Abstract
The aim of this study was to characterize the conformational neutralizing epitopes of the major capsid protein of human papillomavirus type 31. Analysis of the epitopes was performed by competitive epitope mapping using 15 anti-HPV31 and by reactivity analysis using a HPV31 mutant with an insertion of a seven-amino acid motif within the FG loop of the capsid protein. Fine mapping of neutralizing conformational epitopes on HPV L1 was analyzed by a new approach using a system displaying a combinatorial library of constrained peptides exposed on E. coli flagella. The findings demonstrate that the HPV31 FG loop is dense in neutralizing epitopes and suggest that HPV31 MAbs bind to overlapping but distinct epitopes on the central part of the FG loop, in agreement with the exposure of the FG loop on the surface of HPV VLPs, and thus confirming that neutralizing antibodies are mainly located on the tip of capsomeres. In addition, we identified a crossreacting and partially crossneutralizing conformational epitope on the relatively well conserved N-terminal part of the FG loop. Moreover, our findings support the hypothesis that there is no correlation between neutralization and the ability of MAbs to inhibit VLP binding to heparan sulfate, and confirm that the blocking of virus attachment to the extracellular matrix is an important mechanism of neutralization.
Keywords: HPV31, L1 protein, conformational epitope, bacterial surface display, neutralization
Introduction
Human papillomaviruses (HPVs) are epitheliotropic viruses mainly causing benign tumors of the skin and mucosa. However, a subset of sexually transmitted HPVs has been recognized as the etiological agent of cervical carcinoma.1,2 Great progress has been made in the last 15 years toward the development of prophylactic HPV vaccines, and they have recently been shown to induce high levels of clinical protection.3,4 Virus neutralization is defined as the abrogation of virus infectivity by the association of antibodies with the viral particle, and neutralization can be achieved by different mechanisms. The first obvious mechanism is the inhibition of virus attachment to host cells.5–7 However, neutralizing antibodies can act before attachment to the host cell by inhibiting transcytosis through mucosal epithelial cells,8 they can also interfere with postattachment interactions of the virus with its receptors and coreceptors, or they can act after viral endocytosis by negatively affecting virus trafficking.9–11
Papillomaviruses are small nonenveloped viruses with a double-stranded DNA genome of about 8 kb which is encapsidated in a structure consisting of 72 capsomers composed of five L1, the major capsid protein, and L2, the minor capsid protein. Immunization with L1 protein self-assembled into virus-like particles (VLPs) induces the production of high levels of neutralizing antibodies and confers type-specific and long-lasting protection, as demonstrated in animal models.12–14 The antibody responses are typically generated against epitopes found on the external loops of the L1 protein present on the outer VLP surface.15–18 Neutralizing capsid epitopes are important determinants for antibody-mediated immune protection against HPVs, and both linear and conformational epitopes have been identified on the surface of HPV L1 VLPs and pseudovirions.18–21 It is now well established that conformational epitopes are responsible for neutralizing antibody production.22–26 Because neutralizing epitopes of HPVs are conformation-dependent, their amino-acid composition and surface localization have not been fully characterized.
Findings obtained with an animal model have suggested that papillomaviruses require microabrasion in the cervical epithelium for infection of basal cells.27 The virions bind to the exposed basement membrane before migrating to the exposed cervico-vaginal epithelial cells and bind to the host cell receptor(s). Cell surface heparan sulfates (HS) serve as primary attachment receptors, and molecules with structural similarity to HS, such as heparin, are competitive inhibitors of HPV infection.26,28–31 Interaction with HS requires an intact VLP,32,33 with conflicting results indicating that both a conserved domain on the C-terminus part of the L1 protein28,34 and a conformational cluster of lysine residues on the surface of HPV33 virions35 interact with HS. Laminin-5, a protein secreted by the maturing basal epithelium, has also been shown to bind to both VLPs and native HPVs.36 Moreover, keratinocytes expressing α6 integrins are susceptible to infection with HPV prebound with laminin-5, while keratinocytes lacking α6 integrins are resistant to HPV infection,36,37 suggesting that HPV initiates infection by interacting with α6 integrin-expressing basal keratinocytes after binding to laminin-5. HPV31 pseudovirions have been reported to have different entry pathways into cells than those observed with other HPVs associated with cervical cancer,38,39 thus justifying further characterization of the antigenic structure and the mechanisms of neutralization of this particular type.
One of the questions that also remains is the exact location of the epitopes that induce HPV neutralizing antibodies and contribute to protection against infection due to interaction with the viral capsid. These studies may prove useful in future vaccine design and in the investigation of virus-cell interactions. In addition, an understanding of the antigen structure of HPV is crucial for designing HPV-derived gene therapy vectors with reduced immunogenicity. One prerequisite for generating such vectors is greater understanding of viral determinants provoking neutralizing immune responses, to design pseudovirion vectors with deletion or mutation within the conformational epitopes responsible for the production of neutralizing antibodies. These mutated vectors with reduced immunogenicity will allow readministration of these vectors without a dramatic loss of transgene efficacy due to induction of neutralizing antibodies against the vector.
In the study reported here, we investigated a panel of MAbs to HPV31 VLPs using antibody binding competition by surface plasmon resonance to identify the neutralizing epitopes present on the HPV31 L1 protein. The localization of epitopes was then investigated using a L1 protein mutant and five MAbs were fine-mapped using E. coli for display of constrained random peptides on bacterial flagella. We also investigated the L1 sequences involved in HS binding for both HPV16 and HPV31, the competition between HS, extracellular matrix (ECM), and HPV31 MAbs to bind to VLPs, and whether these monoclonal antibodies neutralized HPV31 pseudovirions before or after VLP cell binding.
Results
Epitope mapping of HPV31 L1 MAbs using surface plasmon resonance
Epitope mapping was performed using 15 MAbs raised against HPV31 L1 protein. All competitions between these MAbs were tested. For example [Fig. 1(a)], epitope competition was established by saturating coupled HPV31 L1 VLPs with H31.F16 MAb (crude ascites fluid), and the MAb saturation was verified by injection of the same MAbs (hybridoma supernatant). The low-resonance unit (RU) that occurred after addition of H31.F16 MAb (−52 RU) proved that saturation was achieved. After saturation there was a decrease in RU, due to the dissociation of saturating MAb and this was the reason for the negative RU. MAb binding competition was then established by successively injecting H31.B18, H31.D7, H31.E16, H31.E17, and H31.F7 MAbs (hybridoma supernatants). H31.E16 and H31.F7 MAbs did not bind to HPV31 L1 VLPs (−1 and −13 RU, respectively), suggesting that the epitopes recognized by these two MAbs were similar, or very close, to the epitope recognized by the H31.F16 MAb. In contrast, significant binding to VLPs was observed using H31.B18, H31.D7, and H31.E17 MAbs (171, 523, and 69 RU, respectively), suggesting that these antibodies recognized epitopes which were different from that recognized by the H31.F16 MAb. The biosensor was then regenerated by three injections of 30 mmol/L HCl. This method was used for all the other competition assays.
Figure 1.
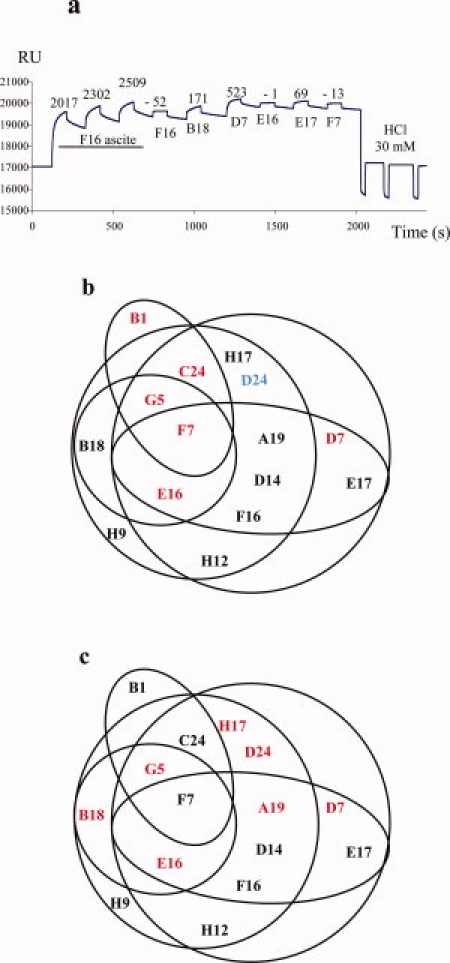
(a) Representative surface plasmon resonance data. Epitope competition was established by saturating coupled HPV31 L1 VLP with H31.F16 MAb (crude ascite fluid), and the MAb saturation was verified by injection of the same MAbs (hybridoma supernatant). MAb binding competition was then established by successively injecting H31.B18, H31.D7, H31.E16, H31.E17, and H31.F7 MAbs (hybridoma supernatants). The biosensor was then regenerated by three injections of 30 mmol/L HCl. Epitope map constructed from the interaction studies between the 15 HPV31 MAbs that recognized conformation epitopes by SPR. (b) Antibodies that neutralized internalization are in black and those that neutralized cell attachment are in red. The H31.D24 antibody was non-neutralizing (in blue). (c) The antibodies that inhibited VLP binding to heparin are in red. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Each of the 15 MAbs investigated competed with at least three others, but none of the MAbs competed with all the other MAbs (data not shown). An epitope map was established using these results [Fig. 1(b)], and epitopes recognized by H31.F7 MAb had a central position in this map. The epitope recognized by this MAb had already been identified on the L1 FG loop.40,41 MAbs competing less with the other MAbs (H31.B18, H31.B1, and H31.H9) were located at the periphery of the epitope map.
Binding of HPV31 MAbs to HPV31/HBc VLPs
The reactivity of MAbs was analyzed using the HPV31 L1/HBc 263/264 mutant and HPV31 L1 wt VLPs to identify whether some of the neutralizing epitopes were located on the FG loop. In addition to HPV31 L1 VLPs produced previously, we constructed a HPV31 L1 mutant by insertion of the hepatitis B core (HBc) motif DPASRE at position 263–264. VLP binding of all the type-specific MAbs was affected by the insertion of the HBc motif at position 263/264. It should be noted that the reactivity of the non-neutralizing H31.D24 MAb, which recognized a linear epitope located at position 271–279 (SVPTDLYIK) was not affected by the insertion. Binding of CamVir-1 MAb that recognized a linear epitope identified outside the FG loop was also not affected by the mutation introduced. The crossneutralizing MAb H31.F7 reacted similarly to both HPV16 and HPV31 wt VLPs, but was affected by insertion of the HBc motif at position 263/264.
Epitope mapping of 5 MAbs using bacterial surface display
Epitope mapping using bacteria for display of peptide libraries provides a new approach for epitope mapping of both monoclonal and polyclonal antibodies.42 One such system, the pFliTrx Bacterial Display system, was used to identify L1 epitopes. The bacterial cell surface display using the pFliTrx vector uses a 12-mer peptide library inserted in a thioredoxin domain to constrain the peptides, allowing the display of conformational epitopes. This thioredoxin domain is itself inserted in the major bacterial flagellar protein of E. coli to be displayed on the surface of bacteria. High-titer MAbs purified from ascites fluid were coated on Nuclon Δ plates for library screening against anti-HPV31 antibodies and in vitro selection rounds were performed on MAbs bound to the plates for the selection of bacteria displaying peptides interacting with the MAbs. For each round, the bacterial library was added to a cell culture dish for positive selection. The unbound bacteria were washed off, and bound bacteria were recovered. After five rounds, single clones were selected and DNA was isolated for sequencing. Sequences were first analyzed using the Dialign2 program. High-scoring matching peptides were selected and then aligned using Dialign2 with the full length HPV31 L1 protein sequence. The HPV31 L1 sequences matching all the selected peptides were retained.
We first used this system with the H31.D24 MAb, which recognized a previously identified linear epitope at position 271–279 (SVPTDLYIK) within the FG loop of HPV31 L1. After bacterial display selection, 24 positive clones were sequenced and analyzed. Six of the 24 peptides selected with H31.D24 MAb matched each other and matched the HPV31 L1 protein (see Fig. 2), the consensus sequence identified being at position 275–279 (DLIYK).
Figure 2.
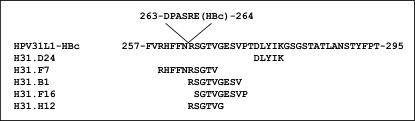
Mapping the HPV31 L1 protein epitopes recognized by a non-neutralizing MAb (H31.D24) and four neutralizing MAbs (H31.B1, H31.F7, H31.F16, and H31.H12) using the Bacterial display method.
Bacterial display was therefore used to investigate the neutralizing conformational epitope recognized by MAb H31.F7, which is crossreactive with HPV16, 18 and 58, and weakly neutralizing for HPV types 16 and 31. Twenty-four positive clones were selected by bacterial display using the H31.F7 MAb. Five of the 24 peptides selected matched each other and matched the HPV31 L1 protein sequence 259–266 (RHFFNRSGTV).
Three type-specific neutralizing MAbs (H31.F16, H31.H12, and H31.B1), which recognized conformational epitopes, were also investigated. Positive clones were selected for each MAb. Five peptides selected with H31.F16 MAb, five with H31.H12 MAb and six peptides selected with H31.B1 MAb matched each other and matched the HPV31 L1 protein sequence SGTVGESVP (265–273) for H31.F16 MAb, RSGTVG sequence (264–269) for H31.H12 MAb and RSGTVGESV sequence (264–272) for H31.B1 MAb.
MAb neutralization of pseudovirions pre- and postattachment
HPV16 and HPV33-specific antibodies have been shown to neutralize before or after attachment to target cells.32,43 The mechanism of viral neutralization by anti-HPV31 MAbs was determined by neutralization assays, the MAbs being added either before addition of the pseudovirions to COS-7 cells [Fig. 3(a)] or 1 h postcell attachment [Fig. 3(b)]. The six MAbs [H31.B1, H31.C24, H31.G5, H31.E16, H31.F7, and H31.D7, Fig. 3(b)] neutralized HPV31 pseudovirions by inhibition of cell attachment because they neutralize before virus attachment but not after pseudovirus binding to the cell [Fig. 3(b)]. The eight other MAbs neutralized HPV31 pseudovirions via a postcell attachment mechanism because they neutralized the pseudovirions before and after cell attachment. It should be noted that the epitope map [Fig. 1(b)] obtained by surface plasmon resonance analysis suggested a cluster of five of these six MAbs that neutralized HPV31 pseudovirions by inhibition of cell attachment.
Figure 3.
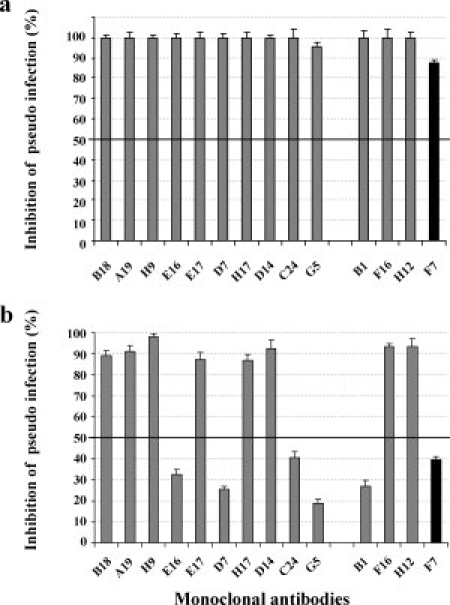
MAb neutralization of pseudovirions pre- and postattachment. (a) Inhibition of HPV31 pseudovirion entry by HPV31 MAbs. HPV31 pseudovirions were preincubated with HPV31 MAbs and then added to cells. (b) Inhibition of HPV31 pseudovirion internalization by HPV31 MAbs. HPV31 pseudovirions were preattached to cells and then HPV31 MAbs were added. The four neutralizing MAbs investigated using the Bacterial display method are grouped at the right of the figure (Gray columns, neutralizing type-specific MAbs; black columns, cross neutralizing MAbs).
VLPs binding to HS and ECM: effects of HPV31 MAbs and L1 C-terminal deletion
The inhibition of VLP binding to heparin by HPV31 MAbs was investigated using an ELISA in which MAbs were added to VLPs before their binding to heparin coated on ELISA plates. The results indicated that the non-neutralizing H31.D24 MAb that recognized a linear epitope strongly inhibited the binding of VLPs to heparin, as only 6 of the 14 MAbs recognized conformational neutralizing epitopes [Fig. 4(a)]. The epitope map obtained by surface plasmon resonance analysis indicated a cluster of some of the MAbs that inhibited VLP binding to heparin [Fig. 1(c)]. There was no clear correlation between MAbs that neutralized the pseudovirions before attachment to the cells and those that inhibited binding to HS. However, three neutralizing antibodies (H31.G5, H31.E16, and H31.D7) demonstrated both abilities (see Fig. 1(b),(c)).
Figure 4.
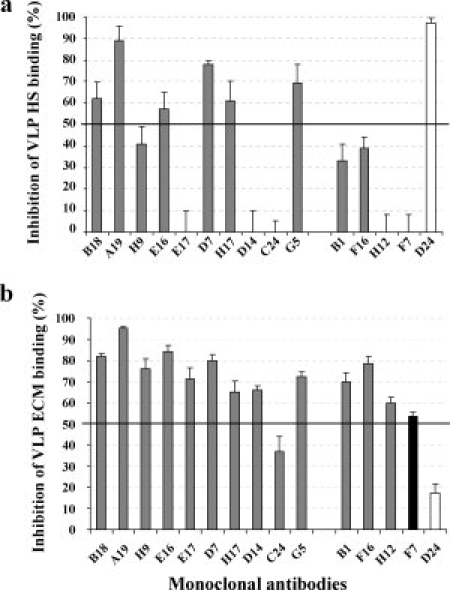
VLP binding to HS and ECM: (a) Inhibition of heparin binding after preincubation of VLPs with MAbs. (b) Inhibition of ECM binding after preincubation of VLPs with MAbs. The five MAbs investigated using the Bacterial display method are grouped at the right of the figure (Gray columns, neutralizing type-specific MAbs; black columns, cross neutralizing MAb; open column, non-neutralizing MAb).
As infection of target cells by HPV is a multistep process involving HSPG, ECM, and an unknown secondary receptor, we investigated the effects of HPV31 MAbs on neutralization of ECM HPV VLP binding.
To investigate further the interaction of VLPs with HPSGs, we used four C-terminal deletion mutants for the analysis of heparin binding to L1 proteins. HPV16 Δ9 and HPV16 Δ31 mutants with C-terminal deletions of 9 and 31 amino acids, respectively, had already been produced,44 and HPV31Δ9 and HPV31Δ31 L1 mutants were similarly produced for the purposes of this study. These HPV31 deletion mutants self-assembled into VLPs (see Fig. 5) as previously observed for the corresponding HPV16 deletion mutants44 and these four C-terminal deleted VLPs bound to heparin in a similar manner to wt VLPs. However, binding to heparin is lost when these VLPs were denatured, confirming that heparin binds to a conformational motif on VLPs, and that this motif is not present in the C-terminal part of the L1 protein (see Fig. 5).
Figure 5.
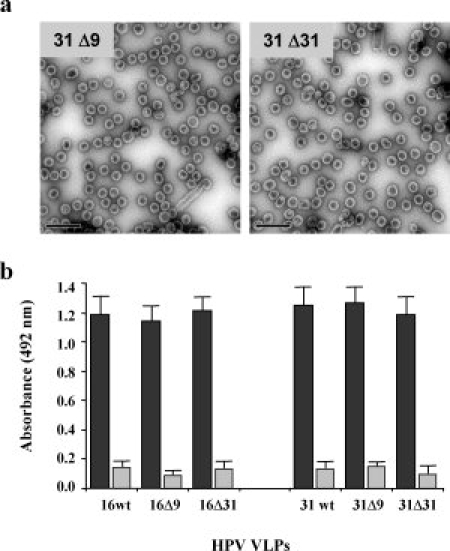
(a) HPV31Δ9 and HPV31Δ31 VLPs observed by transmission electron microscopy (Bars represent 200 nm). (b) Heparin binding of native VLPs (black columns) and denatured VLPs (grey columns) for type 16 and type 31 with or without C-terminal deletions (Δ9 and Δ31).
Inhibition of VLP binding to ECM proteins was investigated by ELISA using Matrigel® as a surrogate for ECM. The results [Fig. 4(b)] indicated inhibition of VLP binding to ECM proteins (>50%) by all the neutralizing antibodies, with the exception of H31.C24 (37% inhibition). The non-neutralizing antibody H31.D24 did not inhibit binding of VLPs to ECM proteins.
Discussion
According to the three-dimensional HPV capsomer model of Chen et al.,45 all conformation-dependent type-specific epitopes identified to date16,18,41,46–48 have been found to reside on the VLP surface within hypervariable loops where the amino-acid sequence is highly divergent between HPV types, with the exception of the epitope recognized by H16.U4 MAb. This epitope was identified between residues 427–445,46 that in the three-dimensional HPV16 capsomer model of Moddis et al.49 are in a loop at the crown of the pentamer within the arm extending into the adjacent pentamer between two L1 subunits. L1 protein binding sites of neutralizing MAbs generated against HPV11 include residues within the DE loop,50,51 and those against HPV6 and HPV33 within BC and EF loops,52 and BC, DE, and FG loops,46 respectively. In addition, L1 residues within the FG loop of HPV16 L116,25,53 and the EF loop of HPV31 L146 have been reported to be involved in the binding of neutralizing antibodies. In addition to type-specific MAbs, crossreacting neutralizing MAbs have rarely been identified21,41,53,54 and had low-neutralization titers. The crossneutralization observed with MAbs was in agreement with the crossneutralization observed with polyclonal mouse antibodies53,55 and with the crossprotection observed in clinical trials.4,56
The results obtained using an HPV31 FG loop insertion mutant confirmed the involvement of the FG loop in the epitopes recognized by all HPV31 neutralizing antibodies investigated. Surface plasmon resonance was used to determine whether different epitope zones exist on the surface of capsomeres, a method successfully used to establish a competition map for HIV gp120 and adenovirus hexon epitopes.57,58 The competition mapping of the 15 anti-HPV31 L1 MAbs was performed by SPR and revealed a high degree of competition between the entire set of MAbs. To rule out the possibility that competition between MAbs was not due to sterical hindrance but to overlapping epitope competition, binding of H31.A6 MAb which recognize an epitope located on the N-terminal part of the EF loop in the vicinity of the FG loop46,59 was investigated. H31.A6 MAb did not compete with the three HPV31 MAbs (H31.D24, H31.F7, and H31.H12) directed against the FG loop that we investigated (data not shown).
The results obtained suggest the existence of only one major epitope region on HPV31 VLPs. This is in agreement with the existence of a major neutralizing epitope on the HPV16 capsid, because H16.V5 MAb has been shown to inhibit the VLP binding of >75% of serum antibodies from naturally infected women,60 and most of the anti-HPV VLP antibodies are directed against epitopes located on the BC, EF, FG, and HI hypervariable loops intertwined across the outermost surface of the pentamers.45
To investigate further the exact position of the epitopes, fine mapping of L1 sequences binding to five MAbs was undertaken using the FliTrx bacterial surface display method, a unique system that displays conformation-constrained random peptides on the bacterial surface. Epitope determination was first validated with the H31.D24 MAb that recognized a linear epitope at position 275–279 (DLYIK), similar to the 271–279 sequence (SVPTDLYIK) identified previously using synthetic peptides.41 This DLYIK motif is also part of the conserved linear epitopes described for other crossreacting MAbs.61–63
The bacterial cell surface display method was then used to determine the conformational epitopes recognized by H31.F16, H31.H12, and H31.B1 neutralizing MAbs and the conformational epitope recognized by the crossreacting and partially crossneutralizing H31.F7 MAb. The type-specific epitopes were identified at positions 264–272 (RSGTVGESV) for H31.B1, 264–269 (RSGTVG) for H31.H12, and 265–273 (SGTVGESVP) for H31.F16 on the FG loop. This result is in agreement with our previous studies40,41 and with ELISA studies using the HPV31 insertion mutant, and confirms that multiple epitopes exist on the FG hypervariable loop, but that not all antibodies interacting with the HPV31 FG loop are neutralizing antibodies.
The epitope recognized by H31.F7 MAb, a crossreacting and partially crossneutralizing MAb, was identified on the N-terminal part of the FG loop of the HPV31 L1 protein at position 259–268 (RHFFNRSGTV) [Fig. 6(a)], and is mostly inaccessible from the surface of the capsomer according to the HPV31 model. This is in agreement with the fact that the early region of the FG loop evidenced identical structures between HPV types and with the hypothesis of Bishop et al.64 on the structural analysis of L1 proteins suggesting that this region of the FG loop must be able to induce crossreactivity between types but be poorly antigenic and/or inaccessible.
Figure 6.
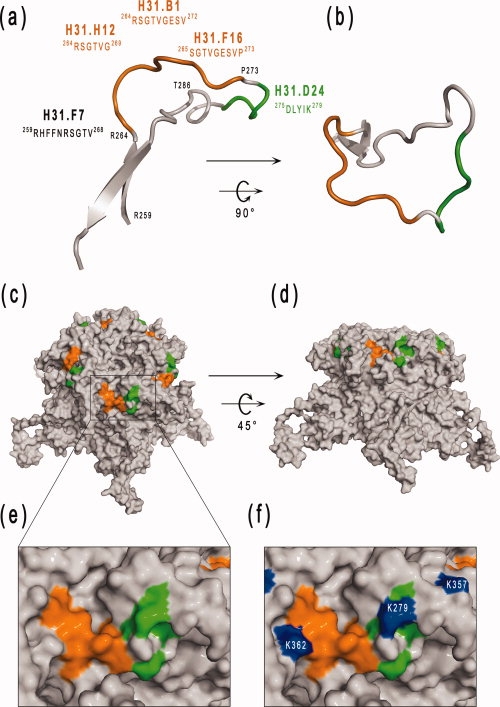
Localization of the epitopes recognized by five monoclonal antibodies on the FG loop of HPV-31. The epitope recognized by H31.D24 is a linear epitope and is colored green. Conformational epitopes recognized by the three neutralizing MAbs (H31.B1, H31.F16, and H31.H12) are colored orange. H31.F7 MAb is partially crossneutralizing and crossreacting. The 3D capsomer structure was reconstructed with SwissPDBViewer using the HPV31 L1 model and the information for noncrystallographic symmetries of the HPV16 VLP [PDB code: 1DZL, (45)]. (a) and (b) position of the epitopes recognized on the FG loop of the HPV31 L1 protein by four MAbs. (b) FG loop in the same orientation as that found in the capsomere shown in (c). (c, d) Solvent-accessible surface of the L1 capsomere of HPV31 showing the location of FG loop epitopes (colored orange and green) in the context of the whole capsomere shown in lateral view (c) or rotated 45° around the x-axis (d). (e, f) Close-up view of the epitope zone on the capsomer corresponding to the boxed area in (c). The Lys residues involved in heparin sulfate binding which overlap the epitope region are colored blue (f). The figure was prepared using the PYMOL program (www.pymol.org). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
For HPV16 and HPV33, it has been shown that various L1 protein surface exposed loops contribute to the induction of neutralizing antibodies to epitopes identified in only one loop and epitopes for which several loops contribute to the binding site,15,46,47 and it has been suggested that noncontiguous regions of L1 could contribute to neutralizing epitopes recognized by MAbs. Our findings were in favor of the HPV31 neutralizing antibodies being mainly directed against the FG loop and the FG loop only contributing to the binding to neutralizing antibodies. The fact that L1 hypervariable loops are in close proximity to each other did not rule out the possibility that mutations or insertions in other loops affect the FG conformational epitopes as observed by Roth et al.47
The interaction experiments between heparin and VLPs indicated the existence of at least two sequences interacting with heparin, one being a linear motif on the C-terminus part of the L1 protein, in agreement with results of Joyce et al.28 and Bousarghin et al.,34 and the second being a conformational binding site, in agreement with recent findings reported by Knappe et al.35 indicating that HS binds to a cluster of three lysine residues on the surface of the VLP. As the C-terminal part of the L1 protein is located inside the capsid,45 this highly positively charged linear sequence was expected to have no role in the binding to HS on the cell surface. We observed that the crossreacting H31.D24 MAb which recognized the DLYIK sequence (275–279) dramatically reduced the ability of HPV31 to bind to heparin, thus confirming the role of Lysine 279 (278 in HPV16 L1) in HS binding. However, this antibody had no neutralizing effect on HPV31 pseudovirions,41,65 and three other neutralizing antibodies that recognized conformational epitopes known to bind to a sequence situated in the N-terminal part of the FG loop had no or limited effect on binding to heparin. H16.V5 and H16.E70 MAbs have been shown not to block interactions with the cell surface. However, these neutralizing antibodies inhibit virus binding to ECM produced from HaCaT cells, and neutralize pseudovirions via a postcell attachment mechanism.43 In agreement with this, we observed that the three type-specific neutralizing antibodies did not compete with HS for binding to VLPs and that only one of them (H31.B1) neutralized by inhibition of the binding of the VLPs to cells. However, all these antibodies compete for binding to the ECM, in contrast to the non-neutralizing H31.D24 MAb. These results suggested that these antibodies recognized an epitope in the vicinity of the conformational basic cluster of lysine involved in HPV16 VLP binding to HS35 [Fig. 6(d)]. It should be noted that the most important lysine of the cluster was present within the epitope recognized by the H31.D24 MAb.
In addition, investigation of pre- and postattachment neutralization of HPV31 pseudovirions by MAbs revealed that six of the HPV31 neutralizing antibodies acted by preventing the cell surface binding of the viral particles, and the other eight neutralizing MAbs interfered with pseudovirions by preventing their internalization. Our findings indicate that, for the entire set of MAbs, there is no correlation between neutralization and the ability of MAbs to bind HS. Lopez et al.66 was recently reported that anti-HPV antibodies detected in natural infection inhibit HPV16 binding to HS, and they observed that those with the highest neutralizing titers were those that inhibited binding to HS.
In conclusion, our findings showed that HPV31-neutralizing MAbs recognize conformational epitopes located on the FG loop of HPV31 L1 protein and that they act either by blocking attachment to target cells or by neutralization of postcell attachment. The precise determination of three type-specific neutralizing epitopes by bacterial surface display confirmed their location on the central part of the FG loop and identified a crossreacting and partially crossneutralizing conformational epitope on the relatively conserved N-terminal part of the FG loop. The solvent-exposed amino acid residues of the FG loop were distributed on both sides of a groove formed along an axis defined by Pro 273 to Leu 287 [Fig. 6(a,b)]. Our findings showed that the right part of the groove contained a linear non-neutralizing epitope recognized by H31.D24 MAb. This is also a hot spot for heparan sulfate binding, thus explaining why H31.D24 and HS compete for binding to this region. The conformational, neutralizing epitopes identified were all located on the left side of the FG loop groove (see Fig. 6). The fact that antibodies directed against these epitopes did not strongly compete with HS may be explained by differences in the MAb binding modes, as previously described for MAbs directed against BPV L1 protein.67 However, some other HPV31 neutralizing MAbs did compete with HS binding, suggesting that either these MAbs have a different tilt when they bind to their respective epitopes, thus interfering with HS binding to a greater or lesser extent, or that they are directed against amino acid residues from both sides of the groove.
H31.D24 MAb bound to the tips of VLPs [Fig. 6(c,d)] and this supports the hypothesis that non-neutralizing epitopes are also present on the surface of HPV particles. The results obtained indicated that the FG hypervariable loop of HPV31 is dense in neutralizing epitopes and suggested that HPV31 MAbs bind to overlapping but distinct epitopes on the central part of the FG loop, in agreement with the exposure of the FG loop on the surface of HPV VLPs, and thus confirming that neutralizing antibodies are mainly located on the tip and crown of the capsomere. The results also support and confirm that the blocking of virus attachment to the ECM is an important neutralization mechanism but not the blocking of virus attachment to HS. In addition, we identified a crossreacting and partially crossneutralizing conformational epitope on the relatively conserved N-terminal part of the FG loop.
Materials and Methods
HPV31 monoclonal antibodies
The HPV31 MAbs used in this study were as previously produced and characterized.41,65 H31.D24 MAb recognize a common linear epitope that has been identified within the FG loop (amino acids 271–279) and H31.F7 MAb recognize a conformational crossneutralizing epitope that has been identified within the N-terminal part of the FG loop.41 Thirteen other MAbs recognize specific conformational neutralizing epitopes. In addition, CamVir-1 monoclonal antibody (CV), which recognizes a common linear epitope on the DE loop,40 was used as control. MAbs investigated using the bacterial cell surface display method were purified from crude ascites fluid by salting out with ammonium sulfate (33% final), then dialyzed against phosphate-buffered saline (PBS) followed by affinity chromatography on Protein AG/Sepharose (Pierce; Immunopure IgG purification kit).
Pseudovirus neutralization
The neutralizing ability of each monoclonal antibody was determined previously.41,65 In addition, we investigated whether the neutralization took place before or after pseudovirion cell surface binding. Tests were performed with HPV31 pseudovirions produced in 293FT cells and neutralization assays were performed using Cos-7 cells cultured in complete Dulbecco's modified Eagle's medium (Invitrogen, DMEM supplemented with 10% FCS, 100 IU/mL penicillin and 100 μg/mL streptomycin) seeded in 96-well plates and incubated for 24 h at 37°C. Assays measuring neutralization before pseudovirion cell surface binding were performed by adding the HPV31 pseudovirions previously preincubated with MAbs to the cells (100 μL) for 1 h at 37°C. Assays measuring neutralization after pseudovirion cell surface binding were performed by adding HPV31 pseudovirions to Cos-7 cells for 1 h at 37°C. After three washings to remove unbound pseudovirions, MAbs were added in 100 μL DMEM. For each assay, the supernatant was removed after 3 h at 37°C, and 200 μL complete DMEM was added. After a further 48 h incubation at 37°C, infectivity was scored by measuring the luciferase expressed by transfected cells using the Firefly Luciferase assay kit (Interchim, Montluçon, France) and luciferase expression was quantified using a Multiskan microplate luminometer (Thermo-Fisher Scientific, Courtaboeuf, France).
MAbs were considered to be neutralizing if luciferase activity was reduced by >80%. Inhibition of pseudovirus binding to the cell was scored if MAbs neutralized the pseudoinfection only when added before pseudovirion binding to the cells. If antibodies neutralized pseudovirions before and after their addition to Cos-7 cells, MAbs were considered to neutralize pseudovirions via a postcell attachment mechanism.
Generation of recombinant L1 proteins and purification of VLPs
The Bac-to-Bac system (Invitrogen, Fisher-Scientific, Illkirch, France) was used for expression of the HPV L1 proteins in Spodoptera frugiperla (Sf21) cells. Baculoviruses encoding the L1 gene of HPV16, HPV31, HPV16 ΔC9, and HPV16 ΔC31 (with a 9 or 31 amino-acid C-terminal deletion, respectively) were generated previously.44,68 HPV31 ΔC9 and HPV 31 ΔC31 truncated genes were amplified by PCR from a full-length HPV31 L1 codon-optimized gene65 using forward (GGATCCCACCATGAGCCTGTGGAGACCCAGC) and reverse primers for ΔC9 (GGAAGCTTATGTGGTGGTGCTGGCGCTGGGGGC) and for ΔC31 (GCAAGCTTAGGCCTGCAGCAGGAACTTTCTGCCC), respectively. PCR products were cloned into pCR TOPO 2.1 by TA cloning. Positive clones were sequenced to verify the absence of unwanted mutations. HPV L1 genes were then cloned into pFastBac1 plasmid previously digested by BamHI and HindIII. Recombinant baculoviruses encoding the different L1 deleted genes were generated using the Bac-to-Bac system (Invitrogen) according to the manufacturer's recommendations.
Sf21 cells maintained in Grace's insect medium (Invitrogen, Cergy Pontoise, France) supplemented with 10% fetal calf serum (FCS, Invitrogen) were infected with the respective recombinant baculoviruses and incubated at 27°C. Three days post infection, cells were harvested and VLPs were purified as previously described.29,40 Briefly, cells were resuspended in PBS containing Nonidet P40 (0.5%), pepstatin A, and leupeptin (1 μg/mL each, Sigma Aldrich, Saint Quentin Fallavier, France), and allowed to stand for 30 min at 4°C. Nuclear lysates were then centrifuged and pellets were resuspended in ice cold PBS containing pepstatin A and leupeptin and then sonicated. Samples were then loaded on a CsCl gradient and centrifuged to equilibrium (22 h, 27,000 rpm in a SW28 rotor, 4°C). CsCl gradient fractions were investigated for density by refractometry and for the presence of L1 protein by electrophoresis in 10% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) and Coomassie blue staining. Positive fractions were pooled, diluted in PBS and pelleted in a Beckman SW 28 rotor (3 h, 28,000 rpm, 4°C). After centrifugation, VLPs were resuspended in 0.15 mol/L NaCl and sonicated by one 5 s burst at 60% maximum power. Total protein content was determined using the MicroBCA kit (Pierce, Ozyme, France).
HPV31 L1 HBc 263/264 mutant VLPs were produced in 293 FT cells. DNA-encoding chimeric L1 protein was obtained by mutagenesis of a codon-optimized HPV31 L1 gene using a two-step PCR protocol. Overlapping PCRs were performed to obtain the DPASRE sequence of the HBc protein at position 263/264 of HPV31 L1 protein. In the first step, one fragment was generated using the optimized HPV31 L1 DNA sequence as template and 5′L1-NheI (CCGCTAGCCACCATGAGCCTGTGGAGACCC) and 3′L1-DPASRE (CTCTCTGCTGGCGGGGTCGTTGAAGAAGTGCCGCACGAA) as primers. Another fragment was amplified using the optimized HPV31 L1 DNA sequence as template and 3′L1-EcoRI (CGGAATTCTATCACTTCTTGGTTTTCTTCC) and 5′L1-DPASRE (GACCCCGCCAGCAGAGAGAGAAGCGGCACCGTGGGCGAG) as primers. These two overlapping fragments were used in the second PCR step as DNA templates using 5′L1-NheI and 3′L1-EcoRI primers. The resulting DNA sequence had an HBc78-83-encoding sequence between L1 bases 263/264 and an NheI restriction site in 5′ and an EcoRI restriction site in 3′. For protein expression, the HPV31 L1 HBc 263/264 gene was cloned into the pIRES mammalian expression vector (BDbiosciences, Clontech). This DNA plasmid (HPV31 L1 HBc 263/264-pIRES) was prepared by classical alkaline lysis and phenol/chloroform extraction and used to transfect 293 FT cells with Fugene6 (Roche Diagnostic, Meylan, France) according to the manufacturer's instructions. The cells were transfected with a total of 0.5 μg of DNA and 1 μL of Fugene6 per cm2 of culture area. 293FT cells were harvested 44 h after transfection and VLPs were purified as mentioned earlier.
The self-assembly of the different HPV-L1 proteins expressed in VLPs was investigated by electron microscopy. For this purpose, VLP preparations were applied to carbon-coated grids, negatively stained with 1.5% uranyl acetate and observed at 50,000× nominal magnification with a JEOL 1010 electron microscope. All the electron micrographs were taken at 30,000× or 50,000×.
Investigation of VLP binding competition of HPV31 MAbs by surface plasmon resonance
Analyses were performed with a Biacore 1000 (Biacore AB, Uppsala, Sweden) equipped with CM3 (carboxymethylated dextran) sensor chips. CM3 sensorchips were treated with 35 μL of 0.05 mol/L 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride and 0.2 mol/L N-hydroxysuccinimide at a flow rate of 5 μL/min and then the HPV31 L1 VLPs (32 μg/mL in 35 μL of PBS buffer) were covalently coupled at a flow rate of 5 μL/min. The residual carboxyl groups were subsequently blocked by 50 μL of 1 mol/L ethanolamine-HCl (pH 8.5) at a flow rate of 10 μL/min. Unbound VLPs were eliminated by injection of 5 μL of HCl-glycine (10 mmol/L, pH 2.2) at a flow rate of 10 μL/min. A flow rate of 20 μL/min was used for all interaction analyses conducted at room temperature. Antibody saturation of the bound HPV31 L1 VLPs was obtained for each MAb by three injections of 30 μL of ascites fluid diluted 1:10 in PBS. The MAb saturation was verified by injection of 30 μL of hybridoma supernatant of the same MAb diluted 1:4 in PBS. Competition was then established by successively injecting five different hybridoma supernatants diluted 1:4 in PBS (30 μL each). The biosensor was then regenerated by three injections of 10 μL of 30 mmol/L HCl, and another cycle of saturation-competition was performed on the same VLPs coupled flow-cell. Several other regeneration buffers (including 2 mol/L NaCl, 10 mmol/L NaOH, and 20, 25, 30, and 50 mmol/L HCl) were investigated. The selected buffer (30 mmol/L HCl) was shown to remove 100% of the bound antibodies, not to remove VLPs from the sensorchip, and not to affect VLPs conformation, because antibodies directed at the conformational epitopes still bound to VLPs as effectively as before the regeneration treatment.
Epitope mapping using bacterial cell surface display
The bacterial cell surface display method using the pFliTrx vector uses a 12-mer peptide library inserted in a thioredoxin domain (TrxA) to constrain the peptides. This thioredoxin domain is itself inserted into the major bacterial flagellar protein (FliC) to be displayed on the surface of E. coli.69,70
The pFliTrx Random Peptide Display Library (Invitrogen, Cergy Pontoise, France) was obtained by inoculation of 1 mL of the peptide library stock solution into 50 mL of IMC Medium (6 g/L Na2HPO4, 3 g/L KH2PO4, 0.5 g/L NaCl, 1 g/L NH4Cl, 0.2% casamino acid, 0.5% glucose, 1 mmol/L MgCl2) containing 100 μg/mL ampicillin and then shaken (225–250 rpm) overnight at 25°C. Peptide expression was induced by adding 100 μg/mL of tryptophan to 1010 bacteria from the overnight culture in 50 mL of IMC Medium containing 100 μg/mL of ampicillin, and then shaken at 25°C for 6 h.
For library screening against anti-HPV31 antibodies, 20 μg of MAb in 1 mL sterile water was coated on a 60-mm plate (Nunclon Δ, Nunc, ATGC, Marne-la-Vallée, France) for 1 h with gentle agitation at 50 rpm on an orbital shaker. After washing with 10 mL of sterile water, 10 mL of Blocking Solution (IMC medium containing 100 μg/mL ampicillin, 1% low-fat dry milk, 150 mmol/L NaCl, 1% α-methyl mannoside) was added and incubated for 1 h under agitation (50 rpm). After decanting the blocking solution, 10 mL of the bacteria cell culture were added with 0.1 g dried milk, 300 μL of 5 mol/L NaCl, 500 μL of 20% α-methyl mannoside (final concentration: 1% nonfat, dried milk, 150 mmol/L NaCl, 1% α-methyl mannoside). After gentle agitation at 50 rpm for 1 min and incubation at room temperature for 1 h, the plates were washed five times (50 rpm for 5 min) with 10 mL of the IMC medium containing 100 μg/mL ampicillin and 1% α-methyl mannoside. The bound bacterial cells were then eluted into the residual volume of washing solution by vortexing the plates for 30 s. They were transferred to a 50 mL culture flask and grown under shaking (225–250 rpm) at 25°C overnight. This panning cycle was repeated four more times and the overnight culture from the fifth panning was streaked onto RMG plates (6 g/L Na2HPO4, 3 g/L KH2PO4, 0.5 g/L NaCl, 1 g/L NH4Cl, 2% Casamino acid, 0.5% glucose, 1 mmol/L MgCl2, 1.5% agar) containing 100 μg/mL ampicillin and incubated overnight at 30°C.
Twenty-four to 30 colonies from the RMG/ampicillin plate were each inoculated into 2 mL RM medium (6 g/L Na2HPO4, 3 g/L KH2PO4, 0,5 g/L NaCl, 1 g/L NH4Cl, 2% Casamino acid, 1% glycerol, 1 mmol/L MgCl2) containing 100 μg/mL ampicillin. After incubation at 30°C overnight with shaking, DNA was extracted by the classical alkaline lysis phenol/chloroform DNA minipreparation. Nucleotide sequence analysis was performed using the FliTrx forward sequencing primer. Sequences were run on an ABI PRISM 3100 Avant DNA sequencer (PerkinElmer, Courtaboeuf, France). Sequences were analyzed using Dialign2.71
Detection of MAb reactivity against wild-type and mutant VLPs by ELISA
Microplate wells (Maxisorp, Nunc) were coated with VLPs. After incubation at 4°C overnight and two washes with PBS-Tween 20 (0.1%), wells were saturated with PBS supplemented with 1% FCS for 1 h at 37°C. Duplicate wells (two tests and one control) were incubated with MAbs diluted in PBS 5X—Tween (1%)—FCS (10%) for 1 h at 37°C. After four washes, peroxidase-conjugated goat anti-mouse Ig Fc (Sigma Aldrich) diluted 1:1,000 in PBS—Tween (1%)—FCS (10%) was added to the wells and incubated for 1 h at 37°C. Then after four washes, 0.4 mg/mL O-phenylene-diamine and 0.03% hydrogen peroxide in 25 mmol/L sodium citrate and 50 mmol/L Na2HPO4 were added. After 30 min, the reaction was stopped with H2SO4 4N and absorbance was read at 490 nm. For data analysis, optical density (OD) values obtained in the absence of the first antibodies were subtracted from the OD values of test samples. The data presented are the means of three to four determinations.
Heparin- and ECM-based enzyme-linked immunosorbent assays
The interaction between VLPs and heparin was tested using an assay derived from the heparin-binding assays described by Giroglou et al.26 for HPV33 VLPs. Microtiter plates (Maxisorp, Nunc) were coated overnight at 4°C with 200 ng per well of heparin-BSA (Sigma) or Matrigel® (BD Biosciences, Le pont de Claix, France) After four washes with PBS containing 0.1% Tween 20, nonspecific binding sites were blocked by incubation for 1 h at 37°C with PBS plus 1% FCS. After washing, 200 ng/well of VLPs diluted in PBS were added. Following incubation at 37°C for 60 min and four washes, anti-HPV31 VLP MAbs diluted 1:1000 in PBS, 0.1% Tween 20, and 10% FCS were added and incubated at 37°C for 60 min. After 1 h incubation at 37°C and four washes, bound antibodies were detected using mouse anti-IgG antibodies covalently linked to horseradish peroxidase. After 1 h incubation at 37°C and four washes, 100 μL of substrate solution containing O-phenylene diamine and H2O2 were added. The reaction was stopped after 30 min by addition of 100 μL 2 mol/L H2SO4 and absorbance was read at 492 nm with an automated plate reader. The absorbance of control wells without VLPs was subtracted from values for test wells. Simultaneously, a second plate was coated with heparin-BSA or Matrigel®. After incubation and washing, 200 ng/well of VLPs preincubated for 1 h at 37°C with MAbs diluted 1:1000 in PBS, 0.1% Tween 20, and 10% FCS were added. Bound antibodies were detected with mouse anti-IgG antibodies covalently linked to horseradish peroxidase as described earlier. Inhibition of VLP binding to heparin or Matrigel was calculated as the reduction in OD observed between adding the MAbs before the interaction of VLPs with heparin and after. The results are expressed as the percentage of reduction in OD.
VLP binding to HS and ECM assays
Microtiter plates (Maxisorp, Nunc) were coated with heparin-BSA as described earlier. After a blocking step, VLPs diluted in PBS were added. Following incubation at 37°C for 60 min and washing, CanVir1 MAbs diluted 1:5000 in PBS, 0.1% Tween 20 and 10% FCS were added and incubated at 37°C for 60 min. Bound antibodies were detected after four washes using mouse anti-IgG antibodies covalently linked to horseradish peroxidase and then the tests were performed as mentioned earlier in the test for the interaction between heparin and MAbs. The results are expressed as OD value.
Visualization of HPV31 epitopes by homology modeling of HPV31 L1 protein
The sequence for HPV31 L1 protein (UniprotKB ID, P17388) was submitted as input to the Swiss-Model modeling tool (http://swissmodel.expasy.org). A template search based on sequence similarity identified HPV16 L1 (PDB code 1DZL and 2R5H), HPV-35 L1 (2R5J), HPV-11 L1 (2R5K), and HPV-18 L1 (2R5I) as possible 3D templates. Because HPV31 L1 is more similar to HPV16 and HPV35 L1 sequences, we selected the corresponding 3D templates (1DZL, 2R5H and 2R5J) to model the L1 structure of HPV31. The model of HPV31 L1 was evaluated using ANOELA (http://swissmodel.expasy.org/anolea/) and found to be correct for further structural analysis of epitope locations. The L1 pentamer of HPV31 was reconstructed with SwissPDBViewer using the HPV31 L1 model and the information for noncrystallographic symmetries (transformation matrices) of the HPV16 VLP (1DZL).45 Atomic coordinates of the pentamer for the HPV31 L1 VLP were saved in PDB format and displayed using the PYMOL program, a molecular graphic visualization tool for macromolecular structures (http://www.pymol.org).
Acknowledgments
The authors thank Neil D. Christensen (Penn State Hershey Medical Center, PA) for providing H16.V5 and H31.A6 MAbs, and Pierre-Yves Sizaret (François Rabelais University, Tours, France) for electron microscopy.
References
- 1.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Munoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 2.Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, Snijders PJ, Meijer CJ. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 3.Ault KA, Future II Study Group Effect of prophylactic human papillomavirus L1 virus-like-particle vaccine on risk of cervical intraepithelial neoplasia grade 2, grade 3, and adenocarcinoma in situ: a combined analysis of four randomised clinical trials. Lancet. 2007;369:1861–1868. doi: 10.1016/S0140-6736(07)60852-6. [DOI] [PubMed] [Google Scholar]
- 4.Paavonen J, Jenkins D, Bosch FX, Naud P, Salmerón J, Wheeler CM, Chow SN, Apter DL, Kitchener HC, Castellsague X, de Carvalho NS, Skinner SR, Harper DM, Hedrick JA, Jaisamrarn U, Limson GA, Dionne M, Quint W, Spiessens B, Peeters P, Struyf F, Wieting SL, Lehtinen MO, Dubin G, HPV PATRICIA Study Group Efficacy of a prophylactic adjuvanted bivalent L1 virus-like-particle vaccine against infection with human papillomavirus types 16 and 18 in young women: an interim analysis of a phase III double-blind, randomised controlled trial. Lancet. 2007;369:2161–2170. doi: 10.1016/S0140-6736(07)60946-5. [DOI] [PubMed] [Google Scholar]
- 5.He RT, Innis BL, Nisalak A, Usawattanakul W, Wang S, Kalayanarooj S, Anderson R. Antibodies that block virus attachment to Vero cells are a major component of the human neutralizing antibody response against dengue virus type 2. J Med Virol. 1995;45:451–461. doi: 10.1002/jmv.1890450417. [DOI] [PubMed] [Google Scholar]
- 6.Klasse PJ, Sattentau QJ. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J Gen Virol. 2002;83:2091–2108. doi: 10.1099/0022-1317-83-9-2091. [DOI] [PubMed] [Google Scholar]
- 7.Law M, Hangartner L. Antibodies against viruses: passive and active immunization. Curr Opin Immunol. 2008;20:486–492. doi: 10.1016/j.coi.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomont N, Hocini H, Gody JC, Bouhlal H, Becquart P, Krief-Bouillet C, Kazatchkine M, Bélec L. Neutralizing monoclonal antibodies to human immunodeficiency virus type 1 do not inhibit viral transcytosis through mucosal epithelial cells. Virology. 2008;370:246–254. doi: 10.1016/j.virol.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Virgin HW, 4th, Mann MA, Tyler KL. Protective antibodies inhibit reovirus internalization and uncoating by intracellular proteases. J Virol. 1994;68:6719–6729. doi: 10.1128/jvi.68.10.6719-6729.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhuge W, Jia F, Mackay G, Kumar A, Narayan O. Antibodies that neutralize SIV(mac)251 in T lymphocytes cause interruption of the viral life cycle in macrophages by preventing nuclear import of viral DNA. Virology. 2001;287:436–445. doi: 10.1006/viro.2001.1053. [DOI] [PubMed] [Google Scholar]
- 11.Varghese R, Mikyas Y, Stewart PL, Ralston R. Postentry neutralization of adenovirus type 5 by an antihexon antibody. J Virol. 2004;78:12320–12332. doi: 10.1128/JVI.78.22.12320-12332.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breitburd F, Kirnbauer R, Hubbert NL, Nonnenmacher B, Trin-Dinh-Desmarquet C, Orth G, Schiller JT, Lowy DR. Immunization with viruslike particles from cottontail rabbit papillomavirus (CRPV) can protect against experimental CRPV infection. J Virol. 1995;69:3959–3963. doi: 10.1128/jvi.69.6.3959-3963.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzich JA, Ghim SJ, Palmer-Hill FJ, White WI, Tamura JK, Bell JA, Newsome JA, Jenson AB, Schlegel R. Systemic immunization with papillomavirus L1 protein completely prevents the development of viral mucosal papillomas. Proc Natl Acad Sci USA. 1995;92:11553–11557. doi: 10.1073/pnas.92.25.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen ND, Reed CA, Cladel NM, Han R, Kreider JW. Immunization with virus-like particles induces long-term protection of rabbits against challenge with cottontail rabbit papillomavirus. J Virol. 1996;70:960–965. doi: 10.1128/jvi.70.2.960-965.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christensen ND, Cladel NM, Reed CA, Budgeon LR, Embers ME, Skulsky DM, McClements WL, Ludmerer SW, Jansen KU. Hybrid papillomavirus L1 molecules assemble into virus-like particles that reconstitute conformational epitopes and induce neutralizing antibodies to distinct HPV types. Virology. 2001;291:324–334. doi: 10.1006/viro.2001.1220. [DOI] [PubMed] [Google Scholar]
- 16.Sadeyen JR, Tourne S, Shkreli M, Sizaret PY, Coursaget P. Insertion of a foreign sequence on capsid surface loops of human papillomavirus type 16 virus-like particles reduced their immunogenicity and delineates a conformational neutralizing epitope. Virology. 2003;309:32–40. doi: 10.1016/s0042-6822(02)00134-4. [DOI] [PubMed] [Google Scholar]
- 17.Orozco JJ, Carter JJ, Koutsky LA, Galloway DA. Humoral immune response recognizes a complex set of epitopes on human papillomavirus type 6 L1 capsomers. J Virol. 2005;79:9503–9514. doi: 10.1128/JVI.79.15.9503-9514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yaegashi N, Jenison SA, Valentine JM, Dunn M, Taichman LB, Baker DA, Galloway DA. Characterization of murine polyclonal antisera and monoclonal antibodies generated against intact and denatured human papillomavirus type 1 virions. J Virol. 1991;65:1578–1583. doi: 10.1128/jvi.65.3.1578-1583.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Volpers C, Sapp M, Snijders PJ, Walboomers JM, Streeck RE. Conformational and linear epitopes on virus-like particles of human papillomavirus type 33 identified by monoclonal antibodies to the minor capsid protein L2. J Gen Virol. 1995;76:2661–2667. doi: 10.1099/0022-1317-76-11-2661. [DOI] [PubMed] [Google Scholar]
- 20.Heino P, Skyldberg B, Lehtinen M, Rantala I, Hagmar B, Kreider JW, Kirnbauer R, Dillner J. Human papillomavirus type 16 capsids expose multiple type-restricted and type-common antigenic epitopes. J Gen Virol. 1995;76:1141–1153. doi: 10.1099/0022-1317-76-5-1141. [DOI] [PubMed] [Google Scholar]
- 21.Christensen ND, Reed CA, Cladel NM, Hall K, Leiserowitz GS. Monoclonal antibodies to HPV-6 L1 virus-like particles identify conformational and linear neutralizing epitopes on HPV-11 in addition to type-specific epitopes on HPV-6. Virology. 1996;224:477–486. doi: 10.1006/viro.1996.0554. [DOI] [PubMed] [Google Scholar]
- 22.Christensen ND, Kirnbauer R, Schiller JT, Ghim SJ, Schlegel R, Jenson AB, Kreider JW. Human papillomavirus types 6 and 11 have antigenically distinct strongly immunogenic conformationally dependent neutralizing epitopes. Virology. 1994;205:329–335. doi: 10.1006/viro.1994.1649. [DOI] [PubMed] [Google Scholar]
- 23.Rose RC, Reichman RC, Bonnez W. Human papillomavirus (HPV) type 11 recombinant virus-like particles induce the formation of neutralizing antibodies and detect HPV-specific antibodies in human sera. J Gen Virol. 1994;75:2075–2079. doi: 10.1099/0022-1317-75-8-2075. [DOI] [PubMed] [Google Scholar]
- 24.White WI, Wilson SD, Bonnez W, Rose RC, Koenig S, Suzich JA. In vitro infection and type-restricted antibody-mediated neutralization of authentic human papillomavirus type 16. J Virol. 1998;72:959–964. doi: 10.1128/jvi.72.2.959-964.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White WI, Wilson SD, Palmer-Hill FJ, Woods RM, Ghim SJ, Hewitt LA, Goldman DM, Burke SJ, Jenson AB, Koenig S, Suzich JA. Characterization of a major neutralizing epitope on human papillomavirus type 16 L1. J Virol. 1999;73:4882–4889. doi: 10.1128/jvi.73.6.4882-4889.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giroglou T, Florin L, Schafer F, Streeck RE, Sapp M. Human papillomavirus infection requires cell surface heparan sulfate. J Virol. 2001;75:1565–1570. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts JN, Buck CB, Thompson CD, Kines R, Bernardo M, Choyke PL, Lowy DR, Schiller JT. Genital transmission of HPV in a mouse model is potentiated by nonoxynol-9 and inhibited by carrageenan. Nat Med. 2007;13:857–861. doi: 10.1038/nm1598. [DOI] [PubMed] [Google Scholar]
- 28.Joyce JG, Tung JS, Przysiecki CT, Cook JC, Lehman ED, Sands JA, Jansen KU, Keller PM. The L1 major capsid protein of human papillomavirus type 11 recombinant virus-like particles interacts with heparin and cell-surface glycosaminoglycans on human keratinocytes. J Biol Chem. 1999;274:5810–5822. doi: 10.1074/jbc.274.9.5810. [DOI] [PubMed] [Google Scholar]
- 29.Combita AL, Touzé A, Bousarghin L, El Mehdaoui S, Sizaret PY, Muñoz N, Coursaget P. Gene transfer using human papillomavirus pseudovirions varies according to virus genotype and required cell surface heparan sulfate. FEMS Microbiol Lett. 2001;204:183–188. doi: 10.1111/j.1574-6968.2001.tb10883.x. [DOI] [PubMed] [Google Scholar]
- 30.Selinka HC, Giroglou T, Sapp M. Analysis of the infectious entry pathway of human papillomavirus type 33 pseudovirions. Virology. 2002;299:279–287. doi: 10.1006/viro.2001.1493. [DOI] [PubMed] [Google Scholar]
- 31.Selinka HC, Florin L, Patel HD, Freitag K, Schmidtke M, Makarov VA, Sapp M. Inhibition of transfer to secondary receptors by heparan sulfate-binding drug or antibody induces noninfectious uptake of human papillomavirus. J Virol. 2007;81:10970–10980. doi: 10.1128/JVI.00998-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selinka HC, Giroglou T, Nowak T, Christensen ND, Sapp M. Further evidence that papillomavirus capsids exist in two distinct conformations. J Virol. 2003;77:12961–12967. doi: 10.1128/JVI.77.24.12961-12967.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rommel O, Dillner J, Fligge C, Bergsdorf C, Wang X, Selinka HC, Sapp M. Heparan sulfate proteoglycans interact exclusively with conformationally intact HPV L1 assemblies: basis for a virus-like particle ELISA. J Med Virol. 2005;75:114–121. doi: 10.1002/jmv.20245. [DOI] [PubMed] [Google Scholar]
- 34.Bousarghin L, Touzé A, Combita-Rojas AL, Coursaget P. Positively charged sequences of human papillomavirus type 16 capsid proteins are sufficient to mediate gene transfer into target cells via the heparan sulfate receptor. J Gen Virol. 2003;84:157–164. doi: 10.1099/vir.0.18789-0. [DOI] [PubMed] [Google Scholar]
- 35.Knappe M, Bodevin S, Selinka HC, Spillmann D, Streeck RE, Chen XS, Lindahl U, Sapp M. Surface-exposed amino acid residues of HPV16 L1 protein mediating interaction with cell surface heparan sulfate. J Biol Chem. 2007;282:27913–27922. doi: 10.1074/jbc.M705127200. [DOI] [PubMed] [Google Scholar]
- 36.Culp TD, Budgeon LR, Marinkovich MP, Meneguzzi G, Christensen ND. Keratinocyte-secreted laminin 5 can function as a transient receptor for human papillomaviruses by binding virions and transferring them to adjacent cells. J Virol. 2006;80:8940–8950. doi: 10.1128/JVI.00724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evander M, Frazer IH, Payne E, Qi YM, Hengst K, McMillan NA. Identification of the alpha6 integrin as a candidate receptor for papillomaviruses. J Virol. 1997;71:2449–2456. doi: 10.1128/jvi.71.3.2449-2456.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bousarghin L, Touzé A, Sizaret PY, Coursaget P. Human papillomavirus types 16, 31, and 58 use different endocytosis pathways to enter cells. J Virol. 2003;77:3846–3850. doi: 10.1128/JVI.77.6.3846-3850.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith JL, Campos SK, Ozbun MA. Human papillomavirus type 31 uses a caveolin 1- and dynamin 2-mediated entry pathway for infection of human keratinocytes. J Virol. 2007;81:9922–9931. doi: 10.1128/JVI.00988-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carpentier GS, Fleury MJ, Touzé A, Sadeyen JR, Tourne S, Sizaret PY, Coursaget P. Mutations on the FG surface loop of human papillomavirus type 16 major capsid protein affect recognition by both type-specific neutralizing antibodies and cross-reactive antibodies. J Med Virol. 2005;77:558–565. doi: 10.1002/jmv.20492. [DOI] [PubMed] [Google Scholar]
- 41.Fleury MJ, Touzé A, Alvarez E, Carpentier G, Clavel C, Vautherot JF, Coursaget P. Identification of type-specific and cross-reactive neutralizing conformational epitopes on the major capsid protein of human papillomavirus type 31. Arch Virol. 2006;151:1511–1523. doi: 10.1007/s00705-006-0734-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rockberg J, Löfblom J, Hjelm B, Uhlén M, Ståhl S. Epitope mapping of antibodies using bacterial surface display. Nat Methods. 2008;5:1039–1045. doi: 10.1038/nmeth.1272. [DOI] [PubMed] [Google Scholar]
- 43.Day PM, Thompson CD, Buck CB, Pang YY, Lowy DR, Schiller JT. Neutralization of human papillomavirus with monoclonal antibodies reveals different mechanisms of inhibition. J Virol. 2007;81:8784–8792. doi: 10.1128/JVI.00552-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Touzé A, Mahé D, El Mehdaoui S, Dupuy C, Combita-Rojas AL, Bousarghin L, Sizaret PY, Coursaget P. The nine C-terminal amino acids of the major capsid protein of the human papillomavirus type 16 are essential for DNA binding and gene transfer capacity. FEMS Microbiol Lett. 2000;189:121–127. doi: 10.1111/j.1574-6968.2000.tb09217.x. [DOI] [PubMed] [Google Scholar]
- 45.Chen XS, Garcea RL, Goldberg I, Casini G, Harrison SC. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol Cell. 2000;5:557–567. doi: 10.1016/s1097-2765(00)80449-9. [DOI] [PubMed] [Google Scholar]
- 46.Carter JJ, Wipf GC, Benki SF, Christensen ND, Galloway DA. Identification of a human papillomavirus type 16-specific epitope on the C-terminal arm of the major capsid protein L1. J Virol. 2003;77:11625–11632. doi: 10.1128/JVI.77.21.11625-11632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roth SD, Sapp M, Streeck RE, Selinka HC. Characterization of neutralizing epitopes within the major capsid protein of human papillomavirus type 33. Virol J. 2006;3:83. doi: 10.1186/1743-422X-3-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pastrana DV, Buck CB, Pang YY, Thompson CD, Castle PE, FitzGerald PC, Krüger Kjaer S, Lowy DR, Schiller JT. Reactivity of human sera in a sensitive, high-throughput pseudovirus-based papillomavirus neutralization assay for HPV16 and HPV18. Virology. 2004;321:205–216. doi: 10.1016/j.virol.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 49.Modis Y, Trus BL, Harrison SC. Atomic model of the papillomavirus capsid. EMBO J. 2002;21:4754–4762. doi: 10.1093/emboj/cdf494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ludmerer SW, Benincasa D, Mark GE, III, Christensen ND. A neutralizing epitope of human papillomavirus type 11 is principally described by a continuous set of residues which overlap a distinct linear, surface-exposed epitope. J Virol. 1997;71:3834–3839. doi: 10.1128/jvi.71.5.3834-3839.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ludmerer SW, Benincasa D, Mark GE., III Two amino acid residues confer type specificity to a neutralizing, conformationally dependent epitope on human papillomavirus type 11. J Virol. 1996;70:4791–4794. doi: 10.1128/jvi.70.7.4791-4794.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McClements WL, Wang XM, Ling JC, Skulsky DM, Christensen ND, Jansen KU, Ludmerer SW. A novel human papillomavirus type 6 neutralizing domain comprising two discrete regions of the major capsid protein L1. Virology. 2001;89:262–268. doi: 10.1006/viro.2001.1146. [DOI] [PubMed] [Google Scholar]
- 53.Combita AL, Touzé A, Bousarghin L, Christensen ND, Coursaget P. Identification of two cross-neutralizing linear epitopes within the L1 major capsid protein of human papillomavirus. J Virol. 2002;76:6480–6486. doi: 10.1128/JVI.76.13.6480-6486.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rizk RZ, Christensen ND, Michael KM, Müller M, Sehr P, Waterboer T, Pawlita M. Reactivity pattern of 92 monoclonal antibodies with 15 human papillomavirus types. J Gen Virol. 2008;89:117–129. doi: 10.1099/vir.0.83145-0. [DOI] [PubMed] [Google Scholar]
- 55.Giroglou T, Sapp M, Lane C, Fligge C, Christensen ND, Streeck RE, Rose RC. Immunological analyses of human papillomavirus capsids. Vaccine. 2001;19:1783–1793. doi: 10.1016/s0264-410x(00)00370-4. [DOI] [PubMed] [Google Scholar]
- 56.Brown DR, Kjaer SK, Sigurdsson K, Iversen OE, Hernandez-Avila M, Wheeler CM, Perez G, Koutsky LA, Tay EH, Garcia P, Ault KA, Garland SM, Leodolter S, Olsson SE, Tang GW, Ferris DG, Paavonen J, Steben M, Bosch FX, Dillner J, Joura EA, Kurman RJ, Majewski S, Muñoz N, Myers ER, Villa LL, Taddeo FJ, Roberts C, Tadesse A, Bryan J, Lupinacci LC, Giacoletti KE, Sings HL, James M, Hesley TM, Barr E. The impact of quadrivalent human papillomavirus (HPV; types 6, 11, 16, and 18) L1 virus-like particle vaccine on infection and disease due to oncogenic nonvaccine HPV types in generally HPV-naive women aged 16–26 years. J Infect Dis. 2009;199:926–935. doi: 10.1086/597307. [DOI] [PubMed] [Google Scholar]
- 57.Moore JP, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J Virol. 1996;70:1863–1872. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pichla-Gollon SL, Drinker M, Zhou X, Xue F, Rux JJ, Gao GP, Wilson JM, Ertl HC, Burnett RM, Bergelson JM. Structure-based identification of a major neutralizing site in an adenovirus hexon. J Virol. 2007;81:1680–1689. doi: 10.1128/JVI.02023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Christensen ND, Dillner J, Eklund C, Carter JJ, Wipf GC, Reed CA, Cladel NM, Galloway DA. Surface conformational and linear epitopes on HPV-16 and HPV-18 L1 virus-like particles as defined by monoclonal antibodies. Virology. 1996;223:174–184. doi: 10.1006/viro.1996.0466. [DOI] [PubMed] [Google Scholar]
- 60.Wang Z, Christensen N, Schiller JT, Dillner J. A monoclonal antibody against intact human papillomavirus type 16 capsids blocks the serological reactivity of most human sera. J Gen Virol. 1997;78:2209–2215. doi: 10.1099/0022-1317-78-9-2209. [DOI] [PubMed] [Google Scholar]
- 61.Cason J, Patel D, Naylor J, Lunney D, Shepherd PS, Best JM, McCance DJ. Identification of immunogenic regions of the major coat protein of human papillomavirus type 16 that contain type-restricted epitopes. J Gen Virol. 1989;70:2973–2987. doi: 10.1099/0022-1317-70-11-2973. [DOI] [PubMed] [Google Scholar]
- 62.Christensen ND, Dillner J, Eklund C, Carter JJ, Wipf GC, Reed CA, Cladel NM, Galloway DA. Surface conformational and linear epitopes on HPV-16 and HPV-18 L1 virus-like particles as defined by monoclonal antibodies. Virology. 1996;223:174–184. doi: 10.1006/viro.1996.0466. [DOI] [PubMed] [Google Scholar]
- 63.Kulski JK, Sadlei JW, Kelsall SR, Cicchini MS, Shellam G, Peng SW, Qi YM, Galloway DA, Zhou J, Frazer IH. Type specific and genotype cross reactive B 20 epitopes of the L1 protein of HPV16 defined by a panel of monoclonal antibodies. Virology. 1998;243:275–282. doi: 10.1006/viro.1997.9011. [DOI] [PubMed] [Google Scholar]
- 64.Bishop B, Dasgupta J, Klein M, Garcea RL, Christensen ND, Zhao R, Chen XS. Crystal structures of four types of human papillomavirus L1 capsid proteins: understanding the specificity of neutralizing monoclonal antibodies. J Biol Chem. 2007;282:31803–31811. doi: 10.1074/jbc.M706380200. [DOI] [PubMed] [Google Scholar]
- 65.Fleury MJ, Touzé A, de Sanjosé S, Bosch FX, Klaustermeiyer J, Coursaget P. Detection of human papillomavirus type 31-neutralizing antibodies from naturally infected patients by an assay based on intracellular assembly of luciferase-expressing pseudovirions. Clin Vaccine Immunol. 2008;15:172–175. doi: 10.1128/CVI.00292-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lopez TV, Cancio C, Cruz-Talonia F, Ruiz B, Sapp M, Rocha-Zavaleta L. Binding of human papillomavirus type 16 to heparan sulfate is inhibited by mucosal antibodies from pateints with low-grade squamous intraepithelial lesions but not from cervical cancer patients. FEMS Immunol Med Microbiol. 2008;54:167–176. doi: 10.1111/j.1574-695X.2008.00484.x. [DOI] [PubMed] [Google Scholar]
- 67.Belnap DM, Olson NH, Cladel NM, Newcomb WW, Brown JC, Kreider JW, Christensen ND, Baker TS. Conserved features in papillomavirus and polyomavirus capsids. J Mol Biol. 1996;259:249–263. doi: 10.1006/jmbi.1996.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Touzé A, El Mehdaoui S, Sizaret PY, Mougin C, Muñoz N, Coursaget P. The L1 major capsid protein of human papillomavirus type 16 variants affects yield of virus-like particles produced in an insect cell expression system. J Clin Microbiol. 1998;36:2046–2051. doi: 10.1128/jcm.36.7.2046-2051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu Z, Murray KS, Van Cleave V, LaVallie ER, Stahl ML, McCoy JM. Expression of thioredoxin random peptide libraries on the Escherichia coli cell surface as functional fusions to flagella system designed for exploring protein-protein interactions. Biotechnology (NY) 1995;13:366–372. doi: 10.1038/nbt0495-366. [DOI] [PubMed] [Google Scholar]
- 70.James LC, Roversi P, Tawfik DS. Antibody multispecificity mediated by conformational diversity. Science. 2003;299:1362–1367. doi: 10.1126/science.1079731. [DOI] [PubMed] [Google Scholar]
- 71.Morgenstern B. DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics. 1999;15:211–218. doi: 10.1093/bioinformatics/15.3.211. [DOI] [PubMed] [Google Scholar]


