Abstract
HEp-2 cell monolayers were cocultured with intracellular Staphylococcus aureus, and changes in gene expression were profiled using DNA microarrays. Intracellular S. aureus affected genes involved in cellular stress responses, signal transduction, inflammation, apoptosis, fibrosis, and cholesterol biosynthesis. Transcription of stress response and signal transduction-related genes including atf3, sgk, map2k1, map2k3, arhb, and arhe was increased. In addition, elevated transcription of proinflammatory genes was observed for tnfa, il1b, il6, il8, cxcl1, ccl20, cox2, and pai1. Genes involved in proapoptosis and fibrosis were also affected at transcriptional level by intracellular S. aureus. Notably, intracellular S. aureus induced strong transcriptional down-regulation of several cholesterol biosynthesis genes. These results suggest that epithelial cells respond to intracellular S. aureus by inducing genes affecting immunity and in repairing damage caused by the organism, and are consistent with the possibility that the organism exploits an intracellular environment to subvert host immunity and promote colonization.
1. Introduction
Staphylococcus aureus (S. aureus), a nosocomial or community-acquired pathogen that colonizes much of the healthy population [1], is an important cause of skin infections, pneumonia, septicemia, endocarditis, osteomyelitis, folliculitis, mastitis, and other infections. The organism also causes toxigenic illnesses such as food poisoning and toxic shock syndrome [2]. Infections caused by S. aureus may be refractory to therapy and become chronic or recur, despite acceptable therapy [3–6].
Several studies showed that S. aureus can become internalized by nonprofessional phagocytes [7–9]; α 5 β 1 integrin is necessary for fibronectin-mediated S. aureus internalization involving staphylococcal fibronectin-binding proteins [10, 11]. Internalization may provide several benefits to S. aureus. It has been proposed that intracellular S. aureus evades exposure to antibiotics [3] and host immunity. It also provides an intracellular milieu which leads to the formation of small-colony variants with decreased metabolic activity and increased antibiotic resistance [12].
Microarray technology has helped elucidate pathogen-host cell interactions and profile the effects on epithelial cells by organisms including, but not limited to, Yersinia enterocolitica [13], Salmonella dublin [14], Shigella flexneri [15], Bordetella pertussis [16], Mycobacterium tuberculosis [17], Pseudomonas aeruginosa [18], Listeria monocytogenes [19], Streptococcus pyogenes [20], and S. aureus [21, 22]. Although the internalization of S. aureus by nonprofessional phagocytes is well documented [5, 7–9, 23–25], the cellular response to intracellular S. aureus has only been partially elucidated [3, 26], focusing mainly on apoptosis [27–33]. The present study assessed global changes in gene expression over an 8-hour time period in epithelial cell monolayers induced by intracellular S. aureus. The data demonstrated that cultured epithelial cells respond to intracellular S. aureus by inducing several classes of genes that could influence the outcome of colonization or infection by this organism in vivo.
2. Materials and Methods
2.1. Cultures
HEp-2 cells [34] were purchased from the American Type Culture Collection (ATCC). Routine maintenance was conducted using complete growth medium (CGM) [10]. S. aureus RN6390 [32, 33, 35] provided by A. Cheung (Dartmouth Medical School) was used to infect HEp-2 cells using established techniques described previously [8, 32, 33, 36]. Briefly, bacteria from 16-hour Todd Hewitt broth cultures were washed three times with phosphate buffered saline (PBS), and resuspended in invasion medium (IM; CGM lacking antibiotics and FBS) to make stocks with approximately 109 colony-forming units (CFU) mL−1. Bacterial stocks were diluted 10-fold in fresh IM; 500 μL of the cell suspension well−1 were used to infect each HEp-2 culture at a multiplicity of infection (MOI) of 10. The cocultures were centrifuged immediately to synchronize monolayer infections and incubated at 37°C for 10 minutes to allow internalization, after 10 minutes, the IM was rapidly replaced with fresh medium containing gentamicin (100 μg mL−1) to kill noninternalized bacteria. Thereafter, the cocultures were incubated (up to 8 hours following S. aureus exposure) and analyzed at various times following exposure to S. aureus as described below.
For growth rate analyses, cells from 16-hour S. aureus RN6390 TH broth cultures (above) were pelleted, washed three times with PBS, and diluted with PBS to 105 CFU mL−1. A 100 μL aliquot was inoculated into 10 mL of TH broth or IM, with or without FBS (without antibiotics). Cultures were incubated with vigorous shaking up to 8 hours. CFU concentrations were determined by a standard plate count method.
2.2. RNA Isolation and Purification
HEp-2 cells were harvested at 2, 4, 6, or 8 hours following addition of bacteria. RNA was isolated using TRIZOL (Invitrogen) according to the manufacturer's instructions and further purified with RNAeasy MinElute Cleanup Kits (Qiagen). RNA samples, quantified using a NanoDrop ND-1000 spectrophotometer (Nanodrop Technologies) and showing OD260:OD280 ratios >1.95 were used for subsequent experiments.
2.3. Microarray Methods and Data Analysis
MWG Human 30K microarrays (MWG) were used according to the manufacturer's instructions. cDNA was synthesized using the BD Atlas PowerScript Fluorescent Labeling Kit (BD) with oligo(dT)12-18 primer (Invitrogen). CyDye Post-Labeling Reactive Dyes (Amersham) were used to fluorescently label the cDNA (Cy3 for cDNA from uninfected cells and Cy5 for cDNA from S. aureus infected cells). Unincorporated dye was removed from labeled cDNA with CHROMA SPIN+TE-30 columns (Clontech). Labeled cDNA was dissolved in salt-based hybridization buffer (MWG), incubated at 95°C (3 minutes), chilled on ice, and hybridized to the microarray chips in the dark for 16–24 hours at 42°C with slow rocking. Arrays were washed and scanned with an Axon 4000A dual channel microarray scanner (Axon) to generate multi-TIFF images which were processed with GenePix Pro 6.0 software (Molecular Devices).
2.4. Quantitative Real-Time PCR (QRT-PCR)
QRT-PCR was used to validate selected microarray data. cDNA was synthesized from 1 μg of RNA using Superscript Π Reverse Transcriptase (Invitrogen). Primers (Table 1), designed using Primer Express 2.0 software (PE Applied Biosystems), were purchased from Integrated DNA Technologies (IDT). Data were analyzed as described previously [37]. The threshold cycle (C T) was calculated as the cycle number at which the ΔRn crossed the baseline. Data were normalized by calculating ΔC T [C T of target − C T of the internal control (β-actin)]. Normalized ΔC T data from S. aureus infected HEp-2 cells were compared to data from uninfected HEp-2 cells by calculating ΔΔC T [ΔC T of S. aureus infected HEp-2 cells − ΔC T of uninfected HEp-2 cells]. Each experiment was conducted thrice for validation, and the mean value is reported.
Table 1.
DNA primers used for QRT-PCR experiments.
| Gene | Forward primers (5′-3′) | Reverse primers (5′-3′) |
|---|---|---|
| atf3 | GATGTCCTCTGCGCTGGAAT | CCTCGGCTTTTGTGATGGA |
| c-fos | GCCCTTTGATGACTTCCTGTTC | GGAGCGGGCTGTCTCAGA |
| c-jun | GCAAAGATGGAAACGACCTTCT | GCTCTCGGACGGGAGGAA |
| junB | CTACACGACTACAAACTCCTGAAACC | CCCCAGGCGCTTTGAGA |
| sgk | GTGCCTGGGAGCTGTCTTGT | GCTGTGTTTCGGCTATAAAAAGG |
| arhb | TCCCAATGTGCCCATCATC | ATGCGGGCCAGCTCTGT |
| map2k3 | CCCTACATGGCCCCTGAGA | TCCAGACGTCGGACTTGACA |
| Il1b | CGAATCTCCGACCACCACTAC | TCCATGGCCACAACAACTGA |
| tnfa | CCTGGTATGAGCCCATCTATCTG | TAGTCGGGCCGATTGATCTC |
| Il6 | AGCCGCCCCACACAGA | TCGAGGATGTACCGAATTTGTTT |
| Il8 | CTGGCCGTGGCTCTCTTG | CTTGGCAAAACTGCACCTTCA |
| ccl20 | TCCTGGCTGCTTTGATGTCA | AAAGTTGCTTGCTGCTTCTGATT |
| cxcl1 | AACATCCAAAGTGTGAACGTGAA | GAGTGTGGCTATGACTTCGGTTT |
| Il10 | CTTGTCTGAGATGATCCAGTTTTACCT | CCTTGATGTCTGGGTCTTGGTT |
| ptgs2 | GGAAGCCTTCTCTAACCTCTCCTATT | AGGGAGTCGGGCAATCATC |
| adm | GGATGTCGCGTCGGAGTTT | TGCTGGACATCCGCAGTTC |
| dkk1 | AAGTACCAGACCATTGACAACTACCA | GGGACTAGCGCAGTACTCATCAGT |
| igfbp1 | CCATCTGATGGCCCCTTCT | CCTTCGAGCCATCATAGGTACTG |
| casp9 | AGGACATGCTGGCTTCGTTT | TTCTAGGGTTGGCTTCGACAA |
| tgfb1 | CCTGGCGATACCTCAGCAA | CCGGTGACATCAAAAGATAACCA |
| thbs1 | TCCGCAAAGTGACTGAAGAGAA | TGAACTCCGTTGTGATAGCATAGG |
| cyr61 | GGTGGAGTTGACGAGAAACAATG | AGGGAGCCGCTTCAGTGA |
| hmgcr | CCCAGTTGTGCGTCTTCCA | TGCGAACCCTTCAGATGTTTC |
| sqle | CGCCCTCTTCTCGGATATTCT | CCGAGCTGCTCCTTATTTTCTG |
| dhcr7 | AGCCGCCCAGCTCTATACCT | TTATGGCAGAAGTCAGGGAGAGA |
| ldlr | GATGAAGTTGGCTGCGTTAATGT | CGCCGCTGTGACACTTGA |
| actb | CGTTGCTATCCAGGCTATGCT | TCACCGGAGTCCATCACGAT |
2.5. Cholesterol Analyses
HEp-2 cells were dislodged with TrypLE Express (Gibco) and collected by centrifugation. Lipids were extracted with chloroform and methanol [38], analyzed and quantified by gas chromatography/mass spectrometry (GC-MS 6890N; Agilent Technologies) and reported as μg/105 cells. Each experiment was conducted at least three times.
2.6. Flow Cytometry
Prior to infection, S. aureus was labeled with 0.5 μM 5- (and-6)-carboxyfluorescein diacetate, succinimidyl ester (CFSE) (Invitrogen) for 10 minutes at 37°C. CFSE-stained S. aureus was washed three times with PBS and used to infect HEp-2 cells as described above. After coculturing for 10 minutes, cells were washed and incubated (15 minutes, 37°C) with S. aureus specific antibody ab37644 (Abcam), followed by goat antimouse IgG conjugated with Cy5 (Southern Biotech) to quantify extracellular bacteria. In parallel experiments to quantify extracellular bacteria, infected monolayers were treated with lysostaphin for 2 hours resulting in loss of the CFSE signal. Confirmation of the effectiveness of lysostaphin treatment was accomplished by treatment with Cy5-conjugated antibody as described above. Cells were harvested and analyzed with a FACSAria flow cytometer (BD), equipped with FACSDiva software (BD).
2.7. Statistical Analyses
GeneSpring version 7.2 (Silicon Genetics) was used to analyze microarray data. For each time point, data from 3–5 separate replicated experiments were obtained and analyzed by 2-way ANOVA (P < .05) to determine their validity, followed by Benjamini and Hochberg false discovery rate correction for each data set [39]. Correction for spot intensity variations among arrays was performed by intensity-dependent normalization and subtraction of background based on negative controls. Normalized mean values were determined for all data points. Microarray data were reported as increased or decreased expression (>1.0 or <1.0, resp.) by dividing the mean Cy5 value (infected HEp-2 cells) by the mean Cy3 value (uninfected HEp-2 cells) for each time point.
3. Results and Discussion
3.1. Experimental Model
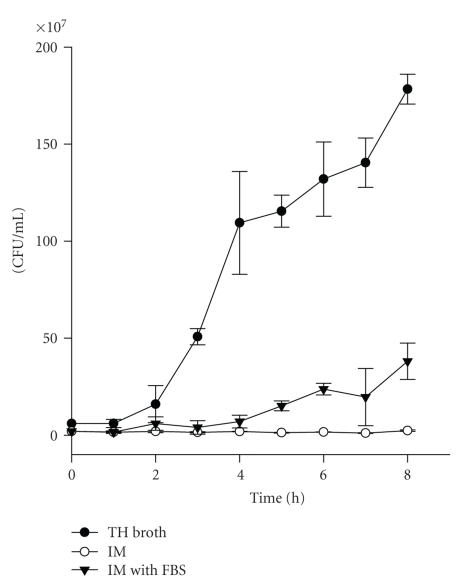
As this study was designed to assess the effects of internalized S. aureus on the HEp-2 pharyngeal epithelial cell line, the influences of extracellular bacteria or their exotoxins produced prior to internalization of S. aureus were minimized by (1) thoroughly washing the inocula; (2) treating cocultures with gentamicin after a very short (10 minutes) extracellular bacterial exposure; (3) conducting the extracellular exposure period in a medium that does not support extracellular growth. Specifically, unlike control cultures in TH broth which supported robust growth, S. aureus RN6390 cultured in IM did not grow, even when incubated for periods of time much longer than the 10 minutes used to infect cells (Figure 1). Furthermore, IM supplemented with FBS supported moderate growth, indicating that a lack of growth in IM alone was not due to inhibitory components.
Figure 1.
Growth analysis of S. aureus RN6390. To assess growth, S. aureus RN6390 was inoculated into different media (TH broth, IM, or IM supplemented with FBS). CFUs were determined hourly by a standard plate count method up to 8 hours, and represented as the mean ± SEM of data acquired from three experiments.
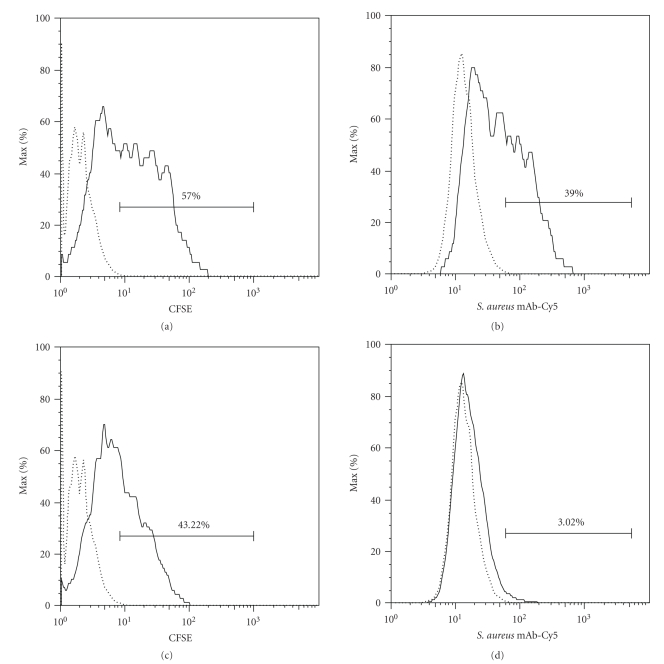
Considering the short exposure of HEp-2 cells to extracellular S. aureus, it was of interest to quantify the percentage of infected HEp-2 cells containing intracellular bacteria. This was accomplished by differential staining of intracellular and extracellular bacteria and by monitoring intracellular CFSE-stained S. aureus following lysostaphin treatment to remove extracellular bacteria. As shown in Figure 2(a), a 10-minute-exposure resulted in monolayers in which approximately 57.0% of the HEp-2 cells contained cell-associated S. aureus (extracellular and/or intracellular), while approximately 39.0% of HEp-2 cells were associated with extracellular bacteria (Figure 2(b)). Lysostaphin treatment which removed nearly all extracellular bacteria (Figure 2(d)) revealed that approximately 43.2% of the HEp-2 cells had intracellular S. aureus (Figure 2(c)).
Figure 2.
Assessment of S. aureus RN6390 internalization using flow cytometry. Dotted lines indicate the uninfected HEp-2 cell control, and solid lines indicate HEp-2 cells infected with CFSE-labeled S. aureus ((a) and (c)) or infected with CFSE-labeled S. aureus followed by labeling Cy5-conjugated mAb specific for S. aureus ((b) and (d)). In panels (a) and (b), HEp-2 cells were infected with CFSE-labeled S. aureus without treatment with lysostaphin. CFSE signal represents HEp-2 cells infected with extracellular and/or intracellular S. aureus (a). Cy5 signal represents HEp-2 cells infected with extracellular S. aureus only (b). In panels (c) and (d), HEp-2 cells were infected with CFSE-labeled S. aureus followed by the treatment with lysostaphin which degrades staphylococcal cell wall causing a loss of CFSE signal by extracellular S. aureus. CFSE signal represents HEp-2 cells infected with intracellular S. aureus only (c). This was confirmed by showing the loss of Cy5 signal in panel (d). Data shown are from a representative experiment which was conducted three times.
3.2. Microarray and QRT-PCR Data Analysis
Intracellular S. aureus altered expression of several classes of HEp-2 genes. Genes with statistically validated altered transcription levels >1.50-fold (increase or decrease) at any of the four-time-points in microarrays are listed in Table 2. To avoid potential pitfalls associated with amplification of mRNA such as inferior reducibility, mRNA was not amplified in this study. The microarray data shown here represented true transcription levels. Although we suspect that relatively low mRNA levels resulted in microarray data for some samples which were not statistically significant (P > .05), data for selected genes of interest were validated by QRT-PCR (summarized in Table 3). Data not shown in Table 2 resulted from signal intensities <50 which were too low to quantify.
Table 2.
Microarray analysis of gene expression changes in infected HEp-2 cell monolayers.
| Category | Gene | Fold change (P value) | |||
|---|---|---|---|---|---|
| 2 h | 4 h | 6 h | 8 h | ||
| Stress response | atf3 | 7.89 (.024) | 1.53 (.113) | 2.00 (.041) | 0.85 (.189) |
| c-fos | 2.59 (.052) | 1.21 (.072) | 0.911 (.302) | 0.843 (.287) | |
| fosB | 2.29 (.009) | 1.14 (.605) | 0.95 (.919) | 1.10 (.101) | |
| c-jun | 1.88 (.011) | 1.40 (.094) | 0.77 (.064) | 1.27 (.035) | |
| junB | 1.98 (.007) | 1.16 (.015) | 1.22 (.015) | 1.03 (.407) | |
| sgk | 4.17 (.018) | 2.16 (.005) | 1.84 (.178) | 2.13 (.066) | |
|
| |||||
| Signal transduction | map2k1 | 1.23 (.835) | 1.52 (.025) | 1.80 (.030) | 2.86 (.041) |
| arhe | 2.61 (.005) | 2.31 (.060) | 1.26 (.370) | 1.70 (.085) | |
| arhb | 2.21 (.044) | 2.18 (.004) | 1.78 (.065) | 1.60 (.022) | |
| ack-1 | 0.67 (.061) | 0.56 (.081) | 0.39 (.004) | 0.51 (.026) | |
| map2k3 | 1.98 (.014) | 2.41 (.003) | 1.96 (.127) | 2.33 (.052) | |
|
| |||||
| Proinflammatory response | cox2 | 3.32 (.012) | 2.71 (.088) | 1.29 (.065) | 2.55 (.064) |
|
| |||||
| Cell proliferation and proapoptosis | dkk1 | 2.17 (.026) | 7.11 (.001) | 3.23 (.012) | 4.52 (.055) |
| klf4 | 2.33 (.001) | 1.78 (.134) | 1.61 (.008) | 1.61 (.104) | |
| klf6 | 2.51 (.019) | 1.50 (.064) | 1.52 (.046) | 1.62 (.090) | |
| Igfbp1 | 2.54 (.062) | 4.32 (.001) | 2.13 (.086) | 11.10 (.030) | |
| Igfbp3 | 0.77 (.664) | 1.80 (.051) | 3.65 (.003) | 2.23 (.088) | |
| casp9 | 1.95 (.015) | 1.57 (.021) | 0.54 (.617) | 0.78 (.666) | |
| bnip3 | 1.25 (.073) | 1.64 (.034) | 2.86 (.002) | 2.47 (.041) | |
| nur77 | 6.17 (.038) | 1.28 (.181) | 0.78 (.114) | 0.87 (.235) | |
|
| |||||
| Profibrotic | tgfbr2 | 1.47 (.011) | 1.67 (.083) | 2.09 (.008) | 2.10 (.010) |
| v-erb-b | 0.96 (.608) | 1.67 (.055) | 1.96 (.005) | 2.11 (.007) | |
| itga5 | 0.88 (.768) | 2.19 (.037) | 2.12 (.033) | 3.20 (.030) | |
| thbs1 | 1.28 (.112) | 2.93 (.001) | 2.54 (.024) | 2.44 (.209) | |
| pai1 | 2.45 (.006) | 1.80 (.040) | 1.27 (.089) | 1.32 (.136) | |
| pai2 | ND | 2.09 (.015) | 4.43 (.001) | 4.13 (.013) | |
| cyr61 | 4.43 (.007) | 3.24 (.001) | 2.33 (.152) | 1.61 (.249) | |
| ctgf | 6.78 (.026) | 2.13 (.141) | ND | ND | |
| nov | 1.46 (.301) | 1.98 (.059) | 2.05 (.002) | 2.15 (.006) | |
|
| |||||
| Cholesterol synthesis | sc4mol | 0.81 (.367) | 0.32 (.001) | 0.36 (.001) | 0.31 (.001) |
| hmgcr | 1.11 (.700) | 0.30 (.002) | 0.21 (.001) | 0.36 (.016) | |
| hsd17b7 | 0.74 (.189) | 0.50 (.001) | 0.30 (.006) | 0.31 (.014) | |
| idi1 | 1.00 (.979) | 0.53 (.018) | 0.33 (.004) | 0.31 (.151) | |
| sqle | 0.90 (.397) | 0.50 (.007) | 0.26 (.001) | 0.31 (.008) | |
| sc5dl | 0.90 (.358) | 0.45 (.017) | 0.23 (.008) | 0.35 (.064) | |
| fdft1 | 0.96 (.551) | 0.59 (.002) | 0.29 (.001) | 0.26 (.010) | |
| dhcr7 | 0.95 (.527) | 0.63 (.010) | 0.51 (.016) | 0.39 (.014) | |
| insig1 | 0.69 (.338) | 0.20 (.001) | 0.26 (.001) | 0.41 (.014) | |
| acas2 | ND | 0.58 (.080) | 0.27 (.001) | 0.37 (.024) | |
| ldlr | 1.09 (.048) | 0.41 (.002) | 0.54 (.017) | 0.68 (.114) | |
ND: Not determined. Data not shown due to low signal intensity (<50).
Table 3.
Validation of selected genes by QRT-PCR.
| Category | Gene | Fold change (P value) | |||
|---|---|---|---|---|---|
| 2 h | 4 h | 6 h | 8 h | ||
| Stress response | atf3 | 15.45 (.001) | 4.46 (.005) | 1.38 (.004) | 1.97 (.027) |
| c-fos | 4.55 (.001) | 1.32 (.005) | 1.47 (.001) | 1.86 (.001) | |
| c-jun | 2.93 (.001) | 1.26 (.001) | 1.77 (.001) | 2.91 (.001) | |
| junb | 3.60 (.001) | 1.26 (.001) | 1.37 (.008) | 2.29 (.004) | |
| sgk | 3.36 (.001) | 1.20 (.001) | 1.87 (.001) | 2.02 (.001) | |
|
| |||||
| Signal transduction | arhb | 2.61 (.002) | 1.80 (.001) | 2.17 (.001) | 1.71 (.019) |
| map2k3 | 1.57 (.001) | 1.89 (.013) | 2.04 (.005) | 2.12 (.001) | |
|
| |||||
| Proinflammatory response | il1b | 3.64 (.001) | 1.58 (.007) | 2.02 (.001) | 1.47 (.002) |
| tnfa | 3.36 (.001) | 1.30 (.009) | 2.21 (.001) | 1.43 (.001) | |
| il6 | 2.65 (.001) | 1.87 (.001) | 2.96 (.001) | 1.55 (.004) | |
| ccl20 | 6.29 (.001) | 5.07 (.002) | 4.53 (.001) | 1.82 (.001) | |
| cxcl1 | 3.82 (.001) | 2.41 (.001) | 2.87 (.001) | 3.05 (.050) | |
| cox2 | 4.16 (.001) | 3.69 (.001) | 3.96 (.001) | 2.95 (.011) | |
|
| |||||
| Cell proliferation and Proapoptosis | dkk1 | 3.43 (.001) | 6.90 (.001) | 4.31 (.001) | 2.28 (.001) |
| igfbp1 | 3.50 (.001) | 6.82 (.045) | 4.20 (.010) | 9.89 (.001) | |
| casp9 | 2.74 (.001) | 1.34 (.005) | 1.42 (.004) | 1.13 (.021) | |
|
| |||||
| Profibrotic | tgfb1 | 1.55 (.002) | 1.20 (.023) | 1.71 (.001) | 2.66 (.001) |
| thbs1 | 1.83 (.001) | 4.55 (.001) | 4.12 (.002) | 4.09 (.001) | |
| cyr61 | 3.31 (.001) | 4.01 (.002) | 2.28 (.016) | 2.44 (.001) | |
|
| |||||
| Cholesterol synthesis | hmgcr | 1.25 (.025) | 0.17 (.001) | 0.15 (.001) | 0.17 (.001) |
| sqle | 1.00 (.005) | 0.30 (.001) | 0.14 (.001) | 0.15 (.001) | |
| dhcr7 | 1.17 (.050) | 0.50 (.001) | 0.27 (.001) | 0.16 (.001) | |
| ldlr | 1.57 (.010) | 0.09 (.001) | 0.12 (.001) | 0.23 (.001) | |
3.3. Stress Response
The adaptor-related protein complex 1 (AP-1) comprises JUN, FOS, and activating transcription factor (ATF) proteins; it regulates a variety of activities including proliferation, apoptosis, and inflammation in response to stress signals, cytokines, growth factors, and microbial infections [40, 41]. Internalization of S. aureus induced a rapid (7.89-fold) increase in atf3 mRNA levels at 2 hours postinfection that rapidly declined thereafter, as measured by microarray analysis (Table 2). QRT-PCR analysis yielded consistent findings (Table 3). Other AP-1 genes, such as c-fos, fosB, c-jun, and junB, were up-regulated as measured by microarray and/or QRT-PCR analysis, albeit less dramatically at 2 hours. Another stress response gene, sgk, encoding serum and glucocorticoid-induced protein kinase (SGK) [42], was up-regulated maximally at 2 hours (Tables 2 and 3). SGK is involved in epithelial sodium transport, and is induced in epithelial cells in response to environmental stimuli and stress [42].
3.4. Signal Transduction
Intracellular S. aureus also affected genes involved in several mitogen-activated protein kinase (MAPK) pathways. MAPK kinase 1 (map2k1) mRNA levels gradually increased and reached a maximum level at 8 hours (Table 2), MAPK kinase 1 activates downstream extracellular signal-regulated protein kinases (ERKs) in the Ras-Raf-MEK-ERK pathway. Two Ras homolog genes, arhe and arhb, were generally up-regulated >1.50-fold throughout the 8-hour-infection (Tables 2 and 3), whereas, the Ras inhibitor gene, ack-1, was down-regulated (Table 2). Thus, up-regulation of map2k1, arhe, and arhb, and down-regulation of the inhibitor ack-1 are consistent with activation of Ras-ERK pathway. Ras proteins are important for cytoskeleton reorganization [43, 44], coinciding with bacterial uptake and intracellular movement. Transcription of another MAPK gene (map2k3), a dual-specific kinase that phosphorylates MAPK14 (p38), was up-regulated >1.50-fold at all four-time-points (Tables 2 and 3). P38 pathway plays an important role in regulating proinflammatory gene expression including tnfa, il1b, and cox2 [43, 44].
Staphylococcal activation of the ERK and P38 pathways in epithelial cells has also been observed in previous studies [45–47]. Activation of ERK and P38 pathways, in epithelial cells, was also seen in other intracellular pathogen infections such as Helicobacter pylori [48] and Salmonella enterica [49].
3.5. Proinflammatory Response
Intracellular bacteria frequently up-regulate several proinflammatory cytokine genes (tnfa, il1b, and il6) and chemokine genes (il8, ccl20, and cxcl1) [15, 49, 50]. Due to the low transcriptional activity of il1b, tnfa, il6, cxcl1, and ccl20 in uninfected HEp2-cells, accurate comparison of these genes was not obtained with microarray analysis. QRT-PCR analysis demonstrated that transcription of il1b, tnfa, il6, cxcl1, and ccl20 genes was up-regulated (Table 3), although only small to moderate increases were observed, compared to previous study [22]. This finding is likely due to differences in types of host cells and in S. aureus strains, and also due to the fact that we investigated only the effects of intracellular staphylococci. For example, human umbilical endothelial cells infected with a clinical S. aureus isolate, were induced expression of several proinflammatory cytokines/chemokines with similar fold changes to our study at transcriptional level. However, it did not induce expression of either tnfa, or ilb, which was different from our study [26]. Similarly, vaginal epithelial cells cocultured simultaneously with intracellular and extracellular S. aureus MNSM, producing toxic shock syndrome toxin-1, for 3 hours showed increases in the transcription of il8, cxcl1, and ccl20 (11.3-fold, 17.1-fold and 207.9-fold, resp.) which were much stronger than our results [22].
Cyclooxygenase-2 gene (cox2), an inducible form of the cyclooxygenase-1 gene (cox1), was up-regulated at all four-time-points in this study (Tables 2 and 3). As an immediate early response gene that is responsible for prostanoid biosynthesis involved in proinflammation, cox2 is expressed in epithelial cells, macrophages, fibroblasts, and vascular endothelial cells [51]. COX2 is induced by IL-1β [52] and lipoteichoic acid from S. aureus [53]. Up-regulation of cox2 transcription was also associated with infection of epithelial cells by gram-negative bacteria: Y. enterocolitica [13] and S. flexneri M90T, probably via LPS [15]. The induction of cox2 expression is not significantly in vaginal epithelial cell cultures infected (intracellular plus extracellular) with the superantigen producing strain S. aureus MNSM (see above) [22], further emphasizing the potentially different effects caused by various S. aureus strains, as well as the systems employed to measure their effects.
3.6. Cell Proliferation and Proapoptosis
Intracellular S. aureus RN6390 affected transcription of several proapoptotic genes. Dickkopf-1 (dkk1), was up-regulated >2.00-fold at all time points examined (Tables 2 and 3). Krüppel-like factors 4 and 6 genes (klf4 and klf6) were up-regulated >2.00-fold at 2 hours postinfection (Table 2). Microarray data showed the gene for caspase-9 (casp9) up-regulated ~2.00-fold at 2 hours (Table 2), and this result was confirmed by QRT-PCR (Table 3). The gene (bnip3) encoding Bcl2/adenovirus E1B 19kDa interacting protein 3, a mitochondrial proapoptotic protein, was up-regulated >2.00-fold at both 6 hours and 8 hours (Table 2). Two insulin-like growth factor binding protein genes (igfbp1 and igfbp3) were up-regulated >1.50-fold at 4 hours, 6 hours, and 8 hours postinfection (Tables 2 and 3). The NR4A1 receptor gene (nur77), which encodes a transcription factor that exhibits proapoptotic properties in T cells [54], was up-regulated ~6-fold at 2 hours (Table 2). These findings were similar to several studies demonstrating that the infection of epithelial cells [8, 28, 32, 33], endothelial cells [29, 30, 55, 56], and osteoblasts [3, 57, 58] with S. aureus can lead to apoptosis. Previous work in our lab had shown the involvement of host caspases 3 and 8 in S. aureus-induced apoptosis [32] and the requirement of the S. aureus virulence gene regulator agr in the induction of epithelial cell apoptosis [33].
3.7. Profibrotic Gene Transcription in HEp-2 Cells
TGFβ1 is a key protein involved in many cell functions including fibrosis formation, regulation of cell cycle, apoptosis, and matrix remodeling [59]. QRT-PCR indicated that tgfβ1 was up-regulated by intracellular S. aureus (Table 3). Intracellular S. aureus also induced transcription of several genes related to TGFβ1, especially in regard to fibrosis formation (Tables 2 and 3). In microarray experiments, transforming growth factor beta receptor 2 gene (tgfβr2) and epidermal growth factor receptor (EGFR) gene (v-erb-b) were up-regulated >1.5-fold after 4 hours (Table 2). Integrin α5 gene (itga5) was gradually up-regulated after 2-hour-infection (Table 2). The gene (thbs1) encoding thrombospondin 1 was up-regulated ~3.00-fold at 4 hours and 2.54-fold at 6 hours in microarray experiments (Table 2), and similarly, with QRT-PCR (Table 3).
Plasminogen activator inhibitor 1 and 2 genes (pai1, pai2) were up-regulated in microarray experiments (Table 2). Studies have shown that TGFβ1 induces plasminogen activator inhibitor 1 (PAI1) expression and demonstrated the requirement for EGFR in this process [60–62]. Both PAI1 and PAI2 are inhibitors of the fibrinolysis system, acting to block the activity of tissue plasminogen activator and urokinase, and preventing the conversion of plasminogen to plasmin. Plasmin is a serine protease that degrades fibrin clots as well as extracellular matrix components. Thus, up-regulation of pai1 and pai2 may reduce extracellular matrix degradation.
The CCN (Cysteine-rich 61, Connective tissue growth factor, and Nephroblastoma overexpressed) family members are cysteine-rich and functionally diverse proteins that are involved in mitosis, apoptosis, adhesion, extracellular matrix production, angiogenesis, and tumor growth [63]. Three genes belonging to the CCN family were up-regulated. Two of those, cyr61 and ctgf, were significantly up-regulated at early time points (Table 2 and 3). The third CCN gene, nov, was significantly up-regulated after 4 hours at transcriptional level (Table 2). An increased transcription of cyr61 and ctgf genes has been shown during epithelial cell infection with Y. enterocolitica [13], S. flexneri [15], and B. pertussis [16]. CYR61, CTGF, and NOV have the capability to bind both fibronectin and α 5 β 1 integrin, similar to IGFBP1 and IGFBP3 [64–68], and are implicated in wound healing [68]. Taken together, up-regulation of these profibrotic genes indicates that intracellular S. aureus might affect the extracellular matrix by stimulating fibrosis and aiding in repair of the damage caused by S. aureus infection.
3.8. Cholesterol Biosynthesis
Intracellular S. aureus caused down-regulated expression of cholesterol biosynthesis enzyme genes, including sterol-c4-methyl oxidase-like (sc4mol), 3-hydroxy-3-methylglutaryl-coenzyme A reductase (hmgcr), hydroxysteroid (17β) dehydrogenase 7 (hsd17b7), isopentenyl-diphosphate delta isomerase (idi1), squalene monooxygenase (sqle), sterol c5-desaturase-like (sc5dl), farnesyl-disphosphate famesyltransferase 1 (fdft1), and 7-dehydrocholesterol reductase (dhcr7). Genes involved in regulation of cholesterol synthesis were also down-regulated. Insulin-induced gene 1 (insig1), encoding a membrane endoplasmic reticulum protein, was down-regulated (0.20-fold at 4 hours, 0.26-fold at 6 hours, and 0.41-fold at 8 hours) (Table 2). Acetyl CoA synthetase gene (acas2) and low-density lipoprotein receptor gene (ldlr) were also transcriptionally down-regulated (Table 2). QRT-PCR data confirmed the down-regulation of hmgcr, sqle, dhcr7, and ldlr (Table 3). Cholesterol quantification with GC-MS also showed that host cells displayed a corresponding decreased cholesterol synthesis after a challenge with intracellular S. aureus (Table 4). Garner et al. showed an essential role for cholesterol in the uptake of S. typhimurium into HeLa cells, demonstrating that the removal of cholesterol caused a greater than 90% decrease in bacterial uptake [69]. Thus, a reduction in cholesterol may be a response to limit the internalization of S. aureus. In addition, a decrease in cholesterol levels could limit the effects of S. aureus exotoxins on the host cell membrane. S. aureus alpha toxin, along with other pore-forming toxins from Streptococcus and Clostridium species, showed reduced activity when cholesterol levels in lipid membranes were decreased [70, 71]. A recent study showed that the golden S. aureus pigment, staphyloxanthin, is synthesized with the same substrates used for cholesterol biosynthesis by host cells [72]. It is unclear at present whether the effect on cholesterol biosynthesis is related to this finding; however, it is conceivable that this effect might represent a host response to affect production of this staphylococcal virulence factor.
Table 4.
Cholesterol quantification [(μg/105 cells) ± SD] in uninfected and infected HEp-2 cell monolayers.
| Cell type | Incubation time (h) | |||
|---|---|---|---|---|
| 2 | 4 | 6 | 8 | |
| Unchallenged HEp-2 cell | 75.82 ± 2.99 | 69.78 ± 5.31 | 64.94 ± 5.00 | 55.91 ± 4.78 |
| Challenged HEp-2 cell | 58.43 ± 3.73 | 51.92 ± 2.87 | 50.74 ± 5.22 | 43.38 ± 3.36 |
| % cholesterol reduction | 22.91 | 25.60 | 21.87 | 22.41 |
|
| ||||
| P value | .005 | .050 | .045 | .026 |
In summary, this study demonstrates that several classes of genes in HEp-2 cells undergo changes in transcriptional expression in response to intracellular S. aureus. We observed that, in the first few hours of intracellular infection, epithelial cells can respond to intracellular S. aureus quickly by inducing early stress response (AP-1 complex) and MAPK pathways (Ras, P38), which consequently stimulate broader responses such as proinflammatory response, apoptosis, and fibrosis. Our data support the belief that the role of epithelial cells in innate immunity is not simply that of a physical barrier against invading pathogens, but it is also actively involved in the induction of more complex host defense mechanisms. Another possibility is that, as a successful pathogen, intracellular S. aureus might lead to host gene expression that facilitates its intracellular survival. This is consistent with induction of Ras-related cytoskeleton reorganization and the fibrosis process. Our results are also consistent with, although not definitive of, a delicate balance between effects which benefit the host and those which are more beneficial to S. aureus. Finally, this study showed that intracellular S. aureus suppressed cholesterol synthesis in epithelial cells. The consequence of this suppression on the pathogenesis of S. aureus is not clearly presented but might be related to recent observations regarding staphylococcal pigment production.
Acknowledgments
This work was supported, in part, by the Idaho Agricultural Experiment Station, and Public Health Service grants, U54-AI-57141, P20-RR016454, and P20-RR15587. The authors are grateful to Darren Schnider for assistance in preparing this manuscript. The first two authors contributed equally to this work.
References
- 1.Kuehnert MJ, Kruszon-Moran D, Hill HA, et al. Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001-2002. The Journal of Infectious Diseases. 2006;193(2):172–179. doi: 10.1086/499632. [DOI] [PubMed] [Google Scholar]
- 2.Bohach GA, Fast DJ, Nelson RD, Schlievert PM. Staphylococcal and streptococcal pyrogenic toxins involved in toxic shock syndrome and related illnesses. Critical Reviews in Microbiology. 1990;17(4):251–272. doi: 10.3109/10408419009105728. [DOI] [PubMed] [Google Scholar]
- 3.Alexander EH, Hudson MC. Factors influencing the internalization of Staphylococcus aureus and impacts on the course of infections in humans. Applied Microbiology and Biotechnology. 2001;56(3-4):361–366. doi: 10.1007/s002530100703. [DOI] [PubMed] [Google Scholar]
- 4.Proctor RA, Kahl B, von Eiff C, Vaudaux PE, Lew DP, Peters G. Staphylococcal small colony variants have novel mechanisms for antibiotic resistance. Clinical Infectious Diseases. 1998;27(supplement 1):S68–S74. doi: 10.1086/514906. [DOI] [PubMed] [Google Scholar]
- 5.Proctor RA, von Eiff C, Kahl BC, et al. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nature Reviews Microbiology. 2006;4(4):295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- 6.Speller DCE, Johnson AP, James D, Marples RR, Charlett A, George RC. Resistance to methicillin and other antibiotics in isolates of Staphylococcus aureus from blood and cerebrospinal fluid, England and Wales, 1989–95. The Lancet. 1997;350(9074):323–325. doi: 10.1016/s0140-6736(97)12148-1. [DOI] [PubMed] [Google Scholar]
- 7.Almeida RA, Matthews KR, Cifrian E, Guidry AJ, Oliver SP. Staphylococcus aureus invasion of bovine mammary epithelial cells. Journal of Dairy Science. 1996;79(6):1021–1026. doi: 10.3168/jds.S0022-0302(96)76454-8. [DOI] [PubMed] [Google Scholar]
- 8.Bayles KW, Wesson CA, Liou LE, Fox LK, Bohach GA, Trumble WR. Intracellular Staphylococcus aureus escapes the endosome and induces apoptosis in epithelial cells. Infection and Immunity. 1998;66(1):336–342. doi: 10.1128/iai.66.1.336-342.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beekhuizen H, van de Gevel JS, Olsson B, van Benten IJ, van Furth R. Infection of human vascular endothelial cells with Staphylococcus aureus induces hyperadhesiveness for human monocytes and granulocytes. The Journal of Immunology. 1997;158(2):774–782. [PubMed] [Google Scholar]
- 10.Dziewanowska K, Carson AR, Patti JM, Deobald CF, Bayles KW, Bohach GA. Staphylococcal fibronectin binding protein interacts with heat shock protein 60 and integrins: role in internalization by epithelial cells. Infection and Immunity. 2000;68(11):6321–6328. doi: 10.1128/iai.68.11.6321-6328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sinha B, François PP, Nüße O, et al. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin α 5 β 1 . Cellular Microbiology. 1999;1(2):101–117. doi: 10.1046/j.1462-5822.1999.00011.x. [DOI] [PubMed] [Google Scholar]
- 12.Vesga O, Groeschel MC, Otten MF, Brar DW, Vann JM, Proctor RA. Staphylococcus aureus small colony variants are induced by the endothelial cell intracellular milieu. The Journal of Infectious Diseases. 1996;173(3):739–742. doi: 10.1093/infdis/173.3.739. [DOI] [PubMed] [Google Scholar]
- 13.Bohn E, Müller S, Lauber J, et al. Gene expression patterns of epithelial cells modulated by pathogenicity factors of Yersinia enterocolitica . Cellular Microbiology. 2004;6(2):129–141. doi: 10.1046/j.1462-5822.2003.00346.x. [DOI] [PubMed] [Google Scholar]
- 14.Eckmann L, Smith JR, Housley MP, Dwinell MB, Kagnoff MF. Analysis by high density cDNA arrays of altered gene expression in human intestinal epithelial cells in response to infection with the invasive enteric bacteria Salmonella . The Journal of Biological Chemistry. 2000;275(19):14084–14094. doi: 10.1074/jbc.275.19.14084. [DOI] [PubMed] [Google Scholar]
- 15.Pédron T, Thibault C, Sansonetti PJ. The invasive phenotype of Shigella flexneri directs a distinct gene expression pattern in the human intestinal epithelial cell line caco-2. The Journal of Biological Chemistry. 2003;278(36):33878–33886. doi: 10.1074/jbc.M303749200. [DOI] [PubMed] [Google Scholar]
- 16.Belcher CE, Drenkow J, Kehoe B, et al. The transcriptional responses of respiratory epithelial cells to Bordetella pertussis reveal host defensive and pathogen counter-defensive strategies. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(25):13847–13852. doi: 10.1073/pnas.230262797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danelishvili L, McGarvey J, Li Y-J, Bermudez LE. Mycobacterium tuberculosis infection causes different levels of apoptosis and necrosis in human macrophages and alveolar epithelial cells. Cellular Microbiology. 2003;5(9):649–660. doi: 10.1046/j.1462-5822.2003.00312.x. [DOI] [PubMed] [Google Scholar]
- 18.Ichikawa JK, Norris A, Bangera MG, et al. Interaction of Pseudomonas aeruginosa with epithelial cells: identification of differentially regulated genes by expression microarray analysis of human cDNAs. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(17):9659–9664. doi: 10.1073/pnas.160140297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baldwin DN, Vanchinathan V, Brown PO, Theriot JA. A gene-expression program reflecting the innate immune response of cultured intestinal epithelial cells to infection by Listeria monocytogenes . Genome Biology. 2003;4(1, article R2):1–14. doi: 10.1186/gb-2002-4-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakagawa I, Nakata M, Kawabata S, Hamada S. Transcriptome analysis and gene expression profiles of early apoptosis-related genes in Streptococcus pyogenes-infected epithelial cells. Cellular Microbiology. 2004;6(10):939–952. doi: 10.1111/j.1462-5822.2004.00412.x. [DOI] [PubMed] [Google Scholar]
- 21.Moreilhon C, Gras D, Hologne C, et al. Live Staphylococcus aureus and bacterial soluble factors induce different transcriptional responses in human airway cells. Physiological Genomics. 2005;20:244–255. doi: 10.1152/physiolgenomics.00135.2004. [DOI] [PubMed] [Google Scholar]
- 22.Peterson ML, Ault K, Kremer MJ, et al. The innate immune system is activated by stimulation of vaginal epithelial cells with Staphylococcus aureus and toxic shock syndrome toxin 1. Infection and Immunity. 2005;73(4):2164–2174. doi: 10.1128/IAI.73.4.2164-2174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dziewanowska K, Patti JM, Deobald CF, Bayles KW, Trumble WR, Bohach GA. Fibronectin binding protein and host cell tyrosine kinase are required for internalization of Staphylococcus aureus by epithelial cells. Infection and Immunity. 1999;67(9):4673–4678. doi: 10.1128/iai.67.9.4673-4678.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellington JK, Elhofy A, Bost KL, Hudson MC. Involvement of mitogen-activated protein kinase pathways in Staphylococcus aureus invasion of normal osteoblasts. Infection and Immunity. 2001;69(9):5235–5242. doi: 10.1128/IAI.69.9.5235-5242.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellington JK, Reilly SS, Ramp WK, Smeltzer MS, Kellam JF, Hudson MC. Mechanisms of Staphylococcus aureus invasion of cultured osteoblasts. Microbial Pathogenesis. 1999;26(6):317–323. doi: 10.1006/mpat.1999.0272. [DOI] [PubMed] [Google Scholar]
- 26.Matussek A, Strindhall J, Stark L, et al. Infection of human endothelial cells with Staphylococcus aureus induces transcription of genes encoding an innate immunity response. Scandinavian Journal of Immunology. 2005;61(6):536–544. doi: 10.1111/j.1365-3083.2005.01597.x. [DOI] [PubMed] [Google Scholar]
- 27.Hess DJ, Henry-Stanley MJ, Erickson EA, Wells CL. Intracellular survival of Staphylococcus aureus within cultured enterocytes. Journal of Surgical Research. 2003;114(1):42–49. doi: 10.1016/s0022-4804(03)00314-7. [DOI] [PubMed] [Google Scholar]
- 28.Kahl BC, Goulian M, van Wamel W, et al. Staphylococcus aureus RN6390 replicates and induces apoptosis in a pulmonary epithelial cell line. Infection and Immunity. 2000;68(9):5385–5392. doi: 10.1128/iai.68.9.5385-5392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menzies BE, Kourteva I. Internalization of Staphylococcus aureus by endothelial cells induces apoptosis. Infection and Immunity. 1998;66(12):5994–5998. doi: 10.1128/iai.66.12.5994-5998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menzies BE, Kourteva I. Staphylococcus aureus α-toxin induces apoptosis in endothelial cells. FEMS Immunology and Medical Microbiology. 2000;29(1):39–45. doi: 10.1111/j.1574-695X.2000.tb01503.x. [DOI] [PubMed] [Google Scholar]
- 31.Murai M, Sakurada J, Seki K, Shinji H, Hirota Y, Masuda S. Apoptosis observed in BALB/3T3 cells having ingested Staphylococcus aureus . Microbiology and Immunology. 1999;43(7):653–661. doi: 10.1111/j.1348-0421.1999.tb02453.x. [DOI] [PubMed] [Google Scholar]
- 32.Wesson CA, Deringer J, Liou LE, Bayles KW, Bohach GA, Trumble WR. Apoptosis induced by Staphylococcus aureus in epithelial cells utilizes a mechanism involving caspases 8 and 3. Infection and Immunity. 2000;68(5):2998–3001. doi: 10.1128/iai.68.5.2998-3001.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wesson CA, Liou LE, Todd KM, Bohach GA, Trumble WR, Bayles KW. Staphylococcus aureus Agr and Sar global regulators influence internalization and induction of apoptosis. Infection and Immunity. 1998;66(11):5238–5243. doi: 10.1128/iai.66.11.5238-5243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore AE, Sabachewsky L, Toolan HW. Culture characteristics of four permanent lines of human cancer cells. Cancer Research. 1955;15(9):598–602. [PubMed] [Google Scholar]
- 35.Cheung AL, Chien Y-T, Bayer AS. Hyperproduction of alpha-hemolysin in a sigB mutant is associated with elevated SarA expression in Staphylococcus aureus . Infection and Immunity. 1999;67(3):1331–1337. doi: 10.1128/iai.67.3.1331-1337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shompole S, Henon KT, Liou LE, Dziewanowska K, Bohach GA, Bayles KW. Biphasic intracellular expression of Staphylococcus aureus virulence factors and evidence for Agr-mediated diffusion sensing. Molecular Microbiology. 2003;49(4):919–927. doi: 10.1046/j.1365-2958.2003.03618.x. [DOI] [PubMed] [Google Scholar]
- 37.Seo KK, Lee SU, Park YH, Davis WC, Fox LK, Bohach GA. Long-term staphylococcal enterotoxin C1 exposure induces soluble factor-mediated immunosuppression by bovine CD4+ and CD8+ T cells. Infection and Immunity. 2007;75(1):260–269. doi: 10.1128/IAI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clark RM, Ferris AM, Fey M, Brown PB, Hundrieser KE, Jensen RG. Changes in the lipids of human milk from 2 to 16 weeks postpartum. Journal of Pediatric Gastroenterology and Nutrition. 1982;1(3):311–315. doi: 10.1097/00005176-198201030-00006. [DOI] [PubMed] [Google Scholar]
- 39.Dette H, Munk A. Optimum allocation of treatments for Welch's test in equivalence assessment. Biometrics. 1997;53(3):1143–1150. [PubMed] [Google Scholar]
- 40.Foletta VC, Segal DH, Cohen DR. Transcriptional regulation in the immune system: all roads lead to AP-1. Journal of Leukocyte Biology. 1998;63(2):139–152. doi: 10.1002/jlb.63.2.139. [DOI] [PubMed] [Google Scholar]
- 41.Wisdom R. AP-1: one switch for many signals. Experimental Cell Research. 1999;253(1):180–185. doi: 10.1006/excr.1999.4685. [DOI] [PubMed] [Google Scholar]
- 42.Leong MLL, Maiyar AC, Kim B, O'Keeffe BA, Firestone GL. Expression of the serum- and glucocorticoid-inducible protein kinase, Sgk, is a cell survival response to multiple types of environmental stress stimuli in mammary epithelial cells. The Journal of Biological Chemistry. 2003;278(8):5871–5882. doi: 10.1074/jbc.M211649200. [DOI] [PubMed] [Google Scholar]
- 43.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298(5600):1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 44.Machesky LM, Hall A. Role of actin polymerization and adhesion to extracellular matrix in Rac- and Rho-induced cytoskeletal reorganization. Journal of Cell Biology. 1997;138(4):913–926. doi: 10.1083/jcb.138.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hannigan M, Zhan L, Ai Y, Huang C-K. The role of p38 MAP kinase in TGF-β1-induced signal transduction in human neutrophils. Biochemical and Biophysical Research Communications. 1998;246(1):55–58. doi: 10.1006/bbrc.1998.8570. [DOI] [PubMed] [Google Scholar]
- 46.Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiological Reviews. 2001;81(2):807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- 47.Raingeaud J, Gupta S, Rogers JS, et al. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. The Journal of Biological Chemistry. 1995;270(13):7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 48.Tummala S, Keates S, Kelly CP. Update on the immunologic basis of Helicobacter pylori gastritis. Current Opinion in Gastroenterology. 2004;20(6):592–597. doi: 10.1097/00001574-200411000-00015. [DOI] [PubMed] [Google Scholar]
- 49.Guiney DG. The role of host cell death in Salmonella infections. Current Topics in Microbiology and Immunology. 2005;289:131–150. doi: 10.1007/3-540-27320-4_6. [DOI] [PubMed] [Google Scholar]
- 50.Eckmann L, Kagnoff MF, Fierer J. Epithelial cells secrete the chemokine interleukin-8 in response to bacterial entry. Infection and Immunity. 1993;61(11):4569–4574. doi: 10.1128/iai.61.11.4569-4574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith WL, Garavito RM, DeWitt DL. Prostaglandin endoperoxide H synthases (cyclooxygenases)-1 and -2. The Journal of Biological Chemistry. 1996;271(52):33157–33160. doi: 10.1074/jbc.271.52.33157. [DOI] [PubMed] [Google Scholar]
- 52.Maier JAM, Hla T, Maciag T. Cyclooxygenase is an immediate-early gene induced by interleukin-1 in human endothelial cells. The Journal of Biological Chemistry. 1990;265(19):10805–10808. [PubMed] [Google Scholar]
- 53.Lin C-H, Kuan I-H, Lee H-M, et al. Induction of cyclooxygenase-2 protein by lipoteichoic acid from Staphylococcus aureus in human pulmonary epithelial cells: involvement of a nuclear factor-κB-dependent pathway. British Journal of Pharmacology. 2001;134(3):543–552. doi: 10.1038/sj.bjp.0704290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woronicz JD, Calnan B, Ngo V, Winoto A. Requirement for the orphan steroid receptor Nur77 in apoptosis of T-cell hybridomas. Nature. 1994;367(6460):277–281. doi: 10.1038/367277a0. [DOI] [PubMed] [Google Scholar]
- 55.Esen M, Schreiner B, Jendrossek V, et al. Mechanisms of Staphylococcus aureus induced apoptosis of human endothelial cells. Apoptosis. 2001;6(6):431–439. doi: 10.1023/a:1012445925628. [DOI] [PubMed] [Google Scholar]
- 56.Haslinger-Löffler B, Kahl BC, Grundmeier M, et al. Multiple virulence factors are required for Staphylococcus aureus-induced apoptosis in endothelial cells. Cellular Microbiology. 2005;7(8):1087–1097. doi: 10.1111/j.1462-5822.2005.00533.x. [DOI] [PubMed] [Google Scholar]
- 57.Alexander EH, Rivera FA, Marriott I, Anguita J, Bost KL, Hudson MC. Staphylococcus aureus—induced tumor necrosis factor—related apoptosis—inducing ligand expression mediates apoptosis and caspase-8 activation in infected osteoblasts. BMC Microbiology. 2003;3, article 5:1–11. doi: 10.1186/1471-2180-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tucker KA, Reilly SS, Leslie CS, Hudson MC. Intracellular Staphylococcus aureus induces apoptosis in mouse osteoblasts. FEMS Microbiology Letters. 2000;186(2):151–156. doi: 10.1111/j.1574-6968.2000.tb09096.x. [DOI] [PubMed] [Google Scholar]
- 59.Lee CG, Kang H-R, Homer RJ, Chupp G, Elias JA. Transgenic modeling of transforming growth factor-β 1: role of apoptosis in fibrosis and alveolar remodeling. Proceedings of the American Thoracic Society. 2006;3(5):418–423. doi: 10.1513/pats.200602-017AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arnoletti JP, Albo D, Granick MS, et al. Thrombospondin and transforming growth factor-beta 1 increase expression of urokinase-type plasminogen activator and plasminogen activator inhibitor-1 in human MDA-MB-231 breast cancer cells. Cancer. 1995;76(6):998–1005. doi: 10.1002/1097-0142(19950915)76:6<998::aid-cncr2820760613>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 61.Kutz SM, Higgins CE, Samarakoon R, et al. TGF-β1-induced PAI-1 expression is E box/USF-dependent and requires EGFR signaling. Experimental Cell Research. 2006;312(7):1093–1105. doi: 10.1016/j.yexcr.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 62.Kutz SM, Hordines J, McKeown-Longo PJ, Higgins PJ. TGF-β1-induced PAI-1 gene expression requires MEK activity and cell-to-substrate adhesion. Journal of Cell Science. 2001;114(21):3905–3914. doi: 10.1242/jcs.114.21.3905. [DOI] [PubMed] [Google Scholar]
- 63.Brigstock DR. The CCN family: a new stimulus package. Journal of Endocrinology. 2003;178(2):169–175. doi: 10.1677/joe.0.1780169. [DOI] [PubMed] [Google Scholar]
- 64.Gleeson LM, Chakraborty C, Mckinnon T, Lala PK. Insulin-like growth factor-binding protein 1 stimulates human trophoblast migration by signaling through α5β1 integrin via mitogen-activated protein kinase pathway. The Journal of Clinical Endocrinology & Metabolism. 2001;86(6):2484–2493. doi: 10.1210/jcem.86.6.7532. [DOI] [PubMed] [Google Scholar]
- 65.Gui Y, Murphy LJ. Insulin-like growth factor (IGF)-binding protein-3 (IGFBP-3) binds to fibronectin (FN): demonstration of IGF-I/IGFBP-3/FN ternary complexes in human plasma. The Journal of Clinical Endocrinology & Metabolism. 2001;86(5):2104–2110. doi: 10.1210/jcem.86.5.7472. [DOI] [PubMed] [Google Scholar]
- 66.Jones JI, Gockerman A, Busby WH, Jr., Wright G, Clemmons DR. Insulin-like growth factor binding protein 1 stimulates cell migration and binds to the α5β1 integrin by means of its Arg-Gly-Asp sequence. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(22):10553–10557. doi: 10.1073/pnas.90.22.10553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leask A, Abraham DJ. All in the CCN family: essential matricellular signaling modulators emerge from the bunker. Journal of Cell Science. 2006;119(23):4803–4810. doi: 10.1242/jcs.03270. [DOI] [PubMed] [Google Scholar]
- 68.Lin CG, Chen C-C, Leu S-J, Grzeszkiewicz TM, Lau LF. Integrin-dependent functions of the angiogenic inducer NOV (CCN3): implication in wound healing. The Journal of Biological Chemistry. 2005;280(9):8229–8237. doi: 10.1074/jbc.M404903200. [DOI] [PubMed] [Google Scholar]
- 69.Garner MJ, Hayward RD, Koronakis V. The Salmonella pathogenicity island 1 secretion system directs cellular cholesterol redistribution during mammalian cell entry and intracellular trafficking. Cellular Microbiology. 2002;4(3):153–165. doi: 10.1046/j.1462-5822.2002.00181.x. [DOI] [PubMed] [Google Scholar]
- 70.Giddings KS, Johnson AE, Tweten RK. Redefining cholesterol's role in the mechanism of the cholesterol-dependent cytolysins. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(20):11315–11320. doi: 10.1073/pnas.2033520100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tomita T, Watanabe M, Yasuda T. Influence of membrane fluidity on the assembly of Staphylococcus aureus α-toxin, a channel-forming protein, in liposome membrane. The Journal of Biological Chemistry. 1992;267(19):13391–13397. [PubMed] [Google Scholar]
- 72.Liu C-I, Liu GY, Song Y, et al. A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science. 2008;319(5868):1391–1394. doi: 10.1126/science.1153018. [DOI] [PMC free article] [PubMed] [Google Scholar]