Abstract
Light is a key environmental signal for most life on earth. Over 5% of Neurospora crassa genes are expressed in response to light stimulation in a temporally regulated cascade that includes several transcription factors. Fungal genomes, including Neurospora's, may encode several different proteins capable of binding chromophores with the ability to harvest light energy as well as proteins that can interact with primary photoreceptors or further propogate the light signal. The best understood photo- receptors are the evolutionarily conserved White Collar proteins, and the related Vivid protein, but fungi may also encode phytochromes, cryptochromes and opsins.
Key words: light, microarray, Neurospora, photoreceptors, wc-1, wc-2, vvd, frq, sub-1, nop-1, ve-1
According to a recent report from the American Society of Microbiology, the fungal kingdom comprises an estimated 1.5 million species, many hundreds of which are known animal or plant pathogens.1 The filamentous fungus Neurospora crassa is a leading research model, including studies aimed at understanding light responses in fungal cells.2–7 Decades of effort from several labs, have established the White Collar complex (WCC) as an essential as well as dominant light signaling component. The heterodimeric transcription factor WCC senses light directly through bound FAD, and binds to the promoters of many light-responsive genes, activating gene expression. We have shown that light regulated expression falls into two distinct temporal classes, both under WCC control. SUB-1, identified as an “early” light-responsive transcription factor, was found to regulate most of the “late” light gene expression. Chromatin-immunoprecipitation (ChIP) and bioinformatics analysis further established the hierarchical relationship between early and late light responses.8 Here, we present a brief summary of recent studies on the molecular components involved in Neurospora photobiology.
Light-Regulated Biology in Neurospora crassa
Light acts as an essential cue to regulate a variety of physiological processes in Neurospora, including the resetting of the circadian clock, biosynthesis of the photo-protective carotenoid pigments, asexual conidiospore formation, perithecial development in the sexual cycle, and the direction of ascospores release.2–7 Underlying this biology is the regulation of many Neurospora genes by light. Microarrays representing the approximately 10,000 genes in Neurospora crassa were used as probes against light induced cDNA. Of the 5,600 detectable genes, 314 (approximately 5.6%) responded to the light stimulus by increasing transcript levels.8 Most of the identified genes (92%) were either early (45%), with peak expression between 15 and 45 minutes, or late (55%), with the induced expression peaking between 45 and 90 minutes after lights on. Genes related to the synthesis of photoprotective pigments (7.1%), vitamins, cofactors, and prosthetic groups (4.7%), secondary metabolism (4.7%), DNA processing (6.3%), cellular signaling (5.5%) and environmental sensing and response (1.6%) were found enriched in the early light response. In contrast, genes involved in carbohydrate metabolism (20%), oxidation of fatty acids (1.9%) and oxygen detoxification reaction (2.5%) were found enriched in the late light response. Within the early group were several transcription factors, most of which show mutant phenotypes during development (see Table 1).
Table 1.
Real and putative light signaling components in Neurospora crassa
| NCU #1 | Gene | Light-sensing chromophore | TF? | Light-related or other phenotypes in mutants | Light-responsive2 | Refs |
| NCU02356.2 | wc-1 | FAD | Yes | Blind to most if not all light responses, carotenogenesis repressed in mycelia | + | 2–8 |
| NCU00902.2 | wc-2 | None | Yes | Blind to most if not all light responses, carotenogenesis repressed in mycelia | No | 2–8 |
| NCU03967.2 | vvd | FAD | No | Affects photoadaptation, excess accumulation of carotenoids | + + + | 8, 18–21 |
| NCU04834.2 | phy-1 | Undetermined | No | Wild-type light responses, k/o wild-type | No | 8, 28 |
| NCU05790.2 | phy-2 | Biliverdin or Phycocyanobilin | No | Wild-type light responses, k/o wild-type | No | 8, 28 |
| NCU00582.2 | cry | FAD | No | Wild-type light responses, k/o wild-type | + + + | 8, 29 |
| NCU10055.2 | nop-1 | Retinal | No | Wild-type light responses, involved in late-stage asexual development | No | 8, 31–33 |
| NCU02265.2 | frq | None | No | Affects amplitude of light induced gene expression | + | 8, 34, 35 |
| NCU01154.2 | sub-1 | None | Yes | Affects some early and most late light responses, submerged protoperithecia in the sexual cycle | + | 8, 36 |
| NCU02713.2 | csp-1 | None | Yes | Wild-type light responses, defective in conidiospore maturation | + | 8, 37 |
| NCU04179.2 | sah-1 | None | Yes | Wild-type light responses, shortened aerial hyphae | + | 8, 36 |
| NCU06407.2 | vad-3 | None | Yes | Wild-type light responses, slowed basal and aerial hyphal extension | + | 8, 36 |
| NCU03643.2 | None | None | Yes | Wild-type light responses, k/o wild-type | + | 8 |
| NCU01731.3 | ve-1 | None | No | Wild-type light responses, shortened aerial hyphae, increased conidiation | No | 40 |
NCU numbers are from Neurospora annotation (http://www.broad.mit.edu/annotation/genome/neurospora/Home.html).
Fold change of mRNA transcripts in response to a white light stimulus; +, less than 10-fold; + +, 10-∼100-fold; + + +, more than 100-fold. The data adapted from ref. 8.
All currently known light responses in Neurospora are restricted to near UV/blue light,5,7 suggesting the presence of a master photoreceptor dedicated to blue light sensing and signal transduction. Extensive genetic screening and analysis has resulted in the isolation of only two fully blind mutants, wc-1 and wc-2,3,9,10 both GATA family zinc finger transcription factors.11 The direct connection between light sensing and gene activation has subsequently been demonstrated both in vitro and in vivo.12–15 The photoreceptor WC-1 forms an obligate complex with WC-2 to bind to specific DNA sequences,16,17 including the promoter of a light-responsive transcription factor, sub-1, the function of which is essential for the late light response.8 Several additional components are or may be involved in the light signaling mechanism (Fig. 1). As the WCC has been reviewed in some detail elsewhere,2–7 our discussion below will focus on the additional players.
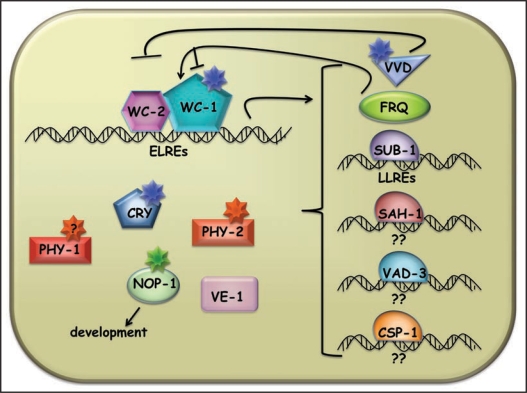
Figure 1.
Established and putative molecular components involved in Neurospora light signaling. WC-1 and WC-2 form a heterodimeric transcription factor (WCC) that binds to early light responsive elements (ELREs). In response to a light signal, transcription is rapidly activated, resulting in the expression of several downstream transcription factors, as well as the VVD and FRQ proteins. The SUB-1 transcription factor is required for expression of most late-light responsive genes, many of which have a specific late light responsive element (LLRE), A/GTGAC/TG/ATCA. VVD acts as a potent repressor of WCC activity on light regulated genes. FRQ may block further activity on some genes while at the same time promotes expression of the WCC. Several proteins can bind chromophores (WC-1, PHY-1, NOP-1, VVD, CRY and maybe PHY-2). Chromophores are shown as stars in the color of light they absorb. Several proteins have no described responses to light in Neurospora (CRY, PHY-1, PHY-2, VE-1) although they do in other fungi and other organisms.
Real and Putative Light Signaling Components in Neurospora crassa
Real and putative light signaling components in Neurospora are summarized in Table 1. After the WCC, VVD has been the next most intensely studied photoreceptor in the fungi. Our study and others have clearly shown that VVD acts as a universal repressor for most if not all light-induced gene expression controlled by the WCC.8,18–21 In vvd mutants, once gene transcription is turned on by the light-activated WCC, transcript levels will remain upregulated for many hours in constant light, so-called “photoadaptation defects”. In contrast, in a wild-type strain, light-induced gene expression is transient, usually returning to pre-induction levels within two to four hours.8 Molecularly, VVD is a small, 21 kD flavin-binding photoreceptor consisting of a LOV (light, oxygen or voltage) domain and N-terminal cap.22 Upon light activation, the formation of a protein-flavin bond in the LOV domain induces a conformational change at the N-terminus which appears to be essential for the light function of VVD.22 Formation of a rapidly exchanging VVD dimer in light has been proposed recently.23 Interestingly, VVD appears to localize exclusively in the cytosol while the majority of WCC is in the nucleus.19,24 This raises the question of how VVD communicates with the WCC to repress light responses and regulate various circadian clock properties.21,25,26 The answer to this question will certainly shed light on the molecular mechanisms of photoadaptation in general.
After completion of the Neurospora genome project,27 two putative red-light photoreceptors (N. crassa phytochrome orthologs phy-1 and phy-2) and one additional blue-light photoreceptor (N. crassa cryptochrome orthologue cry) were identified. Although there is yet no report of red light-regulated biology in Neurospora, a collaborative effort has shown that PHY-2 can covalently bind either biliverdin or phycocyanobilin and is capable of undergoing a photocycle in vitro.28 The cry gene encodes a member of the cryptochrome-DASH family. We have found it capable of binding FAD and MTHF, with both transcript and protein levels strongly induced by blue light in a wc-1 dependent manner.29 However, due to the lack of a detectable phenotype or atypical light responses in the respective knockout strains,8,28,29 the biological function(s) of PHY-1, PHY-2 and CRY remains to be discovered in Neurospora, although function has been reported for homologs in other fungi.3,4,6,30 The opsin, NOP-1, is a putative green-light photoreceptor identified via sequence homology with archaeal rhodopsins.31 NOP-1 has been shown to both bind retinal and undergo a slow photocycle32 and the expression levels of several genes are known to be affected in a knockout strain during late asexual development.33 Our microarray data, not carried past two hours after light stimulus in the knock-out strain, suggest that NOP-1 does not play a role in either early or late light regulated gene induction.8
Instead of sensing light directly, which requires the ability to interact with chromophores that absorb light energy, the FRQ and SUB-1 proteins are indirectly involved in light signaling. Our array analysis has confirmed and extended the role of FRQ in regulating the light function of the WCC by affecting the amplitude of induction for both types of light responses.8 Given that FRQ is an oscillating circadian clock component and has been shown to physically interact with the WCC, previous studies34,35 together with our recent microarray data highlight the clock-modulating effect of the light input pathway. The novel GATA family transcription factor, SUB-1, was identified as essential for regulating a subset of the early and most of the late light responses.8 A previously described phenotype associated with the sub-1 (submerged protoperithecia-1) knockout strain36 might easily be the functional consequence of impaired late light responses (i.e., the formation of protoperithecia in Neurospora is a light-regulated developmental process), which might also hold true for developmental defects seen in knockout strains of other light-responsive transcription factors.36,37
Finally, the veA locus has been shown to be required for both light-regulated development and secondary metabolism in Aspergillus nidulans38,39 and the promoter and coding sequences of the N. crassa ortholog, ve-1, is sufficient to complement the role of veA null mutants in A. nidulans.40 However, unlike its counterpart, VE-1 knockout strains in Neurospora lack light-dependent phenotypes40 and have largely normal gene expression in response to white light (Chen C-H and Loros J, unpublished data) suggesting that ve-1 may not have a significant role in regulating light signals in N. crassa, at least under the conditions tested.
Fungal Light Signaling Components are Conserved
Sequence and functional orthologs of WC-1, WC-2 and most of the other light signaling components are widespread among the fungal kingdom. Recent studies have demonstrated that WC-1- and WC-2-like molecules in various fungal species play an essential role in mediating light signals from the Ascomycota, Basidiomycota and Zygomycota phyla.2–4,6,7 Of broader evolutionary interest, WC-1 and the animal circadian-clock-associated bHLH transcription factors, CYC from insects and BMAL1 and NPAS2 from mammals, share a common ancestor. The bHLH transcription factors do not bind chromophores but, like WC-1 in Neurospora, they are critical for light resetting, as well as the maintenance of circadian rhythms in animals, highlighting the close evolutionary relationship between photobiology and circadian rhythmicity.41,42 Successful work on the WCC in Neurospora has led to fundamental breakthroughs in understanding photobiology in other fungi. We predict that future work on the underlying mechanisms of Neurospora light signaling components will continue to illuminate other light-sensitive eukaryotic cells.
Acknowledgements
This work was supported by grants from the National Institutes of Health to J.J.L. (RO1 GM08336), to Jay C. Dunlap and J.J.L. (PO1GM68087), and by the core grant to the Norris Cotton Cancer Center at Dartmouth. We thank Randy Lambreghts for critical reading of the manuscript and are deeply grateful to the Fungal Genetics Stock Center at the University of Missouri, Kansas City in supporting our work with Neurospora.
Footnotes
Previously published online as a Communicative & Integrative Biology E-publication: http://www.landesbioscience.com/journals/cib/article/8835
References
- 1.Casadevall A, Heitman J, Buckley M. The Fungal Kingdom: Diverse and Essential Roles in Earth's Ecosystem. American Academy of Microbiology. 2008 [PubMed] [Google Scholar]
- 2.Herrera-Estrella A, Horwitz BA. Looking through the eyes of fungi: molecular genetics of photoreception. Mol Microbiol. 2007;64:5–15. doi: 10.1111/j.1365-2958.2007.05632.x. [DOI] [PubMed] [Google Scholar]
- 3.Corrochano LM. Fungal photoreceptors: sensory molecules for fungal development and behaviour. Photochem Photobiol Sci. 2007;6:725–736. doi: 10.1039/b702155k. [DOI] [PubMed] [Google Scholar]
- 4.Purschwitz J, Muller S, Kastner C, Fischer R. Seeing the rainbow: light sensing in fungi. Curr Opin Microbiol. 2006;9:566–571. doi: 10.1016/j.mib.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Dunlap JC, Loros JJ. How fungi keep time: circadian system in Neurospora and other fungi. Curr Opin Microbiol. 2006;9:579–587. doi: 10.1016/j.mib.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Idnurm A, Heitman J. Photosensing fungi: phytochrome in the spotlight. Curr Biol. 2005;15:829–832. doi: 10.1016/j.cub.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Dunlap JC, Loros JJ. Neurospora Photoreceptors. In: Briggs WR, Spudich JL, editors. Handbook of Photosensory Receptors. Vol. 18. Weinheim: Wiley-VCH; 2005. pp. 371–388. [Google Scholar]
- 8.Chen CH, Ringelberg CS, Gross RH, Dunlap JC, Loros JJ. Genome-wide analysis of light-inducible responses reveals hierarchical light signalling in Neurospora. EMBO J. 2009;8:1029–1042. doi: 10.1038/emboj.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee K, Dunlap JC, Loros JJ. Roles for WHITE COLLAR-1 in circadian and general photoperception in Neurospora crassa. Genetics. 2003;163:103–114. doi: 10.1093/genetics/163.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collett MA, Garceau N, Dunlap JC, Loros JJ. Light and clock expression of the Neurospora clock gene frequency is differentially driven by but dependent on WHITE COLLAR-2. Genetics. 2002;160:149–158. doi: 10.1093/genetics/160.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linden H, Ballario P, Macino G. Blue light regulation in Neurospora crassa. Fungal Genet Biol. 1997;22:141–150. doi: 10.1006/fgbi.1997.1013. [DOI] [PubMed] [Google Scholar]
- 12.He Q, Cheng P, Yang Y, Wang L, Gardner KH, Liu Y. White collar-1, a DNA binding transcription factor and a light sensor. Science. 2002;297:840–843. doi: 10.1126/science.1072795. [DOI] [PubMed] [Google Scholar]
- 13.Froehlich AC, Liu Y, Loros JJ, Dunlap JC. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science. 2002;297:815–819. doi: 10.1126/science.1073681. [DOI] [PubMed] [Google Scholar]
- 14.Cheng P, Yang Y, Wang L, He Q, Liu Y. WHITE COLLAR-1, a multifunctional Neurospora protein involved in the circadian feedback loops, light sensing and transcription repression of wc-2. J Biol Chem. 2003;278:3801–3808. doi: 10.1074/jbc.M209592200. [DOI] [PubMed] [Google Scholar]
- 15.He Q, Liu Y. Molecular mechanism of light responses in Neurospora: from light-induced transcription to photoadaptation. Genes Dev. 2005;19:2888–2899. doi: 10.1101/gad.1369605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belden WJ, Loros JJ, Dunlap JC. Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol Cell. 2007;25:587–600. doi: 10.1016/j.molcel.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Froehlich AC, Loros JJ, Dunlap JC. Rhythmic binding of a WHITE COLLAR-containing complex to the frequency promoter is inhibited by FREQUENCY. Proc Natl Acad Sci USA. 2003;100:5914–5919. doi: 10.1073/pnas.1030057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwerdtfeger C, Linden H. Blue light adaptation and desensitization of light signal transduction in Neurospora crassa. Mol Microbiol. 2001;39:1080–1087. doi: 10.1046/j.1365-2958.2001.02306.x. [DOI] [PubMed] [Google Scholar]
- 19.Schwerdtfeger C, Linden H. VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J. 2003;22:4846–4855. doi: 10.1093/emboj/cdg451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shrode LB, Lewis ZA, White LD, Bell-Pedersen D, Ebbole DJ. vvd is required for light adaptation of conidiation-specific genes of Neurospora crassa, but not circadian conidiation. Fungal Genet Biol. 2001;32:169–181. doi: 10.1006/fgbi.2001.1264. [DOI] [PubMed] [Google Scholar]
- 21.Heintzen C, Loros JJ, Dunlap JC. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating and regulates clock resetting. Cell. 2001;104:453–464. doi: 10.1016/s0092-8674(01)00232-x. [DOI] [PubMed] [Google Scholar]
- 22.Zoltowski BD, Schwerdtfeger C, Widom J, Loros JJ, Bilwes AM, Dunlap JC, et al. Conformational switching in the fungal light sensor Vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zoltowski BD, Crane BR. Light activation of the LOV protein vivid generates a rapidly exchanging dimer. Biochemistry. 2008;47:7012–7019. doi: 10.1021/bi8007017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cha J, Chang SS, Huang G, Cheng P, Liu Y. Control of WHITE COLLAR localization by phosphorylation is a critical step in the circadian negative feedback process. EMBO J. 2008;27:3246–3255. doi: 10.1038/emboj.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunt SM, Elvin M, Crosthwaite SK, Heintzen C. The PAS/LOV protein VIVID controls temperature compensation of circadian clock phase and development in Neurospora crassa. Genes Dev. 2007;21:1964–1974. doi: 10.1101/gad.437107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elvin M, Loros JJ, Dunlap JC, Heintzen C. The PAS/LOV protein VIVID supports a rapidly dampened daytime oscillator that facilitates entrainment of the Neurospora circadian clock. Genes Dev. 2005;19:2593–2605. doi: 10.1101/gad.349305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 28.Froehlich AC, Noh B, Vierstra RD, Loros J, Dunlap JC. Genetic and molecular analysis of phytochromes from the filamentous fungus Neurospora crassa. Eukaryot Cell. 2005;4:2140–2152. doi: 10.1128/EC.4.12.2140-2152.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Froehlich AC, Chen C-H, Belden WJ, Loros JJ, Dunlap JC. Genetic and molecular characterization of a cryptochrome from the filamentous fungus Neurospora crassa. 2009 doi: 10.1128/EC.00380-09. In preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veluchamy S, Rollins JA. A CRY-DASH-type photolyase/cryptochrome from Sclerotinia sclerotiorum mediates minor UV-A-specific effects on development. Fungal Genet Biol. 2008;45:1265–1276. doi: 10.1016/j.fgb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 31.Bieszke JA, Braun EL, Bean LE, Kang S, Natvig DO, Borkovich KA. The nop-1 gene of Neurospora crassa encodes a seven transmembrane helix retinal-binding protein homologous to archaeal rhodopsins. Proc Natl Acad Sci USA. 1999;96:8034–8039. doi: 10.1073/pnas.96.14.8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bieszke JA, Spudich EN, Scott KL, Borkovich KA, Spudich JL. A eukaryotic protein, NOP-1, binds retinal to form an archaeal rhodopsin-like photochemically reactive pigment. Biochemistry. 1999;38:14138–14145. doi: 10.1021/bi9916170. [DOI] [PubMed] [Google Scholar]
- 33.Bieszke JA, Li L, Borkovich KA. The fungal opsin gene nop-1 is negatively-regulated by a component of the blue light sensing pathway and influences conidiation-specific gene expression in Neurospora crassa. Curr Genet. 2007;52:149–157. doi: 10.1007/s00294-007-0148-8. [DOI] [PubMed] [Google Scholar]
- 34.Tan Y, Merrow M, Roenneberg T. Photoperiodism in Neurospora crassa. J Biol Rhythms. 2004;19:135–143. doi: 10.1177/0748730404263015. [DOI] [PubMed] [Google Scholar]
- 35.Merrow M, Franchi L, Dragovic Z, Gorl M, Johnson J, Brunner M, et al. Circadian regulation of the light input pathway in Neurospora crassa. EMBO J. 2001;20:307–315. doi: 10.1093/emboj/20.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, et al. A high- throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lambreghts R, Shi M, Belden WJ, Decaprio D, Park D, Henn MR, et al. A high-density single nucleotide polymorphism map for Neurospora crassa. Genetics. 2009;181:767–781. doi: 10.1534/genetics.108.089292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calvo AM. The VeA regulatory system and its role in morphological and chemical development in fungi. Fungal Genet Biol. 2008;45:1053–1061. doi: 10.1016/j.fgb.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 39.Bayram O, Krappmann S, Ni M, Bok JW, Helmstaedt K, Valerius O, et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science. 2008;320:1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- 40.Bayram O, Krappmann S, Seiler S, Vogt N, Braus GH. Neurospora crassa ve-1 affects asexual conidiation. Fungal Genet Biol. 2008;45:127–138. doi: 10.1016/j.fgb.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 41.Tauber E, Last KS, Olive PJ, Kyriacou CP. Clock gene evolution and functional divergence. J Biol Rhythms. 2004;19:445–458. doi: 10.1177/0748730404268775. [DOI] [PubMed] [Google Scholar]
- 42.Lee K, Loros JJ, Dunlap JC. Interconnected feedback loops in the Neurospora circadian system. Science. 2000;289:107–110. doi: 10.1126/science.289.5476.107. [DOI] [PubMed] [Google Scholar]