Abstract
Deinococcus radiodurans (Dr) withstands desiccation, reactive oxygen species, and doses of radiation that would be lethal to most organisms. Deletion of a gene encoding a homolog of mammalian nitric oxide synthase (NOS) severely compromises the recovery of Dr from ultraviolet (UV) radiation damage. The Δnos defect can be complemented with recombinant NOS, rescued by exogenous nitric oxide (NO) and mimicked in the wild-type strain with an NO scavenging compound. UV radiation induces both upregulation of the nos gene and cellular NO production on similar time scales. Growth recovery does not depend on NO being present during UV irradiation, but rather can be manifested by NO addition hours after exposure. Surprisingly, nos deletion does not increase sensitivity to oxidative damage, and hydrogen peroxide does not induce nos expression. However, NOS-derived NO upregulates transcription of obgE, a gene involved in bacterial growth proliferation and stress response. Overexpression of the ObgE GTPase in the Δnos background substantially alleviates the growth defect after radiation damage. Thus, NO acts as a signal for the transcriptional regulation of growth in D. radiodurans.
Keywords: D. radiodurans, nitric oxide synthase, UV radiation
Nitric oxide (NO) is a widespread metabolite, cytotoxic agent, and signaling molecule that reacts directly with a select few biological targets (1, 2). In mammals and other higher organisms, NO participates in a large number of processes, including protection against pathogens, regulation of vascular tension, hormone release, and neuronal signaling (3, 4). In bacteria, NO is a key intermediate in nitrate respiration (denitrification), and has recently been shown to act as a regulatory signal for cell dispersal and nitrosative stress responses (5, 6). In mammals, NO is produced from the oxidation of L-arginine (L-arg) to L-citrulline and is catalyzed by the heme-containing NO synthases (NOSs). Mammalian NOSs (mNOSs) are homodimers that contain two domains: an N-terminal heme oxygenase domain (NOSox) that binds the substrate L-arg and cofactors heme and tetrahydrobiopterin (H4B), and a C-terminal reductase domain (NOSred) that binds FAD, FMN, and NADPH (7–9). Proteins with homology to the mNOSox domain are found in several mainly Gram-positive bacterial genera such as Streptomyces, Bacillus, Staphylococcus, and Deinococcus (10–13). These proteins lack NOSred, but retain structural and catalytic properties similar to their mNOS counterparts (10, 12–15). Only a few studies have explored the functional role of bacterial NOSs. NOSs from certain Streptomyces strains are involved in the nitration of a tryptophanyl moiety of thaxtomin, a dipeptide phytotoxin which interferes with plant cell wall synthesis (11, 16, 17). In contrast, NOS-derived NO appears to protect against oxidative damage in bacilli and staphylococci (18, 19).
In both mammals and plants, NO production is an important response for exposure to ultraviolet (UV) radiation (20–22). Deinococcus radiodurans (Dr) is especially adapted to survive UV radiation, ionizing radiation, desiccation, and oxidative damage (23, 24). Dr adaptation involves multiple protective mechanisms, including efficient homologous recombination among its 8–10 genome copies, a tight nucleoid organization, and unusually high intracellular Mn/Fe ratio, which can support/participate in protection against oxidative damage (23–26). Nevertheless, most of the implicated genes are similar to those found in other organisms, and the repair mechanisms themselves are not unusual (23–26).
We have previously demonstrated through biochemical means that the NOS from the radiation-resistant bacterium Dr (DrNOS) interacts with an unusual auxiliary tryptophanyl tRNA-synthetase (TrpRS II) (27); however, the significance of this association remains unclear (28, 29). Here, we have undertaken a genetic approach in an attempt to discover functions for DrNOS. These studies have revealed that the NO generated by DrNOS aids in the recovery of Dr from UV radiation damage.
Deletion of nos (Δnos) renders Dr more susceptible to UV radiation than the wild-type (wt) strain. The mutant can be rescued by genetic complementation, addition of NO donor compounds, and application of exogenous NO gas. Remarkably, the rescue is effective even when NO is supplemented hours after UV exposure. Furthermore, we observe that the Dr nos gene is induced by UV damage and causes a measurable increase in NO production within the cell. We further show that NO upregulates obgE, a gene for an essential GTPase involved in the regulation of many growth-related processes.
Results
Deletion of nos Affects Growth Recovery after UV Irradiation.
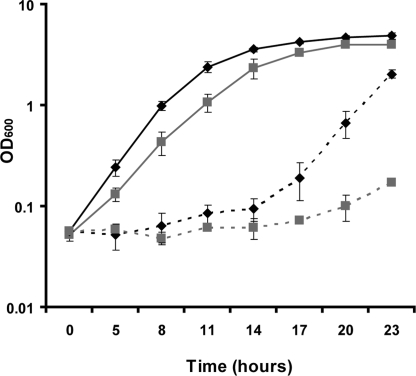
We produced single-gene deletions of nos, trpRS I, and trpRS II and double deletions of nos/trpRS I and nos/trpRS II in Dr strain R1 using allelic replacement. Deletions were confirmed with genomic PCR specific for the target and replacement genes, and with RNA transcript analysis by reverse transcriptase PCR (RT-PCR). The expression levels of the flanking genes do not change significantly under basal and post-irradiation conditions in the Δnos strain compared to the wt (Fig. S1). Δnos exhibited slightly slower growth compared to wt under rich media conditions (Fig. 1). Nonetheless, enhanced growth defects or differences in cell morphology were not observed when Δnos was subjected to a variety of stress conditions, including increased temperature, acidity, salinity, DNA damaging agents (methyl methanesulfonate, bleomycin), and oxidative stress. Furthermore application of hydrogen peroxide (H2O2) at concentrations as high as 1 M did not distinguish the mutant from wt (Fig. S2). However, Δnos did display a striking difference in the ability to grow after a 5 min exposure to polychromatic UV irradiation (30 mW/cm2) (Fig. 1). This UV exposure, which is lethal to non-UV resistant bacteria such as E. coli, was sufficient to cause extensive DNA shearing as resolved by DNA gel analysis and promoted upregulation of key DNA repair genes recA, uvrA, and uvsE (Fig. S3). Compared to wt, Δnos cell density measured by optical density (OD600 nm) was reduced by over 95% and required another 6 h (which corresponds to ≈4 doubling times under rich media conditions) to reach its exponential growth phase after UV radiation (Fig. 1). We quantified the growth recovery of the wt and Δnos strains in terms of colony-forming units (CFUs) by serial dilution of culture suspensions onto TGY/agar plates immediately after UV exposure. This analysis showed that Δnos produces 103–104 fewer CFUs visible to the naked eye, than wt 2 days after plating. Assays for cell viability indicated that irradiation does kill a substantial number of cells (≈40%) in both cases but the number of unlysed cells before and following irradiation was the same for wt and Δnos (Fig. S4). These results show that deletion of the nos gene does not lead to more cell death immediately following radiation but rather results in slower growth recovery.
Fig. 1.
Δnos is more susceptible to UV radiation than wt. Growth curves (monitored by OD600) are represented by black diamonds along a solid line for non-irradiated wt, gray squares along a solid line for non-irradiated Δnos, black diamonds along a dotted line for irradiated wt, and gray squares along a dotted line for irradiated Δnos. Irradiated cultures (OD ≈0.8) were exposed to polychromatic UV radiation (30 mW/cm2) before 1:100 dilution into TGY media. Data are plotted as mean ± SD of three independent experiments.
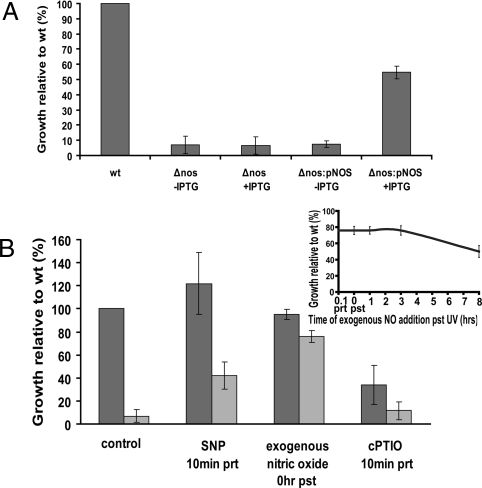
To ensure that the growth defect of Δnos subsequent to UV exposure was mediated by nos, a chloramphenicol-resistant expression plasmid under the control of an IPTG inducible promoter was introduced in the Δnos strain to form Δnos:pNOS. The expression level of nos from induced Δnos:pNOS is higher that of wt without UV exposure, but lower than that of wt with UV exposure (Fig. S5). However, we were able to induce the expression of the nos gene in trans and rescue the growth phenotype of Δnos to levels 55% of wt (Fig. 2A). Expression of recombinant proteins in Dr has only been achieved in a few cases due to the difficult of maintaining exogenous plasmids in the bacterium (30, 31). It should also be noted that selection with antibiotics following irradiation retards growth of uninduced Dr Δnos:pNOS compared Δnos. With these considerations, the complementation results suggest that loss of nos is likely the main reason for the growth defect of the mutant.
Fig. 2.
Nitric oxide plays a critical role in the recovery of Dr growth after UV radiation as monitored by OD600. (A) The complementation strain Δnos:pNOS was generated by introducing an IPTG inducible recombinant nos gene on expression plasmid p11530. Cells were grown to OD ≈0.2 and IPTG was added to induce the nos gene until the cell density reached OD ≈1.0 before irradiation. Uninduced Δnos and Δnos:pNOS served as controls. The cell growth (assessed by OD600) is shown as mean ± SD relative to irradiated wt cells at 22 h after exposure. The complemented strain grows more slowly than Δnos due to the added pressure of antibiotic selection. (B) Both wt (dark gray bars) and Δnos (light gray bars) cells were grown for 22 h with preincubation (prt) or postincubation (pst) of NO donors (SNP and exogenous NO) and NO scavenger (cPTIO) after 5 min of UV radiation. Control cells were grown in the absence of additive compounds. The relative growth at 22 h after UV was compared to that of wt cells, which were given a value of 100. Inset: Degree of NO rescue by the addition of exogenous NO at various times post irradiation. These values are set relative to the growth of wt cells post-irradiation. The data represents an average of three independent experiments ± SD.
In previous work we had found that DrNOS interacted with TrpRS II (27), so the susceptibility to UV radiation of Dr knockout for both TrpRS isoforms (ΔtrpRS I and II) was examined. There was no difference in the growth of ΔtrpRS II after irradiation while ΔtrpRS I showed 40% growth compared to that of wt (Fig. S6). This is not surprising as the catalytic parameters and sequence of TrpRS I indicate that it is the primary TrpRS in Dr (27). However, the viability of the trpRS I knockout did indicate that TrpRS II could act as a functional TrpRS. The double knockout ΔtrpRS IΔtrpRS II could not be obtained and was assumed to be lethal. The double knockouts ΔnosΔtrpRS II and ΔnosΔtrpRS I showed the same growth impairment as Δnos alone (Fig. S6).
Nitric Oxide Rescues Growth of Irradiated Δnos.
To establish a link between the presence of the nos gene and NO production in the susceptibility to UV radiation, we provided NO to Δnos and scavenged it in the wt strain. Pre-incubation with the NO donor 1 mM SNP (sodium nitroprusside) or addition of exogenous NO gas (5 μM final concentration in solution) rescued the growth of Δnos to 42% and 76% of wt levels, respectively (Fig. 2B). The NO donor compound NOC-7 (5 μM) also enhanced growth of Δnos following UV, but showed some detrimental effects on growth in the absence of UV, as did application of Angeli's salt, S-NO donors (e.g., glutathioneS-NO), and NO at concentrations exceeding 10 μM. Full recovery by chemical complementation could be hampered by a number of variables related to cell penetration, availability and the kinetics of NO release by these compounds with Dr. Addition of L-citrulline, the other product of NOS activity, ferrous/ferric cyanide (a non-NO containing derivative of SNP) and spermine (polyamine) did not enhance the growth of Δnos (Fig. S7). Interestingly, NO also rescued the modest growth phenotype of non-irradiated Δnos to within 76% of wt. Thus, the minor defect in Δnos under non-stress conditions is also significant. Conversely, addition of 100 μM NO scavenger cPTIO, [2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide] sensitized wt Dr to UV radiation by reducing growth to 34% after UV exposure and had no effect on growth of Δnos or non-irradiated cells (Fig. S8). These effects are consistent with salvageable concentrations of NO only being present in wt cells after UV exposure (see below); however, reduced growth of the wt after UV exposure could result from loss of function due to less NO or an increase in higher nitrogen oxides caused by reaction of NO with cPTIO. Remarkably, exogenous NO rescued growth of Δnos whether it was added 5–10 min before, during, or up to 8 h after irradiation (Fig. 2B, inset). In contrast, UV-induced oxygen radicals were scavenged within seconds of exposure (Fig. S9). These results suggest that the rescue by NO does not involve its reaction with, or effect on, unstable chemical species generated during UV exposure [e.g., reactive oxygen radicals (ROS)]. Additionally, this is consistent with the observation that the same numbers of wt and Δnos cells were viable after UV and hence the inability of Δnos to proliferate is not caused by UV-induced cell death, but rather a delay in resuming growth post-irradiation.
nos Is Upregulated by UV Irradiation.
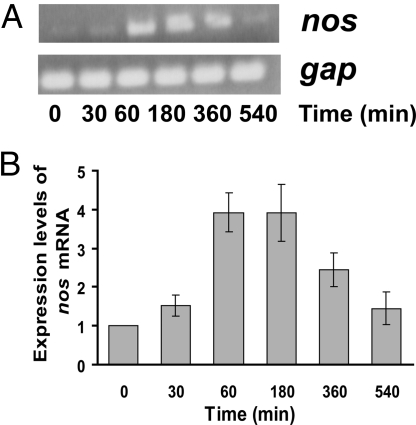
Considering the importance of the nos gene to recovery from UV radiation, we examined its expression pattern following UV treatment. The amount of nos transcript as determined by RT-PCR, substantially increased within 1 h after irradiation (relative to non-irradiated cultures) and remained roughly constant until decreasing 9 h post-irradiation at a time that slightly precedes the onset of log phase growth (Fig. 3). Upregulation of nos was evident as early as 30 min after UV exposure. This pattern of nos expression is similar to that of genes involved in DNA repair during damage responses. For example, when exposed to ionizing radiation, the recA recombinase mRNA level increases 30 min after UV exposure, is highest at 1.5 h and diminishes after 12 h (32). These results strengthen the hypothesis that the cell regulates nos levels in response to damage and requires its product NO in the repair or growth process. Notably, NO availability does not appear to regulate some obvious DNA repair genes associated with UV protection, as we found no major differences between Δnos and wt in the induction of recA, uvrA, or uvsE after UV irradiation (Fig. S3).
Fig. 3.
nos mRNA levels increase after UV irradiation. (A) RNA was extracted from non-irradiated cells (time = 0) and cells that were harvested 30, 60, 180, 360, and 540 min after exposure to UV. RNA was converted to cDNA before PCR with random primers. The top panel shows mRNA from the nos gene, and bottom is from a control gene, gap, glyceraldehyde 3-phosphate dehydrogenase, the expression of which is unaffected by UV and hence serves as a loading control. (B) The nos mRNA expression levels were quantified using image J software and plotted as average ± SD relative to non-irradiated cells, whose mRNA levels were set to 1.
UV Irradiation Induces Nitric Oxide Production.
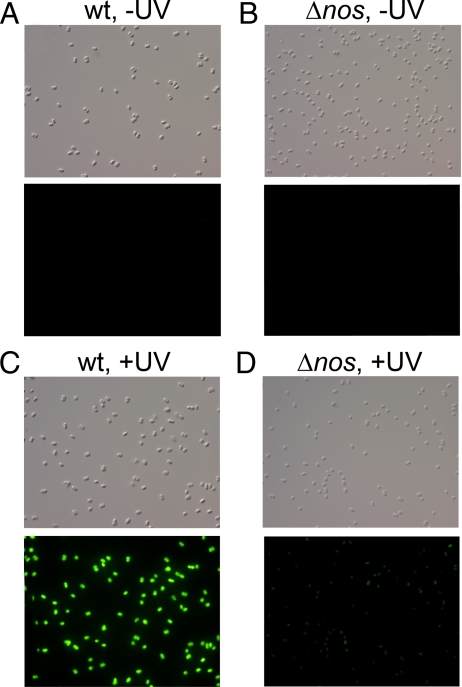
Nitric oxide production was detected in Dr cells using the intracellular highly NO-specific copper fluorescein probe (CuFL) (33). Neither wt (Fig. 4A) nor Δnos cells (Fig. 4B) showed significant CuFL fluorescence in the absence of UV irradiation. However, after irradiation a strong fluorescence from CuFL was detected in the wt background (Fig. 4C) compared to Δnos (Fig. 4D). Moreover, the timing of NO production was consistent with the expression profile of the nos gene, peaking at about 3 h post-irradiation and diminishing approximately 8 h post-irradiation. The low background fluorescence observed in Δnos could be attributed to non-specific reactivity of CuFL with products generated during UV treatment. In control experiments we did find that the fluorescence of CuFL increases slightly in the presence of H2O2 + UV + FeSO4. Much greater background effects were seen with the DAF fluorophore, which is sensitive to oxygen radicals, and like CuFL, is fluorescein based. Overall, the CuFL experiments show that DrNOS produces NO in response to UV radiation.
Fig. 4.
NO production by D. radiodurans after UV radiation requires nos. The cell permeable NO specific probe CuFL was used to detect nitric oxide in unexposed and UV-exposed wt and Δnos cells. Cells were irradiated, washed with PBS, incubated with 10 μM CuFL and photographed after 1 h. The top panel shows the differential interference contrast (DIC) images and the bottom panel the fluorescent images of wt without irradiation (A), Δnos without irradiation (B), wt with irradiation (C), and Δnos with irradiation (D).
NO Induces the Gene for ObgE, a GTPase that Regulates Growth.
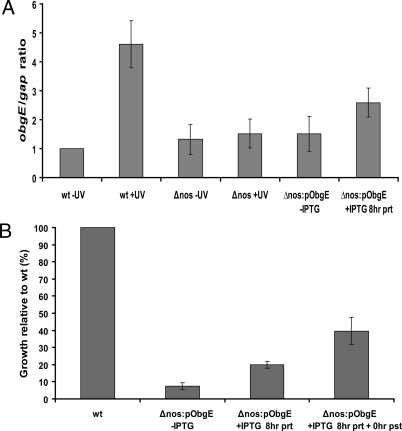
In an attempt to elucidate the mechanism by which DrNOS confers protection, we compared the transcription profiles (details to be reported elsewhere) of wt and Δnos cells with and without UV irradiation using microarrays (34). A number of candidate genes, whose expression levels were significantly increased in wt compared to Δnos after UV exposure were further investigated with quantitative real time PCR. In particular, obgE, which codes for an essential GTP binding protein in many bacteria was elevated 4.7× in irradiated wt cells, but not irradiated Δnos cells (Fig. 5A). Furthermore, treatment of the Δnos with exogenous NO upregulated the obgE gene, although not to the same extent as that observed in the wt with UV irradiation (Fig. S10A). Bacterial obgE genes are often essential and the GTPases they code for play important roles in growth regulation and cell proliferation (35, 36). Introduction of an inducible obgE gene in the Δnos background showed an improvement in bacteria growth recovery following UV treatment (Fig. 5B). This effect was dependent on the extent and level of obgE induction. If obgE expression is induced before UV irradiation the Δnos cells only recover to 20% of wt levels whereas up to 40% recovery was achieved when obgE expression was continuously induced. The obgE mRNA levels increased 2.6× in the overexpressed strain upon a single induction with IPTG (Fig. S10A); less than the 4.7× increase found for wt after UV exposure. Overexpression of obgE in unirradiated Δnos produced no significant change in growth (Fig. S10B). Thus, NO generated from NOS after UV exposure induces the obgE gene and production of the derived GTPase promotes cell growth.
Fig. 5.
obgE mRNA expression levels increase after UV irradiation and rescues Δnos. (A) Quantitative real-time PCR was used to determine the ratios of obgE/gap in wild-type and Δnos cells before and following irradiation, and the induction levels of the overexpression strain Δnos:pObgE. The average ± SEM is shown for three independent experiments. (B) The complementation strain Δnos:pObgE was generated by introducing an IPTG inducible recombinant obgE gene on expression plasmid p11530. Δnos:pObgE cells were grown to OD ≈1.0 and either no IPTG, IPTG only before irradiation or IPTG prior and post-irradiation (throughout growth) was added to induce the ObgE gene. Cells were evaluated at 22 h after UV exposure.
Discussion
This study demonstrates that DrNOS produces NO in its cognate organism Dr. Both Streptomyces NOS (17) and Bacillus anthracis NOS (37) have been shown to produce NO in vivo. However, unlike bacilli, Dr does not appear to contain the biosynthetic enzymes necessary to produce the mammalian NOS cofactor H4B (10, 38). In vitro, DrNOS (as with other bacterial NOSs) can use the alternative reduced pterin tetrahydrofolate (THF), a ubiquitous cofactor that can be generated by Dr (10, 29). Consistent with the binding of an alternative cofactor by bacterial NOSs, structural studies indicate that there is substantial variation in the region of the bacterial enzymes that recognize the pterin side chain (10). DrNOS must also produce NO in the absence of a flavodoxin reductase module, as the Dr genome lacks flavodoxin-like proteins. Work with B. subtilis proteins has demonstrated that a flavodoxin (YkuN), which is similar to the mNOS FMN domain, effectively donates electrons to B. subtilis NOS (39). However, deletion mutants of YkuN were not sensitive to oxidative stress, an assay used to monitor NOS activity, and B. subtilis NOS expressed recombinantly in E. coli still produces NO (19). These data suggest that bacterial NOSs do not require a specific reductase to produce NO. Whether a dedicated reductase is used by DrNOS or not, a flavodoxin-like protein is not necessary to generate NO.
NOS-derived NO enables Dr to better survive UV radiation, but it does not appear to provide protection from the other physiological stresses we have tested. Dr differs from B. subtilis as exposure of cells to NO before H2O2 treatment substantially increased resistance to oxidative damage in the latter (18). This protective effect is thought to result from inhibition of thiol reduction by cysteine S-nitrosation. Free reduced thiols fuel Fenton chemistry, which converts H2O2 into damaging hydroxyl radicals. Blocking free thiols with S-nitrosation may mitigate the Fenton reaction. In addition, NO activates a major B. subtilis catalase which further protects the cell against oxidative stress (18). Furthermore, NOS-derived NO was shown to protect the human pathogen B. anthracis from oxidative damage induced by macrophages (37). Given these observations, it was unexpected that Dr Δnos was not more susceptible to H2O2 than wt and that peroxide treatment did not induce nos gene expression (Fig. S2). Also, the fact that applications of NO up to 8 h post-irradiation, when oxygen radicals are no longer present in cells, induced growth reinforced the fact that NO is more than a general protector against oxidative stress in Dr. Our data suggest that in Dr, NO serves to initiate recovery or remove some impediment to growth in latent cells. As Dr has high intracellular Mn/Fe levels and highly active superoxide dismutases which protect the cells from oxidative damage (23, 40), NO may simply not be needed in this capacity.
UV radiation damages DNA by directly cross-linking pyrimidine bases and by generating radical species (often oxygen based) that can participate in a plethora of reactions, including DNA strand cleavage (41). In Dr, NO production does not upregulate the recombinase recA gene, the nucleotide-excision repair uvrA gene or the UV damage endonuclease uvsE gene, which all appear to be induced normally in the Δnos mutant. Why then is NOS-derived NO induced during UV exposure? And how does it aid in growth recovery? Although many mechanisms may be ultimately involved, we show here that NOS derived NO does upregulate the obgE gene. The functions of the ObgE GTPases are not well understood, but where investigated, they have been shown to impact a number of processes affecting growth. For example, in B. subtilis, ObgE participates in the regulation of DNA replication, the activation of the stress-response transcription factor σB, the monitoring of intracellular GTP levels and proper ribosome function (35). In E. coli, ObgE acts as a checkpoint control for chromosome segregation and subsequent cell cycle processes (36). In C. crescentus, the ObgE homolog, CgtA, is essential for cell viability and its gene expression is enhanced after UV irradiation of cells (35). In humans, the expression of the ObgE homolog Gbp45 correlates with cell proliferation (42). In Dr, not only is obgE upregulated by UV irradiation through NOS activity, but overexpression of ObgE substantially overcomes the growth defect caused by the Δnos mutant. Unlike complementation with nos, where induction before UV exposure is sufficient to maximize the effect on recovery, induction of obgE is required throughout the recovery and growth period to achieve the greatest benefit. IPTG-driven expression of obgE in the complemented strain is reduced compared to obgE induction in the wt after UV. Thus, the incomplete rescue of Δnos with pObgE may stem from either insufficient levels of ObgE, and/or other defects also caused by the loss of NO production. Nonetheless, NO acts to ultimately regulate gene expression important to damage recovery and cell proliferation in Dr, in part through the growth regulator ObgE. The timing of nos induction post-irradiation suggests that rather than playing a direct role in protection against UV radiation or preventing the damage it generates, NO signals to the cell to resume growth related processes after damage is well under repair or perhaps completed. ObgE has been implicated in the regulation of both protein production and DNA replication, either or both processes could be downstream targets of the NO signal.
Notably, many NO-responsive transcriptional regulators and sensor kinase systems have now been characterized in other bacteria (43, 44), and thus it is a strong possibility that NO could act as a regulatory signal in Dr. Dr has seven transcriptional regulators of the MerR class, (possibly an ortholog of SoxR), and two members of the LysR family, (possibly an ortholog of OxyR), paralogs of which are NO-responsive in other organisms (45).
It is possible that there are other NO-mediated responses in addition to obgE expression. NO can directly react with metallo-cofactors of transcription factors and other proteins and also lead to S-nitrosation of cysteine residues. This latter mechanism is known to regulate mammalian phosphatases, kinases, and transcription factors such as HIF-1 and NFκB (46). In mammals, UV irradiation increases inducible NOS (iNOS) levels in macrophages. NO released by iNOS S-nitrosates a specific cysteine residue on HIF-1α, which plays a key role in various inflammatory diseases and wound healing (20). Additionally, the mRNA expression of iNOS increases after UV-A radiation in human skin endothelial cells in the absence of cytokines (21). It may be more than coincidence that UV radiation elevates NO in both mammals and Dr through NOS induction.
Despite its importance in UV radiation recovery, NO may fulfill other functions in Dr. The nos gene is expressed during normal growth and the NOS protein is produced (27). Furthermore, the Δnos mutant shows slightly reduced growth in rich media. This defect is rescued by NO, but not by obgE overexpression. Thus, NO confers a growth advantage to Dr under normal conditions through a process that does not appear to involve regulation of ObgE. A continual benefit from NO would provide a constant selective pressure to maintain the nos gene.
So far the known functions of bacterial NOSs appear quite diverse, not unlike the varied roles played by the animal NOSs. In certain Streptomyces strains NOS participates in the nitration of a tryptophanyl moiety of the thaxtomin phytotoxins (11). However, NOS produces NO in excess of that needed for plant toxin synthesis and the excess NO diffuses from the cell (17). This feature of NO production may assist pathogenesis because NO is also a plant signaling molecule that plays a role in the growth of new root shoots, which are prime sites for bacterial infection (17, 47). Although NO protection against oxidative stress in bacilli and staphylococci involves changes to reduced thiol availability (18), it may also involve other factors, such as upregulation of stress-response factors or growth regulators. Further investigations into the NO-mediated survival mechanism of Dr may yet reveal commonalities in the above mechanisms as well as provide insight into UV radiation responses by other organisms.
Methods
Bacterial Strains and Growth Conditions.
Bacterial strain wild-type (wt) D. radiodurans R1 was obtained from the American Type Culture Collection (13939). Cells were grown in TGY (0.5% tryptone, 0.3% yeast extract and 0.1% glucose) at 30 °C or plated on TGY with 1.5% Bactoagar (Difco).
UV Treatment.
Two milliliter cells in a 3.5-mL quartz cuvette [optical density at 600 nm (OD600) 0.8–1.0] were exposed to polychromatic UV radiation (200–500 nm, 30 mW/cm2) from a mercury/xenon lamp for 5 min. Irradiated cells were then diluted 1:100 in fresh TGY and OD600 measured as a function of time. Cells were also plated and single colonies were counted at 24 and 48 h. Overall we found rates of growth as measured by optical density a more robust method for quantifying recovery from UV damage than colony counts. This is largely because it is difficult to completely kill Dr and as such even highly irradiated cultures will eventually produce colonies. To test the effects of NO donors and scavengers, wt cells were pre-incubated with 100 μM cPTIO [2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide] or 1 mM SNP (sodium nitroprusside) 10 min before UV irradiation. To evaluate the effect of NO addition post-irradiation, NO was bubbled until a final concentration of 5 μM into 1:100 diluted cells already exposed to UV irradiation after various lag times; cell density was then measured at 22 h post-exposure. The NO gas was passed anaerobically from a nitric oxide gas cylinder (Sigma) through concentrated NaOH to remove higher order nitrogen oxides before bubbling into buffer solution or media; aliquotes were taken and evaluated for NO concentration with the hemoglobin assay (48). For the overexpression experiments, the wt, Δnos, and complementation strains Δnos:pNOS, Δnos:pObgE were grown to OD600 0.2 induced when necessary with 10 mM IPTG, and grown for 9 h or OD600 1.0 before irradiation for 5 min. The cells were diluted 1:100 in TGY and allowed to recover for approximately 22 h. At least three, and in some cases more than eight, independent experiments were performed for each condition. BacLight (Molecular Probes) was used to assess the percent of viable cells before and following irradiation according to manufacturer's protocol.
Gene Disruption.
The gene nos was disrupted by targeted mutagenesis using techniques previously described (49). Briefly, the streptomycin resistance gene fused to the Dr katA promoter was cloned from the plasmid TNK103 (49). Genomic DNA sequences 1 kb upstream and downstream of nos (DR2597) were appended to the drug cassette by overlap extension. This process added HindIII and XhoI sites for cloning into Litmus-28 (New England Biolabs). The resulting plasmid was transformed into Dr R1 by electroporation. Recombinant cells of Δnos were selected on TGY plates containing 8 μg/mL streptomycin. The transformants were serially plated, isolated, and re-plated at least eight times on streptomycin. Disruption was followed by isolation of genomic DNA and PCR analysis for the native nos gene and the disruption cassette. Final isolates were tested by RT-PCR to confirm the lack of nos transcript. A similar protocol was followed to generate knockout mutants for the trpRS I (DR0558) and trpRS II (DR1093) genes. Plasmid TNK104, which contains the katA promoter fused to a hygromycin resistance gene, was used instead of TNK103 to generate ΔtrpRS I and ΔtrpRS II. Double knockouts of ΔnosΔtrpRS I and ΔnosΔtrpRS II were constructed by the recombination of individual plasmids made above in the Δnos strain containing the antibiotic resistance sequence and then selecting the clones on both streptomycin and hygromycin background.
Genetic Complementation.
A plasmid to express recombinant NOS in Dr R1 was constructed from the E. coli shuttle vector p11530, which contains a Pspac IPTG-inducible promoter and camR antibiotic marker (50). NOS gene fragment was amplified by PCR from pet15_NOS template (the coding sequence of which was initially isolated from genomic DNA) (10) and cloned into p11530 using PdiI and XhoI restriction sites generating pNOS. The plasmid was electroporated into the Δnos strain to form Δnos:pNOS and transformants were selected on TGY plates containing 3 μg/mL chloramphenicol. Reintroduction of the nos gene was confirmed by PCR. The same protocol and plasmid were used for overexpression of obgE in Δnos.
mRNA Expression.
Cells (10 mL, OD ≈0.8) were grown at 30 °C, UV-irradiated for 5 min, and harvested after 30, 60, 180, 360, and 540 min [control cells (t = 0) were treated similarly without irradiation]. Cells were resuspended in 100 μL Tris-EDTA and lysed by vortexing with 25 μL glass beads (Sigma). Total RNA was extracted from non-irradiated and irradiated cells using the RNEasy kit (Qiagen). DNA (0.5 μg) from each sample was treated with DNase-I (Promega), and converted to cDNA with First Strand Synthesis (Invitrogen) using random hexameric oligonucleotides following the manufacturer's protocol. PCR amplification of approximately 350 bp was carried out using 2 μL cDNA as template. Fluorescent imager and ImageJ software was used to quantify band intensities. Results are representative of at least three independent experiments. To measure obgE expression, the cDNA obtained above was subjected to quantitative real-time PCR (QRT-PCR) by the use of QuantiFast SYBR Green PCR Kit (Invitrogen) following manufacturer's protocol and data acquired using Applied Biosystems 7500 Real-Time PCR System.
Fluorescence Microscopy.
NO production was detected from cells with the cell permeable NO specific probe Cu(II)Fluorescein ligand (CuFL) (Strem Chemicals Inc.) as described (33). Cu(II)-fluorescein was freshly prepared by mixing the FL ligand (1 mM in DMSO) with CuCl2 (1 mM) in a 1:1 ratio. Cells grown to an OD600 ≈3 were washed with PBS (PBS) to remove TGY. Cells were UV irradiated for 5 min and then incubated with 10 μM CuFL for 1 h at room temperature. Cells were then washed with 1 mL PBS to remove excess CuFL and observed under the microscope. Images were obtained at the PCIC supported by TRIAD Foundation (NSF DBI-0618969) using an Olympus SZX-12 stereo fluorescence microscope equipped with a 63× objective lens in water, a I3 wavelength filter [excitation (450–490 nm) emission (515 nm)], and the Optronics MagnaFire acquisition software.
Microarrays.
Ten milliliters Dr wt and Δnos were harvested 1 h after exposure to UV irradiation for 5 min. Controls cells without UV irradiation were harvested similarly and RNA was extracted as mentioned previously. Microarray design and usage were carried out as previously described (34). Genes that showed a 2× difference (wt − Δnos, post-irradiation) in three experiments were selected for further analysis by RT-PCR.
Supplementary Material
Acknowledgments.
We thank J. Battista for the antibiotic plasmids pTNK103 and pTNK104; A. Bailone and M. Lidstrom for the complementation plasmid p11530; A. Holland for protocols, and R. Loria, J. Shapleigh, J. Peters, and members of the B.R.C. laboratory for helpful comments and discussions.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.J.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907262106/DCSupplemental.
References
- 1.Ignarro LJ. Nitric oxide as a unique signaling molecule in the vascular system: A historical overview. J Physiol Pharmacol. 2002;53:503–514. [PubMed] [Google Scholar]
- 2.Moncada S, Palmer RM, Higgs EA. Nitric oxide: Physiology, pathophysiology, and pharmacology. Pharmacol Rev. 1991;43:109–142. [PubMed] [Google Scholar]
- 3.Kerwin JF, Jr, Lancaster JR, Jr, Feldman PL. Nitric oxide: A new paradigm for second messengers. J Med Chem. 1995;38:4343–4362. doi: 10.1021/jm00022a001. [DOI] [PubMed] [Google Scholar]
- 4.Lipton SA. Physiology - Nitric oxide and respiration. Nature. 2001;413:118–121. doi: 10.1038/35093186. [DOI] [PubMed] [Google Scholar]
- 5.Barraud N, et al. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol. 2006;188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poole RK. Nitric oxide and nitrosative stress tolerance in bacteria. Biochem Soc Trans. 2005;33:176–180. doi: 10.1042/BST0330176. [DOI] [PubMed] [Google Scholar]
- 7.Alderton WK, Cooper CE, Knowles RG. Nitric oxide synthases: Structure, function and inhibition. Biochem J. 2001;357:593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rousseau DL, Li D, Couture M, Yeh SR. Ligand-protein interactions in nitric oxide synthase. J Inorg Biochem. 2005;99:306–323. doi: 10.1016/j.jinorgbio.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Griffith OW, Stuehr DJ. Nitric oxide synthases: Properties and catalytic mechanism. Annu Rev Physiol. 1995;57:707–736. doi: 10.1146/annurev.ph.57.030195.003423. [DOI] [PubMed] [Google Scholar]
- 10.Adak S, et al. Cloning, expression, and characterization of a nitric oxide synthase protein from Deinococcus radiodurans. Proc Natl Acad Sci USA. 2002;99:107–112. doi: 10.1073/pnas.012470099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kers JA, et al. Nitration of a peptide phytotoxin by bacterial nitric oxide synthase. Nature. 2004;429:79–82. doi: 10.1038/nature02504. [DOI] [PubMed] [Google Scholar]
- 12.Pant K, Bilwes AM, Adak S, Stuehr DJ, Crane BR. Structure of a nitric oxide synthase heme protein from Bacillus subtilis. Biochemistry. 2002;41:11071–11079. doi: 10.1021/bi0263715. [DOI] [PubMed] [Google Scholar]
- 13.Sudhamsu J, Crane BR. Bacterial nitric oxide synthases: What are they good for? Trends Microbiol. 2009;17:212–218. doi: 10.1016/j.tim.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Sudhamsu J, Crane BR. Structure and reactivity of a thermostable prokaryotic nitric-oxide synthase that forms a long-lived oxy-heme complex. J Biol Chem. 2006;281:9623–9632. doi: 10.1074/jbc.M510062200. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Raman CS, Martasek P, Masters BS, Poulos TL. Crystallographic studies on endothelial nitric oxide synthase complexed with nitric oxide and mechanism-based inhibitors. Biochemistry. 2001;40:5399–5406. doi: 10.1021/bi002658v. [DOI] [PubMed] [Google Scholar]
- 16.Fry BA, Loria R. Thaxtomin A: Evidence for a plant cell wall target. Phys Mol Plant Path. 2002;60:1–8. [Google Scholar]
- 17.Johnson EG, et al. Plant-pathogenic Streptomyces species produce nitric oxide synthase-derived nitric oxide in response to host signals. Chem Biol. 2008;15:43–50. doi: 10.1016/j.chembiol.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 18.Gusarov I, Nudler E. NO-mediated cytoprotection: Instant adaptation to oxidative stress in bacteria. Proc Natl Acad Sci USA. 2005;102:13855–13860. doi: 10.1073/pnas.0504307102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gusarov I, et al. Bacterial nitric-oxide synthases operate without a dedicated redox partner. J Biol Chem. 2008;283:13140–13147. doi: 10.1074/jbc.M710178200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li F, et al. Regulation of HIF-1alpha stability through S-nitrosylation. Mol Cell. 2007;26:63–74. doi: 10.1016/j.molcel.2007.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suschek CV, Bruch-Gerharz D, Kleinert H, Forstermann U, Kolb-Bachofen V. Ultraviolet A1 radiation induces nitric oxide synthase-2 expression in human skin endothelial cells in the absence of proinflammatory cytokines. J Invest Dermatol. 2001;117:1200–1205. doi: 10.1046/j.0022-202x.2001.01502.x. [DOI] [PubMed] [Google Scholar]
- 22.Qu Y. Nitric oxide functions as a signal in ultraviolet-B induced inhibition of pea stems elongation. Plant Science. 2006;170:994–1000. [Google Scholar]
- 23.Daly MJ, et al. Protein oxidation implicated as the primary determinant of bacterial radioresistance. Plos Biology. 2007;5:769–779. doi: 10.1371/journal.pbio.0050092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox MM, Battista JR. Deinococcus radiodurans - the consummate survivor. Nat Rev Microbiol. 2005;3:882–892. doi: 10.1038/nrmicro1264. [DOI] [PubMed] [Google Scholar]
- 25.Minsky A, Shimoni E, Englander J. Ring-like nucleoids and DNA repair through error-free nonhomologous end joining in Deinococcus radiodurans. J Bacteriol. 2006;188:6047–6051. doi: 10.1128/JB.01951-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zahradka K, et al. Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature. 2006;443:569–573. doi: 10.1038/nature05160. [DOI] [PubMed] [Google Scholar]
- 27.Buddha MR, Keery KM, Crane BR. An unusual tryptophanyl tRNA synthetase interacts with nitric oxide synthase in Deinococcus radiodurans. Proc Natl Acad Sci USA. 2004;101:15881–15886. doi: 10.1073/pnas.0405483101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buddha MR, Crane BR. Structure and activity of an aminoacyl-tRNA synthetase that charges tRNA with nitro-tryptophan. Nat Struc Mol Biol. 2005;12:274–275. doi: 10.1038/nsmb907. [DOI] [PubMed] [Google Scholar]
- 29.Reece SY, Woodward JJ, Marletta MA. Synthesis of nitric oxide by the NOS-like protein from Deinococcus radiodurans: A direct role for tetrahydrofolate. Biochemistry. 2009 doi: 10.1021/bi900385g. [DOI] [PubMed] [Google Scholar]
- 30.Meima R, Lidstrom ME. Characterization of the minimal replicon of a cryptic Deinococcus radiodurans SARK plasmid and development of versatile Escherichia coli-D-radiodurans shuttle vectors. App Environ Microbiol. 2000;66:3856–3867. doi: 10.1128/aem.66.9.3856-3867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Servinsky MD, Julin DA. Effect of a recD mutation on DNA damage resistance and transformation in Deinococcus radiodurans. J Bacteriol. 2007;189:5101–5107. doi: 10.1128/JB.00409-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu YQ, et al. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc Natl Acad Sci USA. 2003;100:4191–4196. doi: 10.1073/pnas.0630387100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim MH, Xu D, Lippard SJ. Visualization of nitric oxide in living cells by a copper-based fluorescent probe. Nat Chem Biol. 2006;2:375–380. doi: 10.1038/nchembio794. [DOI] [PubMed] [Google Scholar]
- 34.Chen H, et al. A novel OxyR sensor and regulator of hydrogen peroxide stress with one cysteine residue in Deinococcus radiodurans. PLoS ONE. 2008;3:e1602. doi: 10.1371/journal.pone.0001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Czyz A, Wegrzyn G. The Obg subfamily of bacterial GTP-binding proteins: Essential proteins of largely unknown functions that are evolutionarily conserved from bacteria to humans. Acta Biochim Pol. 2005;52:35–43. [PubMed] [Google Scholar]
- 36.Foti JJ, Schienda J, Sutera VA, Jr, Lovett ST. A bacterial G protein-mediated response to replication arrest. Mol Cell. 2005;17:549–560. doi: 10.1016/j.molcel.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 37.Shatalin K, et al. Bacillus anthracis-derived nitric oxide is essential for pathogen virulence and survival in macrophages. Proc Natl Sci USA. 2008;105:1009–1013. doi: 10.1073/pnas.0710950105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adak S, Wang Q, Stuehr DJ. Arginine conversion to nitroxide by tetrahydrobiopterin-free neuronal nitric-oxide synthase. Implications for mechanism. J Biol Chem. 2000;275:33554–33561. doi: 10.1074/jbc.M004337200. [DOI] [PubMed] [Google Scholar]
- 39.Wang ZQ, et al. Bacterial flavodoxins support nitric oxide production by Bacillus subtilis nitric-oxide synthase. J Biol Chem. 2007;282:2196–2202. doi: 10.1074/jbc.M608206200. [DOI] [PubMed] [Google Scholar]
- 40.Lipton MS, et al. Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc Natl Sci USA. 2002;99:11049–11054. doi: 10.1073/pnas.172170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goosen N, Moolenaar GF. Repair of UV damage in bacteria. DNA Repair (Amst) 2007;7:353–379. doi: 10.1016/j.dnarep.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 42.Kira Y, Nishikawa M. The identification and characterization of a new GTP-binding protein (Gbp45) involved in cell proliferation and death related to mitochondrial function. Cell Mol Biol Lett. 2008;13:570–584. doi: 10.2478/s11658-008-0023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spiro S. Regulators of bacterial responses to nitric oxide. REMS Microbiol Rev. 2007;31:193–211. doi: 10.1111/j.1574-6976.2006.00061.x. [DOI] [PubMed] [Google Scholar]
- 44.Price MS, Chao LY, Marletta MA. Shewanella oneidensis MR-1 H-NOX regulation of a histidine kinase by nitric oxide. Biochemistry. 2007;46:13677–13683. doi: 10.1021/bi7019035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghosal D, et al. How radiation kills cells: Survival of Deinococcus radiodurans and Shewanella oneidensis under oxidative stress. FEMS Microbiol Rev. 2005;29:361–375. doi: 10.1016/j.femsre.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 46.Marshall HE, Merchant K, Stamler JS. Nitrosation and oxidation in the regulation of gene expression. FASEB J. 2000;14:1889–1900. doi: 10.1096/fj.00.011rev. [DOI] [PubMed] [Google Scholar]
- 47.Neill SJ, Desikan R, Hancock JT. Nitric oxide signaling in plants. New Phytol. 2003;159:11–35. doi: 10.1046/j.1469-8137.2003.00804.x. [DOI] [PubMed] [Google Scholar]
- 48.Murphy ME, Noack E. Nitric oxide assay using hemoglobin method. Methods Enzymol. 1994;233:240–250. doi: 10.1016/s0076-6879(94)33027-1. [DOI] [PubMed] [Google Scholar]
- 49.Funayama T, et al. Identification and disruption analysis of the recN gene in the extremely radioresistant bacterium Deinococcus radiodurans. Mutat Res. 1999;435:151–161. doi: 10.1016/s0921-8777(99)00044-0. [DOI] [PubMed] [Google Scholar]
- 50.Lecointe F, Coste G, Sommer S, Ballone A. Vectors for regulated gene expression in the radioresistant bacterium Deinococcus radiodurans. Gene. 2004;336:25–35. doi: 10.1016/j.gene.2004.04.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.