Abstract
Microsporidia are obligate intracellular pathogens mainly infecting both vertebrate and invertebrate hosts. The group comprises approximately 150 genera with 1,200 species. Due to sequence divergence phylogenic reconstructions that are solely based on DNA sequence have been unprecise for these pathogens. Our previous study identified that three microsporidian genomes contained a putative sex-related locus similar to that of zygomycetes. In a comparison of genome architecture of the microsporidia to other fungi, Rhizopus oryzae, a zygomycete fungus, shared more common gene clusters with Encephalitozoon cuniculi, a microsporidian. This provides evidence supporting the hypothesis that microsporidia and zygomycete fungi may share a more recent common ancestor than other fungal lineages. Genetic recombination is an important outcome of sexual development. We describe genetic markers which will enable tests of whether sex occurs within E. cuniculi populations by analyzing tandem repeat DNA regions in three different isolates. Taken together, the phylogenetic relationship of microsporidia to fungi and the presence of a sex-related locus in their genomes suggest the microsporidia may have an extant sexual cycle. In addition, we describe recently reported evidence of horizontal gene transfer from Chlamydia to the E. cuniculi genome and show that these two obligate intracellular pathogens can infect the same host cells.
Key words: fungi, zygomycete, E. cuniculi, Chlamydia, sexual reproduction
Microsporidia are obligate intracellular pathogens that infect vertebrate and invertebrate hosts.1 Several species infect humans, with the most common symptom of infection being self-limited diarrhea in immunocompetent hosts and chronic diarrhea in immunocompromised hosts, including AIDS patients. In addition to gastrointestinal tract involvement, microsporidia can infect virtually any organ system and cases of encephalitis, ocular infection, sinusitis, myositis and disseminated infection are well described in the literature.2 Although the microsporidian phylum contains a large number of species (∼1,200) in 150 genera,1 their phylogenetic placement has long been debated. They have been considered basal eukaryotes due to their apparent absence of mitochondria; however, with the discovery of the mitosome it is now appreciated that microsporidia have a relic mitochrondria.3–8 Evidence has been presented that microsporidia are closely related to fungi.9–12 Studies were conflicting, however, as to whether microsporidia were more closely related to ascomycetes or basidiomycetes or zygomycetes within the kingdom Fungi. Moreover, in the fungal tree of life project, the relationship of the Microsporidia to basidiomycetes, ascomycetes and zygomycetes could not be rejected in phylogenetic analysis based on sequences of two genes, RBP1 and RBP2 and an alignment with a basal fungal lineage, the Rozella group, was instead advanced.13 Overall, the phylogenetic placement of microsporidia has been a conundrum. This is due to the fact that these pathogens have rapidly evolving, highly divergent DNA sequences and have lost whole suites of genes from their genomes.4,14,15 Sequence-based phylogenetic reconstruction has been insufficient to explain their relationship to fungi, and furthermore if they are true fungi, to define which group of fungi they are more closely related to.
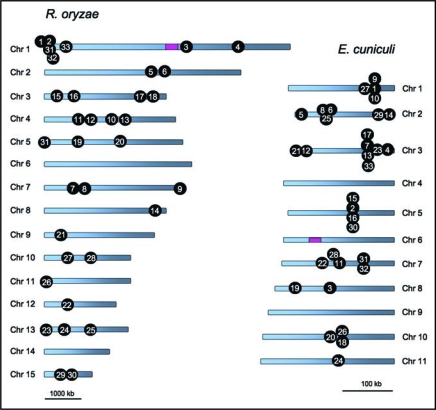
In a recent study we identified that the genomes of three microsporidia, Encephalitozoon cuniculi, Enterocytozoon bieneusi and Antonospora locustae contain a gene cluster with an architecture similar to the sex locus of zygomycetes.16 The gene cluster encodes a putative triose phosphate transporter (TPT), high mobility group protein (HMG), and RNA helicase (A. locustae has an unlinked RNA helicase gene), which is not found in other fungal species with known genomes in the Basidiomycete, Ascomycete and Chytridiomycete phyla.16,17 In addition, we utilized a whole genome comparison to analyze whether the gene order between these two groups is related. In a comparison of the Rhizopus oryzae and E. cuniculi genomes, significantly more gene clusters (33 gene clusters) were detected, which is not the case with other fungal lineages.16 The gene clusters conserved in Rhizopus oryzae and E. cuniculi are distributed throughout the genomes of both species and do not appear to have specific sub-chromosomal locations (Fig. 1).
Figure 1.
Relaxed syntenies shared between the R. oryzae and E. cuniculi genomes. The numbers on each genome indicate the common shared gene clusters. Pink boxes indicate the sex and sex-related locus. Note the size of the scale bar differs between the species.
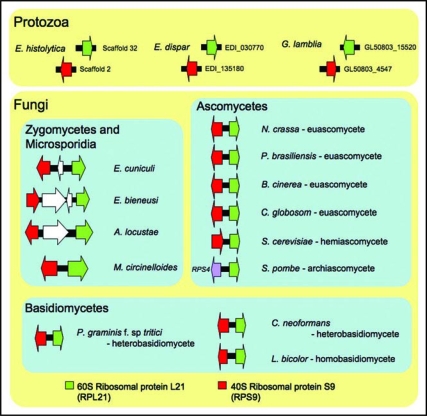
Microsporidial genomes share a synteny of the RPL21 and RPS9 ribosomal genes with the kingdom Fungi further supporting that microsporidia are true fungi (Fig. 2).16 The Rpl21 protein is a component of the large subunit of the ribosome and functions in 18S rRNA maturation.18 The Rps9 protein is known to be involved in dissociation of mRNAs from the ribosomal complex as part of translation termination.19 The RPL21 and RPS9 genes are unlinked in other more diverged lineages, including the protozoans, Entamoeba histolytica, Entamoeba dispar and Giardia lamblia (Fig. 2). This synteny can serve as a syntenic barcode only found in fungal genomes. Interestingly, in the archiascomycete fungus Schizosaccharomyces pombe, the RPS9 and RPL21 genes are not syntenic and instead the RPS4 and RPS21 genes are linked (see Fig. 2).
Figure 2.
The RPL21 and RPS9 gene cluster is fungal-specific. The synteny is conserved in euascomycetes (i.e., Neurospora crassa, Paracoccidioides brasiliensis, Botrytis cinerea, Chaetomium globosum) and hemi-ascomycete (i.e., Saccharomyces cerevisiae), homobasidiomycete (i.e., Laccaria bicolor) and heterobasidiomycetes (i.e., Puccinia graminis f. sp. tritici and Cryptococcus neoformans) as well as in Mucor circinelloides and the three microsporidia. However, the genes are not syntenic and are completely unlinked in three protozoan, Entamoeba histolytica, Entamoeba dispar and Giardia lamblia genomes. The genome of Schizosaccharomyces pombe encodes RPS9 unlinked to RPL21, and instead RPS4 forms a gene cluster with RPL21. Gene sizes are not to scale.
These sets of data based on the genome architecture/gene order independent of DNA or amino acid sequences provide evidence that the Microsporidia are true fungi that shared a common ancestor with the zygomycete phylum. Remarkably, this phylum then harbors fungi with two diametrically opposed genome trajectories: expansion to 45 to 65 Mb genomes versus genome compaction and streamlining to small 3 to 6 Mb genomes and an obligate intracellular life cycle.
In considering the evidence that the Microsporidia are fungi that harbor a putative sex-related locus, it is possible that these pathogens may have an extant sexual cycle. Their unique chance to have sex would be during co-infection of the host by two genetically distinct isolates or they could be self-fertile and sex could occur during solo infection. It is possible that the alternation of spore sizes and different developmental cycles seen in various hosts for some of the microsporidia represent stages in sexual reproduction.20 It does not appear that Microsporidia develop sexual development specific structures like zygospores associated with other zygomycete fungi. Microsporidial sex could be similar to that of Giardia ssp, the human parasitic protozoan. The sexual cycle of G. intestinalis occurs during infection of host cells.21 Their modified sexual cycle involves nuclear fusion (karyogamy), expression of key meiotic homologs, and genetic exchange and segregation between nuclei. One of our goals to verify whether E. cuniculi has an extant sexual cycle is to test whether recombinant progeny are produced following co-infection with genetically distinct strains. Given the lack of a transformation method or introduced genetic markers (such as drug resistance genes), an approach is required to define naturally occurring genetic variation in different isolates.
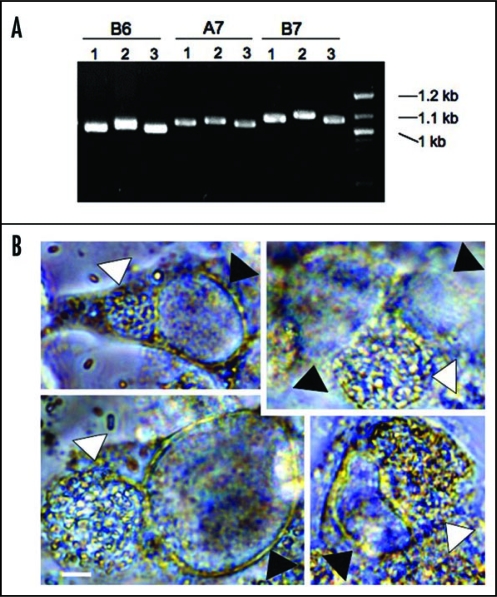
We targeted DNA tandem repeats (TRs) regions because it is possible that different isolates may harbor variation in the number or sequence of tandem repeats (TRs). We applied a web-based software program (http://tandem.bu.edu/trf/trf.html)22 to identify TRs in the reference E. cuniculi genome (GB-M1 strain sequence at NCBI).4 Two TRs were chosen per chromosome. E. cuniculi has 11 chromosomes, and therefore 22 TRs were selected in this study. All 22 TR areas were subjected to PCR analysis from three different isolates, EC1, EC2 and EC3.23 As expected, many TRs displayed size variation in electrophoresis (examples are shown in Fig. 3). Several examples were sequenced, confirming the variable numbers and polymorphic TRs in three different isolates. Overall these findings suggest that TR DNA areas can be applied as genetic markers in E. cuniculi.
Figure 3.
Genetic variation of tandem repeat DNAs in different E. cuniculi isolates and co-infection of E. cuniculi and C. trachomatis. (A) Examples of TR DNAs. Three E. cuniculi isolates (1 is for EC1, 2 for EC2 and 3 for EC3) have different sized banding patterns. B6, A7 and B7 are TR areas chosen with the Tandem Repeat Finder (see the text). (B) E. cuniculi and C. trachomatis infect the same host cell forming two independent parasitophorous vacuoles. White arrowheads indicate E. cuniculi vacuoles and black arrowheads indicate C. trachomatis vacuoles. Scale = 5 µm.
Interestingly, in the sequenced microsporidian genomes there is evidence for the presence of several bacterial genes, particularly genes encoding bacterial-like ATP transporters to obtain ATP.4,24,25 Horizontal gene transfer from bacteria to microsporidia is strongly supported because of sequence similarity between the microsporidian and bacterial transporter genes.26 The E. cuniculi genome contains four Chlamydia-like ATP transporter genes that were horizontally transferred.4 These encode ATP transporters localized to the plasma membrane and in the mitosome, enabling ATP to be transported from the host. For horizontal gene transfer (HGT) to occur between these two organisms, it is necessary that they should share a niche. Chlamydia ssp are obligate intracellular bacterial pathogens that cause sexually transmitted diseases (STDs) in humans.27 Both pathogens are known to form parasitophorous vacuoles during their infection cycles.1,28 It is, therefore, possible that the two pathogens might meet in the same host cell. We tested this hypothesis. Rabbit kidney (RK13) cells were infected by E. cuniculi strains and subsequently Chlamydia trachomatis cells were added to the culture. We observed that these two pathogens infected the same cells. These pathogens, however, did not occupy the same parasitophorous vacuoles based on light microscopic observations that reveal the presence of two different vacuoles: one contains the larger sized spores of E. cuniculi and the other smaller sized particles of Chlamydia in each vacuole (see Fig. 3). However, it is possible that rare C. trachomatis cells might not be visualized between the significantly larger microsporidian cells in the microsporidian parasitophorous vacuole. Further analysis employing fluorescent microscopy will be required to fully test this possibility. The results at least suggest that both E. cuniculi and C. trachomatis can share a niche and therefore HGT from bacteria to microsporidia could have occurred in shared host cells. HGT from Chlamydia to microsporidia most likely was a consequence of the uptake of nucleic acid from Chlamydia by the microsporidia in the same cells. This is similar to the exchange of DNA that has been appreciated to occur in environmental samples of diverse microbial communities in soil. The exchange may have been due to rare cases of both organisms being present in the same vacuoles, but this is not a requirement. Given that four genes have been acquired via HGT from Chlamydia, it is also possible that microsporidia have acquired additional genes by HGT from other microbes or host cells.
In conclusion, our hypothesis is that microsporidia are true fungi and may have an extant sexual cycle during infection of host cells. The next step will be experiments to test for the existence of a sexual cycle in the microsporidian, E. cuniculi. Our studies further revealed that E. cuniculi and C. trachomatis can infect the same cell simultaneously, providing a plausible route by which ancient horizontal gene transfer from Chlamydia ssp to E. cuniculi might have remodeled the microsporidian genome and enabled acquisition of ATP from the host.
Acknowledgements
We are indebted to Raphael Valdivia and Alex Saka for providing a C. trachomatis strain and advice on its culture. This work was supported by NIH/NIAID grant AI50113.
Footnotes
Previously published online as a Communicative & Integrative Biology E-publication: http://www.landesbioscience.com/journals/cib/article/8846
References
- 1.Keeling PJ, Fast NM. Microsporidia: Biology and evolution of highly reduced intracellular parasites. Annu Rev Microbiol. 2002;56:93. doi: 10.1146/annurev.micro.56.012302.160854. [DOI] [PubMed] [Google Scholar]
- 2.Didier ES, Weiss LM. Microsporidiosis: Current status. Curr Opin Infect Dis. 2006;19:485–492. doi: 10.1097/01.qco.0000244055.46382.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vossbrinck CR, Maddox JV, Friedman S, Debrunner-Vossbrinck BA, Woese CR. Ribosomal-RNA sequence suggests microsporidia are extremely ancient eukaryotes. Nature. 1987;326:411–414. doi: 10.1038/326411a0. [DOI] [PubMed] [Google Scholar]
- 4.Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 5.Tsaousis AD, Kunji ERS, Goldberg AV, Lucocq JM, Hirt RP, Embley TM. A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi. Nature. 2008;453:553–556. doi: 10.1038/nature06903. [DOI] [PubMed] [Google Scholar]
- 6.Williams BAP, Hirt RP, Lucocq JM, Embley TM. A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature. 2002;418:865–869. doi: 10.1038/nature00949. [DOI] [PubMed] [Google Scholar]
- 7.Goldberg AV, Molik S, Tsaousis AD, Neumann K, Kuhnke G, Delbac F, et al. Localization and functionality of microsporidian iron-sulphur cluster assembly proteins. Nature. 2008;452:624–628. doi: 10.1038/nature06606. [DOI] [PubMed] [Google Scholar]
- 8.Williams BAP, Cali ANN, Takvorian PM, Keeling PJ. Distinct localization patterns of two putative mitochondrial proteins in the microsporidian Encephalitozoon cuniculi. J Eukaryot Microbiol. 2008;55:131–133. doi: 10.1111/j.1550-7408.2008.00315.x. [DOI] [PubMed] [Google Scholar]
- 9.Hirt RP, Logsdon JM, Healy B, Dorey MW, Doolittle WF, Embley TM. Microsporidia are related to fungi: evidence from the largest subunit of RNA polymerase II and other proteins. Proc Natl Acad Sci USA. 1999;96:580–585. doi: 10.1073/pnas.96.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keeling PJ, Luker MA, Palmer JD. Evidence from beta-tubulin phylogeny that microsporidia evolved from within the fungi. Mol Biol Evol. 2000;17:23–31. doi: 10.1093/oxfordjournals.molbev.a026235. [DOI] [PubMed] [Google Scholar]
- 11.Keeling PJ. Congruent evidence from alpha-tubulin and beta-tubulin gene phylogenies for a zygomycete origin of microsporidia. Fungal Genet Biol. 2003;38:298–309. doi: 10.1016/s1087-1845(02)00537-6. [DOI] [PubMed] [Google Scholar]
- 12.Gill EE, Fast NM. Assessing the microsporidia-fungi relationship: combined phylogenetic analysis of eight genes. Gene. 2006;375:103–109. doi: 10.1016/j.gene.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 13.James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- 14.Slamovits CH, Fast NM, Law JS, Keeling PJ. Genome compaction and stability in microsporidian intracellular parasites. Curr Biol. 2004;14:891–896. doi: 10.1016/j.cub.2004.04.041. [DOI] [PubMed] [Google Scholar]
- 15.Thomarat F, Vivarès CP, Gouy M. Phylogenetic analysis of the complete genome sequence of Encephalitozoon cuniculi supports the fungal origin of microsporidia and reveals a high frequency of fast-evolving genes. J Mol Evol. 2004;59:780–791. doi: 10.1007/s00239-004-2673-0. [DOI] [PubMed] [Google Scholar]
- 16.Lee SC, Corradi N, Byrnes EJ, Torres-Martinez S, Dietrich FS, Keeling PJ, et al. Microsporidia evolved from ancestral sexual fungi. Curr Biol. 2008;18:1675–1679. doi: 10.1016/j.cub.2008.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Idnurm A, Walton FJ, Floyd A, Heitman J. Identification of the sex genes in an early diverged fungus. Nature. 2008;451:193–196. doi: 10.1038/nature06453. [DOI] [PubMed] [Google Scholar]
- 18.Tabb-Massey A, Caffrey JM, Logsden P, Taylor S, Trent JO, Ellis SR. Ribosomal proteins Rps0 and Rps21 of Saccharomyces cerevisiae have overlapping functions in the maturation of the 3′ end of 18S rRNA. Nucleic Acids Res. 2003;31:6798–6805. doi: 10.1093/nar/gkg899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pnueli L, Arava Y. Genome-wide polysomal analysis of a yeast strain with mutated ribosomal protein S9. BMC Genomics. 2007;8:285. doi: 10.1186/1471-2164-8-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wittner M, Weiss LM. The Microsporidia and microsporidiosis. Washington D.C.: ASM Press; 1999. [Google Scholar]
- 21.Poxleitner MK, Carpenter ML, Mancuso JJ, Wang C-JR, Dawson SC, Cande WZ. Evidence for karyogamy and exchange of genetic material in the binucleate intestinal parasite Giardia intestinalis. Science. 2008;319:1530–1533. doi: 10.1126/science.1153752. [DOI] [PubMed] [Google Scholar]
- 22.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao L, Li L, Visvesvara GS, Moura H, Didier ES, Lal AA. Genotyping Encephalitozoon cuniculi by multilocus analyses of genes with repetitive sequences. J Clin Microbiol. 2001;39:2248–2253. doi: 10.1128/JCM.39.6.2248-2253.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slamovits CH, Keeling PJ. Class II photolyase in a microsporidian intracellular parasite. J Mol Biol. 2004;341:713–721. doi: 10.1016/j.jmb.2004.06.032. [DOI] [PubMed] [Google Scholar]
- 25.Fast NM, Law JS, Williams BAP, Keeling PJ. Bacterial catalase in the microsporidian Nosema locustae: implications for microsporidian metabolism and genome evolution. Eukaryot Cell. 2003;2:1069–1075. doi: 10.1128/EC.2.5.1069-1075.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolf YI, Aravind L, Koonin EV. Rickettsiae and Chlamydiae: evidence of horizontal gene transfer and gene exchange. Trends Genet. 1999;15:173–175. doi: 10.1016/s0168-9525(99)01704-7. [DOI] [PubMed] [Google Scholar]
- 27.Schachter J. In: Chlamydia: intracellular biology, pathogenesis and immunity. Stephens RS, editor. Washington D.C.: ASM; 1999. pp. 139–169. [Google Scholar]
- 28.Fields KA, Hackstadt T. The Chlamydial inclusion: escape from the endocytic pathway. Annu Rev Cell Dev Biol. 2002;18:221. doi: 10.1146/annurev.cellbio.18.012502.105845. [DOI] [PubMed] [Google Scholar]