nm) were analysed by western blotting using secondary antibodies conjugated to quantum dots (Invitrogen, Carlsbad, CA, USA). The band intensities for diluted samples (quantum dot fluorescence was detected by FLA-9000; FujiFilm, Tokyo, Japan) were compared with a standard curve, and the level of proteins relative to the control was determined.
Pulse–chase experiments. Yeast cells were grown exponentially at 30°C in a minimal medium lacking methionine and cysteine uracil. Yeast cells (10 ml) were labelled with 100 μCi [35S] methionine and cysteine (PerkinElmer NEG072, Waltham, MA, USA) for 1 min, followed by the addition of cold amino acids (final density of 40 μg/ml). At indicated times, cells were collected and cell extracts were prepared by Complete Lysis-Y (Roche, NJ, USA). Cell extracts were incubated with an anti-FLAG M2 resin in IXA-100 buffer (Inada & Aiba, 2005), and then washed three times and eluted with 0.4 mg/ml FLAG peptide. Samples to undergo immunoprecipitation were separated by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS–PAGE). The radioactivity of precipitated proteins was measured using Typhoon9400 (GE, Healthcare, NJ, USA).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank Dr Allan Jacobson and Dr John Hershey for critically reading this paper and for their valuable comments and suggestions. We also thank Dr Toshiya Endo, Dr Tohru Yoshihisa, Dr Takumi Kamura and Dr Satoru Mimura for yeast strain plasmids and helpful discussions. This study was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to T.I.).
Footnotes
The authors declare that they have no conflict of interest.
References
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A (2004) A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 [DOI] [PubMed] [Google Scholar]
- Bakalova R, Zhelev Z, Ohba H, Baba Y (2005) Quantum dot-based western blot technology for ultrasensitive detection of tracer proteins. J Am Chem Soc 127: 9328–9329 [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E (2007) A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J 26: 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Parker R (2003) Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell 113: 533–545 [DOI] [PubMed] [Google Scholar]
- Cosson B, Couturier A, Chabelskaya S, Kiktev D, Inge-Vechtomov S, Philippe M, Zhouravleva G (2002) Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol Cell Biol 22: 3301–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova LN, Kuroha K, Tatematsu T, Inada T (2009) Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem 284: 10343–10352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimuro M, Sawada H, Yokosawa H (1994) Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett 349: 173–180 [DOI] [PubMed] [Google Scholar]
- Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE (2004) Nonsense-mediated decay approaches the clinic. Nat Genet 36: 801–808 [DOI] [PubMed] [Google Scholar]
- Inada T, Aiba H (2005) Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J 24: 1584–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE (2008) Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell 133: 314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito-Harashima S, Kuroha K, Tatematsu T, Inada T (2007) Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev 21: 519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE (2008) Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 27: 736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S (2006) Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz S, Kerenyi Z, Merai Z, Bartos I, Palfy T, Barta E, Silhavy D (2006) Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res 34: 6147–6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman D, Plasterk RH, Johnstone IL, Caceres JF (2007) Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev 21: 1075–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE (2004) Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 5: 89–99 [DOI] [PubMed] [Google Scholar]
- Mazroui R, Di Marco S, Kaufman RJ, Gallouzi IE (2007) Inhibition of the ubiquitin–proteasome system induces stress granule formation. Mol Biol Cell 18: 2603–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaux S, van Hoof A, Baker KE (2008) Nonsense-mediated mRNA decay in yeast does not require PAB1 or a poly(A) tail. Mol Cell 29: 134–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz SW, Brown AH, Jacobson A (1993) mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev 7: 1737–1754 [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ (2005) Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF–Cdc34. Cell 123: 1107–1120 [DOI] [PubMed] [Google Scholar]
- Saeki Y, Isono E, Shimada M, Kawahara H, Yokosawa H, Toh EA (2005) Knocking out ubiquitin proteasome system function in vivo and in vitro with genetically encodable tandem ubiquitin. Methods Enzymol 399: 64–74 [DOI] [PubMed] [Google Scholar]
- Schwartz AM, Komarova TV, Skulachev MV, Zvereva AS, Dorokhov YL, Atabekov JG (2006) Stability of plant mRNAs depends on the length of the 3′-untranslated region. Biochemistry (Mosc) 71: 1377–1384 [DOI] [PubMed] [Google Scholar]
- Sheth U, Parker R (2006) Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125: 1095–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Rebbapragada I, Lykke-Andersen J (2008) A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol 6: e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Araki Y, Ohya Y, Sakuno T, Hoshino S, Kontani K, Nishina H, Katada T (2008) Upf1 potentially serves as a RING-related E3 ubiquitin ligase via its association with Upf3 in yeast. RNA 14: 1950–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Hoshino S, Imataka H, Sonenberg N, Katada T (2002) A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/Poly(A)-dependent translation. J Biol Chem 277: 50286–50292 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information