Abstract
The regulation of gene expression programmes is essential for the generation of diverse cell types during development and for adaptation to environmental signals. RNA polymerase II (RNAPII) transcribes genetic information and coordinates the recruitment of accessory proteins that are responsible for the establishment of active chromatin states and transcript maturation. RNAPII is post-translationally modified at active genes during transcription initiation, elongation and termination, and thereby recruits specific histone and RNA modifiers. RNAPII complexes are also located at silent genes in promoter-proximal paused configurations that provide dynamic transcriptional regulation downstream from initiation. In embryonic stem cells, silent developmental regulator genes that are repressed by Polycomb are associated with a form of RNAPII that can elongate through coding regions but that lacks the post-translational modifications that are important for coupling RNA synthesis to co-transcriptional maturation. Here, we discuss the mechanisms through which the transcription of silent genes might be dissociated from productive expression, and the sophisticated interplay between the transcriptional machinery, Polycomb repression and RNA processing.
Keywords: RNA polymerase II, C-terminal domain, chromatin bivalency, transcriptional priming, Polycomb silencing
See Glossary for abbreviations used in this article.
Glossary.
- 2A-HUB
H2A histone ubiquitin ligase
carboxy-terminal domain
major histocompatibility complex, class II, DRα
DRB-sensitivity inducing factor
embryonic stem
transcription factor IIF-associated CTD phosphatase
homeobox
histone methyltransferase
interferon-γ
negative elongation factor
protein interacting with NIMA-1
Polycomb repressive complex
positive transcription elongation factor b
RNA polymerase II subunit B1
RE-1 silencing transcription factor
really interesting new gene
regulator of transcription 1
small CTD phosphatase
suppressor of variegation, enhancer of zeste and trithorax 1
suppressor of sua7 protein 2
TATA-binding protein
transcription factor IIH
- CTD
carboxy-terminal domain
major histocompatibility complex, class II, DRα
DRB-sensitivity inducing factor
embryonic stem
transcription factor IIF-associated CTD phosphatase
homeobox
histone methyltransferase
interferon-γ
negative elongation factor
protein interacting with NIMA-1
Polycomb repressive complex
positive transcription elongation factor b
RNA polymerase II subunit B1
RE-1 silencing transcription factor
really interesting new gene
regulator of transcription 1
small CTD phosphatase
suppressor of variegation, enhancer of zeste and trithorax 1
suppressor of sua7 protein 2
TATA-binding protein
transcription factor IIH
- DRA
major histocompatibility complex, class II, DRα
DRB-sensitivity inducing factor
embryonic stem
transcription factor IIF-associated CTD phosphatase
homeobox
histone methyltransferase
interferon-γ
negative elongation factor
protein interacting with NIMA-1
Polycomb repressive complex
positive transcription elongation factor b
RNA polymerase II subunit B1
RE-1 silencing transcription factor
really interesting new gene
regulator of transcription 1
small CTD phosphatase
suppressor of variegation, enhancer of zeste and trithorax 1
suppressor of sua7 protein 2
TATA-binding protein
transcription factor IIH
- DSIF
DRB-sensitivity inducing factor
embryonic stem
transcription factor IIF-associated CTD phosphatase
homeobox
histone methyltransferase
interferon-γ
negative elongation factor
protein interacting with NIMA-1
Polycomb repressive complex
positive transcription elongation factor b
RNA polymerase II subunit B1
RE-1 silencing transcription factor
really interesting new gene
regulator of transcription 1
small CTD phosphatase
suppressor of variegation, enhancer of zeste and trithorax 1
suppressor of sua7 protein 2
TATA-binding protein
transcription factor IIH
- ES
embryonic stem
transcription factor IIF-associated CTD phosphatase
homeobox
histone methyltransferase
interferon-γ
negative elongation factor
protein interacting with NIMA-1
Polycomb repressive complex
positive transcription elongation factor b
RNA polymerase II subunit B1
RE-1 silencing transcription factor
really interesting new gene
regulator of transcription 1
small CTD phosphatase
suppressor of variegation, enhancer of zeste and trithorax 1
suppressor of sua7 protein 2
TATA-binding protein
transcription factor IIH
- Fcp1
transcription factor IIF-associated CTD phosphatase
homeobox
histone methyltransferase
interferon-γ
negative elongation factor
protein interacting with NIMA-1
Polycomb repressive complex
positive transcription elongation factor b
RNA polymerase II subunit B1
RE-1 silencing transcription factor
really interesting new gene
regulator of transcription 1
small CTD phosphatase
suppressor of variegation, enhancer of zeste and trithorax 1
suppressor of sua7 protein 2
TATA-binding protein
transcription factor IIH
- Hox
homeobox
histone methyltransferase
interferon-γ
negative elongation factor
protein interacting with NIMA-1
Polycomb repressive complex
positive transcription elongation factor b
RNA polymerase II subunit B1
RE-1 silencing transcription factor
really interesting new gene
regulator of transcription 1
small CTD phosphatase
suppressor of variegation, enhancer of zeste and trithorax 1
suppressor of sua7 protein 2
TATA-binding protein
transcription factor IIH
- HMT
histone methyltransferase
interferon-γ
negative elongation factor
protein interacting with NIMA-1
Polycomb repressive complex
positive transcription elongation factor b
RNA polymerase II subunit B1
RE-1 silencing transcription factor
really interesting new gene
regulator of transcription 1
small CTD phosphatase
suppressor of variegation, enhancer of zeste and trithorax 1
suppressor of sua7 protein 2
TATA-binding protein
transcription factor IIH
- IFN-γ
interferon-γ
negative elongation factor
protein interacting with NIMA-1
Polycomb repressive complex
positive transcription elongation factor b
RNA polymerase II subunit B1
RE-1 silencing transcription factor
really interesting new gene
regulator of transcription 1
small CTD phosphatase
suppressor of variegation, enhancer of zeste and trithorax 1
suppressor of sua7 protein 2
TATA-binding protein
transcription factor IIH
- NELF
negative elongation factor
protein interacting with NIMA-1
Polycomb repressive complex
positive transcription elongation factor b
RNA polymerase II subunit B1
RE-1 silencing transcription factor
really interesting new gene
regulator of transcription 1
small CTD phosphatase
suppressor of variegation, enhancer of zeste and trithorax 1
suppressor of sua7 protein 2
TATA-binding protein
transcription factor IIH
- Pin1
protein interacting with NIMA-1
Polycomb repressive complex
positive transcription elongation factor b
RNA polymerase II subunit B1
RE-1 silencing transcription factor
really interesting new gene
regulator of transcription 1
small CTD phosphatase
suppressor of variegation, enhancer of zeste and trithorax 1
suppressor of sua7 protein 2
TATA-binding protein
transcription factor IIH
- PRC
Polycomb repressive complex
positive transcription elongation factor b
RNA polymerase II subunit B1
RE-1 silencing transcription factor
really interesting new gene
regulator of transcription 1
small CTD phosphatase
suppressor of variegation, enhancer of zeste and trithorax 1
suppressor of sua7 protein 2
TATA-binding protein
transcription factor IIH
- P-TEFb
positive transcription elongation factor b
RNA polymerase II subunit B1
RE-1 silencing transcription factor
really interesting new gene
regulator of transcription 1
small CTD phosphatase
suppressor of variegation, enhancer of zeste and trithorax 1
suppressor of sua7 protein 2
TATA-binding protein
transcription factor IIH
- RPB1
RNA polymerase II subunit B1
RE-1 silencing transcription factor
really interesting new gene
regulator of transcription 1
small CTD phosphatase
suppressor of variegation, enhancer of zeste and trithorax 1
suppressor of sua7 protein 2
TATA-binding protein
transcription factor IIH
- REST
RE-1 silencing transcription factor
really interesting new gene
regulator of transcription 1
small CTD phosphatase
suppressor of variegation, enhancer of zeste and trithorax 1
suppressor of sua7 protein 2
TATA-binding protein
transcription factor IIH
- Ring
really interesting new gene
regulator of transcription 1
small CTD phosphatase
suppressor of variegation, enhancer of zeste and trithorax 1
suppressor of sua7 protein 2
TATA-binding protein
transcription factor IIH
- Rtr1
regulator of transcription 1
small CTD phosphatase
suppressor of variegation, enhancer of zeste and trithorax 1
suppressor of sua7 protein 2
TATA-binding protein
transcription factor IIH
- SCP
small CTD phosphatase
suppressor of variegation, enhancer of zeste and trithorax 1
suppressor of sua7 protein 2
TATA-binding protein
transcription factor IIH
- Set1
suppressor of variegation, enhancer of zeste and trithorax 1
suppressor of sua7 protein 2
TATA-binding protein
transcription factor IIH
- Ssu72
suppressor of sua7 protein 2
TATA-binding protein
transcription factor IIH
- TBP
TATA-binding protein
transcription factor IIH
- TFIIH
transcription factor IIH
Introduction
RNA polymerase II (RNAPII) transcribes all protein-coding genes and many non-coding RNAs. RPB1, the largest subunit of RNAPII, contains an unusual CTD that comprises repeats of the heptapeptide consensus sequence N-Tyr 1-Ser 2-Pro 3-Thr 4-Ser 5-Pro 6-Ser 7-C, of which there are 52 copies in mammals (Corden et al, 1985). All of the amino acids in these repeats are potential targets for post-translational modification—such as phosphorylation, glycosylation and cis–trans isomerization—and these modifications dictate the functional state of the polymerase. In addition, lysine residues in the several non-consensus CTD repeats that are found in mammals can act as targets for ubiquitination (Li et al, 2007). Finally, in response to DNA damage or drug-induced RNAPII stalling, RNAPII is ubiquitinated outside the CTD, which is crucial for its proteasomal degradation (Bregman et al, 1996; Somesh et al, 2005).
Although the roles and extent of these RNAPII CTD modifications are far from understood, their vast combinatorial potential generates a complex regulatory code (Buratowski, 2003). CTD phosphorylation during the transcription cycle helps to recruit factors that modulate chromatin states and RNA processing, thereby integrating transcription with genome architecture and RNA stability (Phatnani & Greenleaf, 2006). RNAPII is also present across silent regions of the genome, including inactive genes (Guenther et al, 2007; Hargreaves et al, 2009; Muse et al, 2007), enhancer regions of silent genes (reviewed by Szutorisz et al, 2005), and Polycomb-repressed chromatin (Chopra et al, 2009; Dellino et al, 2004; Stock et al, 2007). Its widespread presence across the genome suggests that RNAPII regulation is crucial for controlling the status of gene expression and has significant consequences for cell response pathways.
RNAPII regulation at active genes
RNAPII is recruited to gene promoters in a hypo-phosphorylated state (Fig 1). Its escape from the promoter requires TFIIH-mediated phosphorylation of Ser 5 (Ser 5P; Komarnitsky et al, 2000), which recruits the RNA capping machinery to modify the 5' end of nascent RNAs (Ho & Shuman, 1999) and, in yeast, the Set1 HMT to methylate histone H3K4 (Fig 2A; Ng et al, 2003). During the early stages of the transcription cycle, instability of the RNAPII complex can cause transcription to be aborted after only a few nucleotides (Fig 1). After promoter escape, RNAPII proceeds to intrinsic pausing sites where it can be halted by negative factors. The onset of productive elongation requires P-TEFb, which phosphorylates Ser 2 residues of the RNAPII CTD to produce stable elongation complexes (Fig 1B; Marshall et al, 1996). In both yeast and mammals, Ser 2P integrates productive elongation with chromatin remodelling through the recruitment of H3K36 HMTs, which promote chromatin that is compatible with elongation (Fig 2A; Krogan et al, 2003; Li et al, 2005). Ser 2P also links RNA synthesis with co-transcriptional RNA processing by helping to recruit splicing and polyadenylation factors (Fig 2A; Proudfoot et al, 2002).
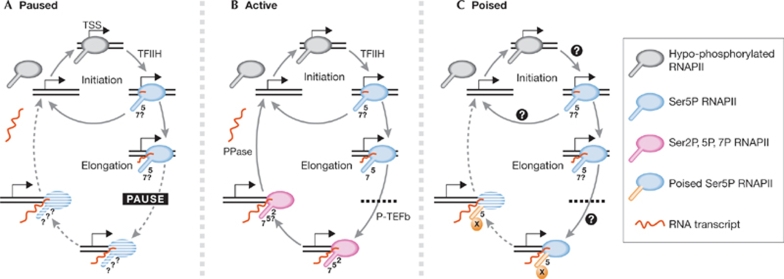
Figure 1.
RNA polymerase II phosphorylation during paused, active and poised transcription cycles. RNAPII is depicted at genes with distinct transcriptional states. (A) Hypo-phosphorylated RNAPII is recruited to the promoter, where TFIIH phosphorylates Ser 5 residues. The initiating RNAPII is unstable and could abort transcription after 2–10 nucleotides (top cycle). RNAPII then progresses to the pause site (10–50 nucleotides) where it is halted by negative elongation factors. Recent evidence suggests that all paused genes produce full-length transcripts at low levels (dashed arrows and cross-hatching of RNAPII indicate rare events). (B) At active genes, a fraction of RNAPII probably undergoes pausing. Phosphorylation of Ser 2 residues by P-TEFb allows elongation. At the 3' end of coding regions, RNAPII terminates and dissociates from its full-length transcript and the DNA template. Phosphatases are involved in RNAPII dissociation and recycling. (C) Poised RNAPII becomes phosphorylated at Ser 5 residues. It is not known whether poised RNAPII undergoes abortive cycles of initiation (top cycle) or pausing. The onset of elongation occurs in the absence of Ser 2P and RNA transcripts are produced at low levels. The CTD of poised RNAPII has a configuration (marked by a cross) that is not compatible with antibody 8WG16 binding and might involve as yet undefined CTD modifications or structural changes. CTD, carboxy-terminal domain; PPase, phosphatase; P-TEFb, positive transcription elongation factor b; RNAPII, RNA polymerase II; TFIIH, transcription factor IIH; TSS, transcription start site.
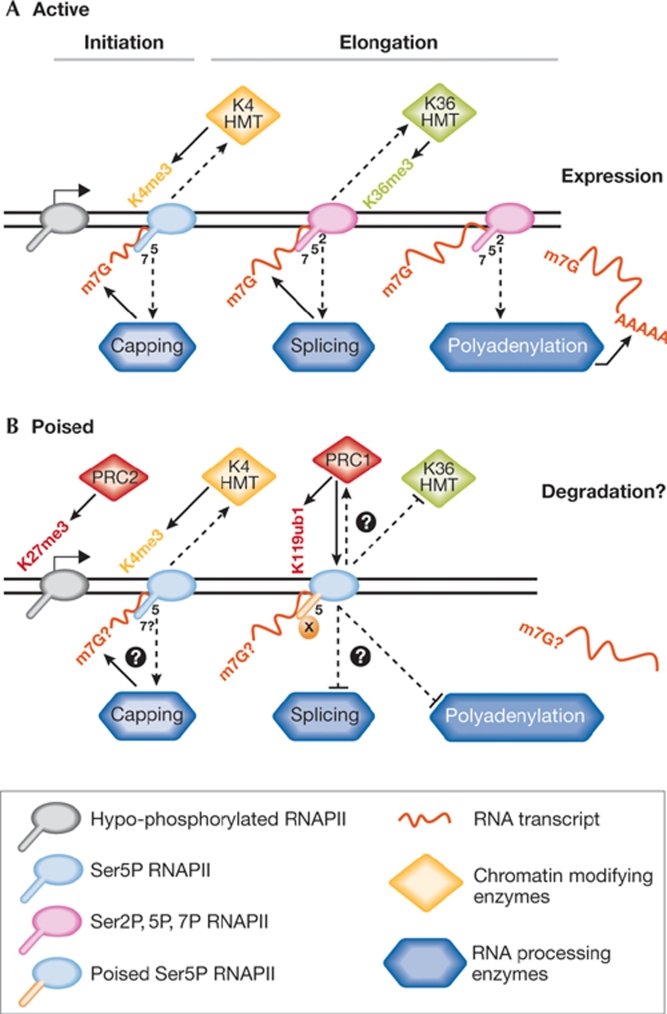
Figure 2.
Integration of carboxy-terminal domain modifications with chromatin structure, RNA processing and Polycomb repression. Distinct modifications of the CTD assist in recruiting different chromatin modifying enzymes and RNA processing factors. (A) At active gene promoters, the phosphorylation of RNAPII at Ser 5 (Ser 5P) recruits HMTs to methylate H3K4 and the RNA capping machinery to add an m7G cap to nascent RNAs. Ser 2P creates an elongating polymerase that recruits the HMTs responsible for trimethylation of H3K36 and RNA processing factors. The mRNA, which is released after termination, is stabilized by its cap and poly(A) tail, thereby promoting mRNA transport and protein expression. (B) At poised bivalent promoters, Ser 5P residues recruit H3K4 HMTs and potentially the capping machinery. PRC2-mediated H3K27 trimethylation is also present at the promoters of poised genes, and PRC1-mediated H2AK119 monoubiquitination tracks RNAPII across the gene, preventing gene expression. RNAPII escape from promoter regions occurs in the absence of Ser 2P, which probably explains the lack of H3K36 trimethylation in coding regions. A deficiency of Ser 2P could hinder the recruitment of RNA processing machinery, leading to the degradation of immature transcripts. CTD, carboxy-terminal domain; HMT, histone methyltransferase; m7G, 7-methyl-guanosine; PRC, Polycomb repressive complex; RNAPII, RNA polymerase II.
RNAPII termination involves the release of RNA transcripts and the dissociation of transcription complexes from the DNA template, and is functionally correlated with transcript polyadenylation (Richard & Manley, 2009). CTD phosphatases are required to recycle RNAPII and produce its hypo-phosphorylated, initiation-competent form (Fig 1). In yeast, Fcp1 removes phosphates from Ser 2 residues (Cho et al, 2001), whereas Ssu72 (Krishnamurthy et al, 2004) and Rtr1 (Mosley et al, 2009) target Ser 5P. Small CTD phosphatases (SCP1–3), recently characterized in mammals, cooperate with the REST chromatin remodelling complex, conferring the potential to regulate RNAPII at specific genes (Yeo et al, 2005).
The post-translational modification of CTD residues other than Ser 2 and Ser 5 is less understood. Phosphorylated Ser 7 residues have been identified at promoter and coding regions of actively transcribing genes in a human tumour cell line (Chapman et al, 2007). Ser 7P is essential for the transcription of small nuclear RNAs (Egloff et al, 2007), but its genome-wide influence on protein-coding gene expression requires clarification. Similar to Ser 5, Ser 7 is phosphorylated by TFIIH, and so could also be required for the early stages of transcription (Akhtar et al, 2009). Interestingly, Ser 7P might influence the extent of Ser 5 phosphorylation (Chapman et al, 2007), thereby helping to modulate the degree of transcription-coupled chromatin remodelling or RNA processing. An additional CTD regulator is the phospho-Ser/Thr-Pro-directed isomerase Pin1, which catalyses the conversion between proline cis–trans configurations, and therefore alters CTD folding (Lu & Zhou, 2007). Mammalian Pin1 regulates RNAPII phosphorylation and activity during the early stages of the transcription cycle and might have a role in RNA processing (Xu & Manley, 2007).
Promoter-proximal pausing of RNAPII
The promoter-proximal regions of many inactive genes are associated with Ser 5P RNAPII in the absence of detectable expression. This phenomenon is known as promoter-proximal pausing (Fig 1A), a state in which RNAPII is transcriptionally engaged but becomes stalled by negative elongation factors (Core & Lis, 2008). Promoter-proximal pausing of RNAPII was first shown to occur at the Drosophila melanogaster heat-shock gene hsp70 (Gilmour & Lis, 1986), where the recruitment of RNAPII before gene activation allows transcription to commence rapidly on heat shock (Boehm et al, 2003).
RNAPII has been detected in human cells at inactive genes such as the proto-oncogene c-MYC (Eick et al, 1987) and the major histocompatibility complex class II DRA gene, where transcription can be rapidly induced by IFN-γ (Spilianakis et al, 2003). Similarly, basal levels of p53 in human cancer cells allow the recruitment of RNAPII to the non-induced p21 gene before its conversion to an elongating complex after activation (Espinosa et al, 2003).
Pausing affects initiating RNAPII molecules to different extents, either preventing all but very infrequent full-length transcription (Fig 1A), or regulating the recurrent release of RNAPII into productive elongation at active genes (Fig 1B; Core & Lis, 2008). Recent genome-wide profiling of RNAPII in human fibroblasts and ES cells, as well as in Drosophila cells, has shown the widespread presence of low levels of RNAPII bound at inactive gene promoters (Guenther et al, 2007; Kim et al, 2005; Muse et al, 2007; Zeitlinger et al, 2007). These studies do not provide evidence for the transcriptional competence of bound RNAPII, although here we define paused genes by their association with promoter-proximal RNAPII in the absence of overt transcription. Global sequencing of nascent RNAs that are labelled by Br-UTP incorporation in nuclei purified from human cells, has revealed transcriptionally engaged RNAPII at virtually all genes (Core et al, 2008). The sensitivity of this technology allowed the detection of full-length RNAs from paused genes, suggesting that a low level of RNAPII escape occurs from promoter-proximal regions. Such findings indicate that regulating the release of RNAPII from promoter-proximal pausing is a universally important control point that primes genes for future activation, for example by promoting open chromatin states (Gilchrist et al, 2008; Lee et al, 2008).
Pausing factors such as NELF and DSIF prevent the exit of RNAPII from promoter-proximal regions into productive elongation (Cheng & Price, 2007). The negative actions of NELF and DSIF are reverted by P-TEFb-dependent phosphorylation, which induces the dissociation of NELF and the activation of DSIF as a positive elongation factor (Fujinaga et al, 2004; Yamada et al, 2006). The presence of abundant NELF at active gene promoters in Drosophila (Lee et al, 2008) supports the view that pausing is an important checkpoint that regulates RNA expression levels at active genes (Fig 1B).
Poised RNAPII at Polycomb-repressed genes
Polycomb-mediated repression is crucial for maintaining the silent state of genes in many cell types (Schwartz & Pirrotta, 2008). Two major Polycomb repressive complexes, PRC1 and PRC2, promote gene silencing through the modification of chromatin structure, by inducing histone H2AK119 monoubiquitination and H3K27 trimethylation, respectively (Cao et al, 2002; Wang et al, 2004). PRCs can also induce chromatin condensation indirectly by influencing DNA methylation (Sakamoto et al, 2007; Vire et al, 2006). However, PRC-mediated repression does not prevent the transcriptional apparatus from accessing promoters (Breiling et al, 2001; Chopra et al, 2009; Dellino et al, 2004; Stock et al, 2007), suggesting that silencing could involve a direct interference with the transcriptional machinery, in addition to chromatin remodelling.
The binding of general transcription factors, including TBP, to Polycomb-repressed homeotic gene promoters has been observed in Drosophila cells (Breiling et al, 2001). Moreover, Drosophila Polycomb proteins associate with general transcription factors in vitro (Saurin et al, 2001). In transgenic Drosophila embryos, in which a Polycomb response element was placed upstream from Hsp26, Polycomb proteins did not prevent the recruitment of the transcriptional machinery to the promoter, but instead repressed the onset of transcription (Dellino et al, 2004). Furthermore, PRC-bound chromatin can maintain an open conformation in Drosophila cells (Mito et al, 2007) and transcriptional activity has been detected at PRC-bound genes in several systems (Bracken et al, 2006; Breiling et al, 2004; Chopra et al, 2009; Stock et al, 2007; Tolhuis et al, 2006), including at the Polycomb genes (Schwartz et al, 2006). Therefore, the presence of RNAPII at Polycomb-silenced genes could be important to modulate their level of expression or to allow later expression. In this respect, the importance of RNAPII regulation at PRC-repressed genes has recently been shown in mouse ES cells (Stock et al, 2007).
Developmental regulator genes are maintained in a silent state in ES cells by PRCs to prevent the activation of differentiation programmes (Boyer et al, 2006; Endoh et al, 2008). Surprisingly, many developmental genes are marked by histone modifications that are characteristic of the active state (H3K4 trimethylation and H3K9 acetylation), as well as repressive modifications that are catalysed by Polycomb proteins (H3K27 trimethylation and H2AK119 monoubiquitination; Sidebar A; Azuara et al, 2006; Bernstein et al, 2006; Stock et al, 2007). Despite PRC repression, the promoters of genes with bivalent chromatin marks are bound by similar amounts of RNAPII as active genes, and RNAPII can be detected at least 8 kb downstream from transcription start sites (Fig 1C; Stock et al, 2007). Here, we refer to the specific RNAPII configuration found in ES cells at PRC-silenced genes as ‘poised' because of its important role in potentiating genes for activation during development, but we anticipate that similar findings will be described in other cell types.
Sidebar A | RNA polymerase II and histone modification profiles across genes in paused, active and poised states.
Distinct RNAPII complexes at paused, active and poised genes can be identified by mapping the binding of antibodies that recognize specific modifications along transcription units by chromatin immunoprecipitation (ChIP). ChIP is performed on cell populations, and so these profiles capture RNAPII distribution simultaneously across the gene in many cells. The profiles below are thought to be representative of mammalian genes, although many variations on these will be observed across the genome and in different cell types.
Paused genes (A) contain low levels of Ser 5P limited to promoter-proximal regions. (B) Actively transcribing genes are associated with Ser 5P enrichment peaking at the promoter and extending through the gene, whereas (C) at poised PRC-repressed genes, Ser 5P is highly enriched within approximately 8 kb of the TSS. Consistent with a role in processive elongation, Ser 2P is present throughout the coding regions of active genes, but is absent from paused and poised complexes. Antibody 8WG16, which recognizes non-phosphorylated Ser 2 residues, binds to RNAPII at promoters and into coding regions of active genes, but not towards the 3′ end of genes. 8WG16 enrichment is restrained to promoter-proximal regions of paused genes, whereas very low levels, or a lack of binding, characterize poised genes.
Histone modification profiles also distinguish each transcriptional state: H3K4 trimethylation marks paused, active and poised genes, whereas repressive H3K27 trimethylation is observed only at Polycomb-repressed poised genes. H3K36 trimethylation is enriched at active genes but not at paused or poised genes, consistent with a role in productive elongation. PRC, Polycomb repressive complex; RNAPII, RNA polymerase II; TSS, transcription start site.
Low levels of α-amanitin-sensitive transcripts are produced from bivalent genes, and transcripts are polymerized and spliced to at least the first exon junction (Stock et al, 2007). This contrasts with the transcriptional activity of paused polymerases, which mainly synthesize short transcripts of a 10–50 nucleotides of base pairs (Rougvie & Lis, 1988). RNAPII at bivalent genes is characterized by an unusual set of post-translational modifications. It contains high levels of Ser 5P but no detectable Ser 2P in spite of elongation. In addition, the binding of an antibody that preferentially recognizes non-phosphorylated Ser 2 residues (8WG16) is unexpectedly low or even absent (Sidebar A; Stock et al, 2007). The lack of recognition of poised RNAPII by antibodies that bind to phosphorylated (H5) or non-phosphorylated (8WG16) Ser 2 residues indicates that these residues are inaccessible owing to another CTD modification or conformational change (Fig 3). The poised RNAPII complexes present at PRC-silenced genes differ from the paused complexes present at rapidly inducible genes, which are characterized by limited RNAPII binding that is restricted to promoter-proximal locations, low levels of Ser 5P and binding by 8WG16 (Sidebar A; Boehm et al, 2003; Espinosa et al, 2003; Guenther et al, 2007; Kim et al, 2005; Spilianakis et al, 2003). A recent study of PRC-repressed Hox genes in Drosophila embryos identified Ser 5P RNAPII complexes—which could be detected by 8WG16—at promoters but not at downstream coding regions (Chopra et al, 2009), raising the possibility that the poised state of RNAPII at PRC-bound genes is specific to mammals.
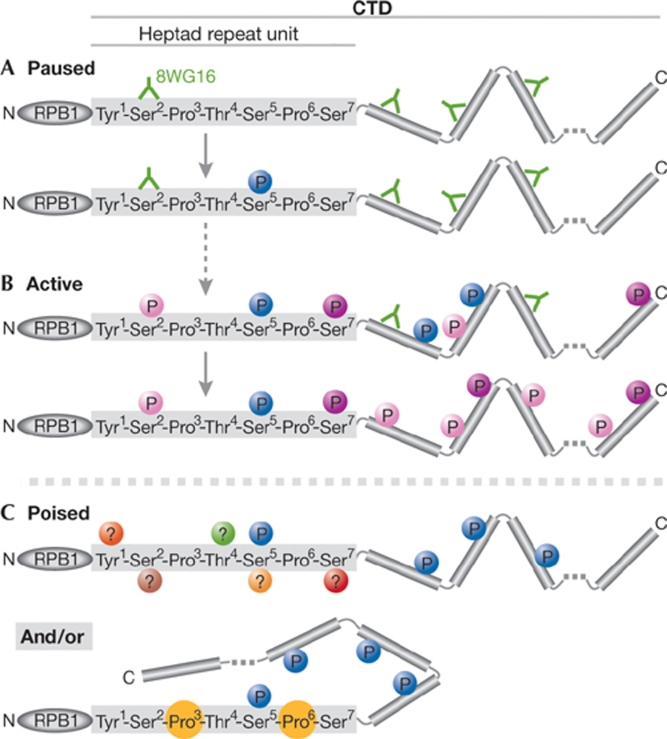
Figure 3.
RNA polymerase II carboxy-terminal domain modifications influence 8WG16 antibody recognition. The CTD of RPB1 comprises 52 repeats of the heptad consensus sequence that is indicated in the figure. Antibody 8WG16 recognizes unphosphorylated Ser 2 residues and has been used as a marker for total RNAPII (that is, all isoforms), on the assumption that not all CTD repeats are simultaneously phosphorylated. However, the detection of RNAPII by 8WG16 is affected by CTD phosphorylation (Doyle et al, 2002; Patturajan et al, 1998; Stock et al, 2007; Xie et al, 2006) and, therefore, the use of this antibody underestimates RNAPII presence. Modifications to the CTD (depicted by different coloured circles) influence its detection, configuration and the factors it recruits. (A) RNAPII is recruited to gene promoters with a hypo-phosphorylated CTD, which is accessible to 8WG16 antibody binding. Promoter escape coincides with phosphorylation of Ser 5 residues and the CTD retains its recognition by 8GW16. (B) RNAPII at active promoters is associated with Ser 5P and Ser 7P. Phosphorylation of Ser 2 residues converts RNAPII into a productively elongating complex. Ser 2P and Ser 7P are refractory to 8WG16 binding within the same heptad (D. Eick, personal communication). As the transcription unit is traversed, Ser 5 phosphorylation diminishes by the action of phosphatases, whereas Ser 2P and Ser 7P increase, thereby obscuring 8WG16 binding sites. (C) Poised RNAPII is highly phosphorylated on Ser 5 residues and shows little (or no) recognition by 8WG16. Additional modifications of CTD residues could obscure 8WG16 binding to poised RNAPII. Alternatively, poised RNAPII could adopt an unusual structure that obscures 8WG16 epitopes, possibly through proline isomerization, which is known to influence CTD conformation. CTD, carboxy-terminal domain; RNAPII, RNA polymerase II; RPB1, RNA polymerase II subunit B1.
The absence of Ser 2P at poised RNAPII complexes is likely to disrupt co-transcriptional RNA maturation and histone modification to help maintain the silent state, whereas binding and transcription by RNAPII counteract Polycomb-mediated chromatin condensation (Fig 2B). Low levels of mRNA from bivalent genes can be detected in microarray analyses (Guenther et al, 2007; Mikkelsen et al, 2007), suggesting that transcript maturation occurs to a limited extent. H3K36 trimethylation—a histone modification that is associated with productive elongation—is not detected in coding regions of bivalent genes (E.B., H. Kimura and A.P., unpublished observations), which is consistent with the disruption of factor recruitment. Similarly, priming of inducible primary response genes in macrophages is characterized by RNAPII transcription through coding regions in the absence of RNA splicing (Hargreaves et al, 2009).
The mechanisms that prevent the phosphorylation of RNAPII Ser 2 at primed genes could involve uncharacterized CTD modifications or conformations that uncouple transcription from productive gene expression (Figs 1,2,3). A CTD modification might prevent Ser 2 phosphorylation by obscuring target sites, by affecting the structure of the CTD or by inducing the recruitment of specific factors. Conversely, the activity of CTD phosphatases that target specific genes—such as SCPs—could actively deplete Ser 2P. The modulation of the configuration of the CTD by isomerases such as Pin1 might regulate RNAPII poising, but their role at bivalent genes is yet to be explored.
Interplay between PRCs and poised RNAPII
In the prevailing model of PRC regulation, PRC2 is recruited to specific genomic locations and catalyses H3K27 trimethylation, thereby creating binding sites for PRC1, which then ubiquitinates H2AK119 (Cao et al, 2002). However, the distribution of the PRC1 ubiquitin ligase Ring1B and monoubiquitinated H2AK119 across individual bivalent loci in mouse ES cells unexpectedly parallels the occupancy of Ser 5P RNAPII more closely than that of PRC2 or H3K27 trimethylation (Stock et al, 2007). This suggests a direct role for PRC1 or H2AK119 monoubiquitination in restraining or recruiting poised RNAPII, or a dependency of Ring1B recruitment on RNAPII.
The removal of the catalytic PRC1 components Ring1A and Ring1B in ES cells causes the depletion of H2AK119 monoubiquitination and a derepression of bivalent genes ( Jorgensen et al, 2006; Stock et al, 2007). The CTD conformation of RNAPII at poised genes also changes in unexpected ways (Stock et al, 2007). Derepression is not accompanied by changes in Ser 5P levels, implying that the recruitment of RNAPII to promoters is not enhanced in the absence of PRC1. Ser 2P levels are not increased, suggesting that derepression occurs without the conversion of restrained, poised polymerases into typical, Ser 2P-dependent elongation complexes. Ring1 depletion does, however, enhance 8WG16 binding to levels similar to those at active genes, raising the possibility of a mechanistic interaction between the conformation of the CTD and the recruitment of Ring1B or H2A monoubiquitination. By contrast, the depletion of 2A-HUB—an H2A ubiquitin ligase that represses chemokine genes in a monocyte–macrophage cell line—correlates with increased Ser 2P at derepressed genes, indicating a release into Ser 2P-dependent elongation (Zhou et al, 2008).
The correlation between Ring1B and RNAPII binding at PRC-repressed genes in ES cells, and the change in RNAPII on Ring1B depletion (Stock et al, 2007), raise the intriguing possibility that PRC1 might directly modify RNAPII or crucial poising components to restrict RNAPII processivity at developmental regulator genes (Fig 2B). Interestingly, non-consensus CTD repeats—which are present in mammals but not in yeast—have lysine residues, which are potential targets for ubiquitination or methylation, instead of the canonical Ser 7. We propose that the direct modification of RNAPII CTD lysines by PRCs could integrate Polycomb silencing with the establishment of a poised RNAPII configuration. PRC1 components have been shown to be absent from a subset of bivalent genes, adding yet another level of complexity to the regulation of genes that control developmental processes (Ku et al, 2008).
Conclusion
The modification of RNAPII confers robust control of gene expression levels by regulating the selective recruitment of chromatin and RNA processing factors. Preloading RNAPII at inactive genes primes them for subsequent activation without leading to immediate expression. RNAPII can be restrained at promoter-proximal regions in the absence of modifications that are important for the onset of productive elongation. Alternatively, polymerases can transcribe through coding regions without the modifications that are necessary for the recruitment of chromatin modifiers or the RNA processing machinery. At developmental regulator genes in ES cells, PRCs control the poised state of RNAPII by influencing chromatin modification and RNAPII configuration. Antisense RNA transcription around promoters is widespread across the mouse ES cell genome (Seila et al, 2008) and represents an additional layer of gene expression regulation that is enabled by RNAPII. The identification of regulators of RNAPII states across the genome and of their mechanisms of action and functional interactions with chromatin modifiers, PRCs and RNA processors, will be important for understanding the mechanisms of gene regulation that control the responses to environmental stimuli and programme gene expression during cell commitment (Sidebar B).
Sidebar B | In need of answers.
What are the enzymatic activities (kinases, phosphatases, other) that establish the different RNAPII states?
What are the key differences between RNAPII pausing at quickly inducible genes and RNAPII poising at PRC-dependent developmental regulator genes?
What is the processing status of RNAs produced by paused and poised RNAPII complexes?
What are the kinetics of transcriptional pausing and poising?
What are the mechanistic relationships between PRC activity and RNAPII function?
Are there additional modifications of CTD residues, what is their function and how are they controlled? What are the roles of Ser 7 phosphorylation? Are non-canonical CTD residues —which are found in higher eukaryotes— also modified?
Acknowledgments
We are grateful to J.K. Stock, C. Ferrai, I. de Santiago, M. Chotalia and A. Möller for critical reading of the manuscript, and to D. Eick and H. Kimura for sharing reagents and unpublished data. We thank the Medical Research Council (UK) for funding.
References
- Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, Ansari AZ (2009) TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell 34: 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V et al. (2006) Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8: 532–538 [DOI] [PubMed] [Google Scholar]
- Bernstein BE et al. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Boehm AK, Saunders A, Werner J, Lis JT (2003) Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol 23: 7628–7637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA et al. (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K (2006) Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20: 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman DB, Halaban R, van Gool AJ, Henning KA, Friedberg EC, Warren SL (1996) UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc Natl Acad Sci USA 93: 11586–11590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiling A, Turner BM, Bianchi ME, Orlando V (2001) General transcription factors bind promoters repressed by Polycomb group proteins. Nature 412: 651–655 [DOI] [PubMed] [Google Scholar]
- Breiling A, O′Neill LP, D′Eliseo D, Turner BM, Orlando V (2004) Epigenome changes in active and inactive Polycomb-group-controlled regions. EMBO Rep 5: 976–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S (2003) The CTD code. Nat Struct Biol 10: 679–680 [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D (2007) Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318: 1780–1782 [DOI] [PubMed] [Google Scholar]
- Cheng B, Price DH (2007) Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J Biol Chem 282: 21901–21912 [DOI] [PubMed] [Google Scholar]
- Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S (2001) Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev 15: 3319–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra VS, Hong JW, Levine M (2009) Regulation of Hox gene activity by transcriptional elongation in Drosophila. Curr Biol 19: 688–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden JL, Cadena DL, Ahearn JM Jr, Dahmus ME (1985) A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc Natl Acad Sci USA 82: 7934–7938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Lis JT (2008) Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science 319: 1791–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT (2008) Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellino GI, Schwartz YB, Farkas G, McCabe D, Elgin SC, Pirrotta V (2004) Polycomb silencing blocks transcription initiation. Mol Cell 13: 887–893 [DOI] [PubMed] [Google Scholar]
- Doyle O, Corden JL, Murphy C, Gall JG (2002) The distribution of RNA polymerase II largest subunit (RPB1) in the Xenopus germinal vesicle. J Struct Biol 140: 154–166 [DOI] [PubMed] [Google Scholar]
- Egloff S, O′Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S (2007) Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 318: 1777–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D, Berger R, Polack A, Bornkamm GW (1987) Transcription of c-myc in human mononuclear cells is regulated by an elongation block. Oncogene 2: 61–65 [PubMed] [Google Scholar]
- Endoh M et al. (2008) Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development 135: 1513–1524 [DOI] [PubMed] [Google Scholar]
- Espinosa JM, Verdun RE, Emerson BM (2003) p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell 12: 1015–1027 [DOI] [PubMed] [Google Scholar]
- Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM (2004) Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol 24: 787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K (2008) NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev 22: 1921–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DS, Lis JT (1986) RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol 6: 3984–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA (2007) A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R (2009) Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138: 129–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CK, Shuman S (1999) Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell 3: 405–411 [DOI] [PubMed] [Google Scholar]
- Jorgensen HF, Giadrossi S, Casanova M, Endoh M, Koseki H, Brockdorff N, Fisher AG (2006) Stem cells primed for action: polycomb repressive complexes restrain the expression of lineage-specific regulators in embryonic stem cells. Cell Cycle 5: 1411–1414 [DOI] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B (2005) A high-resolution map of active promoters in the human genome. Nature 436: 876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M (2004) Ssu72 is an RNA polymerase II CTD phosphatase. Mol Cell 14: 387–394 [DOI] [PubMed] [Google Scholar]
- Krogan NJ et al. (2003) Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol 23: 4207–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M et al. (2008) Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS (2008) NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol 28: 3290–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhang Z, Wang B, Zhang J, Zhao Y, Jin Y (2007) Wwp2-mediated ubiquitination of the RNA polymerase II large subunit in mouse embryonic pluripotent stem cells. Mol Cell Biol 27: 5296–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Phatnani HP, Guan Z, Sage H, Greenleaf AL, Zhou P (2005) Solution structure of the Set2–Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc Natl Acad Sci USA 102: 17636–17641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KP, Zhou XZ (2007) The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol 8: 904–916 [DOI] [PubMed] [Google Scholar]
- Marshall NF, Peng J, Xie Z, Price DH (1996) Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem 271: 27176–27183 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S (2007) Histone replacement marks the boundaries of cis-regulatory domains. Science 315: 1408–1411 [DOI] [PubMed] [Google Scholar]
- Mosley AL, Pattenden SG, Carey M, Venkatesh S, Gilmore JM, Florens L, Workman JL, Washburn MP (2009) Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol Cell 34: 168–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K (2007) RNA polymerase is poised for activation across the genome. Nat Genet 39: 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- Patturajan M, Schulte RJ, Sefton BM, Berezney R, Vincent M, Bensaude O, Warren SL, Corden JL (1998) Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem 273: 4689–4694 [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL (2006) Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev 20: 2922–2936 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ (2002) Integrating mRNA processing with transcription. Cell 108: 501–512 [DOI] [PubMed] [Google Scholar]
- Richard P, Manley JL (2009) Transcription termination by nuclear RNA polymerases. Genes Dev 23: 1247–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT (1988) The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54: 795–804 [DOI] [PubMed] [Google Scholar]
- Sakamoto Y, Watanabe S, Ichimura T, Kawasuji M, Koseki H, Baba H, Nakao M (2007) Overlapping roles of the methylated DNA-binding protein MBD1 and polycomb group proteins in transcriptional repression of HOXA genes and heterochromatin foci formation. J Biol Chem 282: 16391–16400 [DOI] [PubMed] [Google Scholar]
- Saurin AJ, Shao Z, Erdjument-Bromage H, Tempst P, Kingston RE (2001) A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412: 655–660 [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V (2006) Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet 38: 700–705 [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V (2008) Polycomb complexes and epigenetic states. Curr Opin Cell Biol 20: 266–273 [DOI] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA (2008) Divergent transcription from active promoters. Science 322: 1849–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somesh BP, Reid J, Liu WF, Sogaard TM, Erdjument-Bromage H, Tempst P, Svejstrup JQ (2005) Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell 121: 913–923 [DOI] [PubMed] [Google Scholar]
- Spilianakis C, Kretsovali A, Agalioti T, Makatounakis T, Thanos D, Papamatheakis J (2003) CIITA regulates transcription onset via Ser5-phosphorylation of RNA Pol II. EMBO J 22: 5125–5136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A (2007) Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol 9: 1428–1435 [DOI] [PubMed] [Google Scholar]
- Szutorisz H, Dillon N, Tora L (2005) The role of enhancers as centres for general transcription factor recruitment. Trends Biochem Sci 30: 593–599 [DOI] [PubMed] [Google Scholar]
- Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, van Lohuizen M (2006) Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet 38: 694–699 [DOI] [PubMed] [Google Scholar]
- Vire E et al. (2006) The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439: 871–874 [DOI] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y (2004) Role of histone H2A ubiquitination in Polycomb silencing. Nature 431: 873–878 [DOI] [PubMed] [Google Scholar]
- Xie SQ, Martin S, Guillot PV, Bentley DL, Pombo A (2006) Splicing speckles are not reservoirs of RNA polymerase II, but contain an inactive form, phosphorylated on serine2 residues of the C-terminal domain. Mol Biol Cell 17: 1723–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YX, Manley JL (2007) Pin1 modulates RNA polymerase II activity during the transcription cycle. Genes Dev 21: 2950–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H (2006) P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell 21: 227–237 [DOI] [PubMed] [Google Scholar]
- Yeo M, Lee SK, Lee B, Ruiz EC, Pfaff SL, Gill GN (2005) Small CTD phosphatases function in silencing neuronal gene expression. Science 307: 596–600 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA (2007) RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet 39: 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG (2008) Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell 29: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]



 Ana Pombo and Emily Brookes
Ana Pombo and Emily Brookes