When a single DNA molecule is stretched beyond its normal B-form contour length, it will undergo one of two types of conformational transitions at high or low force, depending on its attachment and ability to rotate. The structure of the DNA during these transitions is difficult to measure with traditional structural methods because it only occurs on a single molecule. More than a decade after its discovery, van Mameren et al. (1) have, in this issue of PNAS, directly visualized the mechanics of DNA overstretching for the first time by combining single-molecule force spectroscopy and fluorescence imaging. They show that, regardless of whether or not the molecule can freely rotate, when DNA is stretched to high force it will melt, as the work done by the force to increase the length of the DNA converts double-stranded duplex DNA into ssDNA.
A typical force-extension curve for torsionally relaxed DNA is shown in Fig. 1. As the DNA extension reaches its B-form contour length of 0.34 nm/bp, the force required to stretch the molecule increases rapidly as the double helix is straightened out and resists further increase in length. At ≈65 pN, very little additional force is required to increase the DNA length to ≈1.7 times its contour length. Upon initial inspection, the transition appears to be from dsDNA to ssDNA. However, when the transition was discovered, several puzzling facts appeared to preclude a melting model (2). The most important fact was that the two strands do not come apart when the transition reaches the contour length of ssDNA. Instead, the strands do not separate until forces >100 pN are exerted. Therefore, the transition was originally attributed to a conversion from B-form dsDNA to a new form of dsDNA termed “S-DNA,” in which the neighboring base pairs unstack but the base-pairing remains (3). This hypothesis was supported by early molecular dynamics simulations that reproduced a transition to a ladder-like form of DNA, although the predicted force was much higher than that observed experimentally (4).
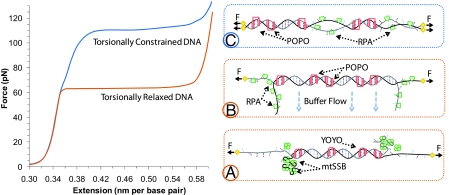
Fig. 1.
Schematic diagram of DNA force-induced melting transitions. The top curve (blue) shows the transition that occurs when DNA is attached by both strands and is therefore torsionally constrained (unable to rotate), and the bottom curve (orange) is observed in experiments in which the DNA is torsionally relaxed (able to rotate). (A) Illustration of force-induced melting detected with mtSSB (green), which wraps ssDNA. Here, the dsDNA remains in the middle of the molecule, labeled by the intercalator YOYO (red). The ssDNA is wrapped from the ends, resulting in a bright fluorescent spot at the location of the moving front of DNA melting. (B) Illustration of DNA force-induced melting detected by dsDNA regions labeled with POPO (red) and ssDNA appearing on the ends labeled by RPA (green). When buffer flows perpendicular to the molecule, the relaxed ssDNA extends out into solution. (C) Model for torsionally constrained force-induced melting. Alternating regions of intercalating POPO or ssDNA binding RPA show melting in domains throughout the molecule.
Rouzina and Bloomfield (5, 6) proposed a theoretical model for force-induced melting that quantitatively explained the observed overstretching transition. They also proposed that the additional transition at high forces represented the breaking of the last base pairs holding the strands together, which is necessarily nonequilibrium and higher force. The force-induced melting model predicted that solution conditions that alter the stability of DNA in thermal melting should also correspondingly alter the overstretching transition. Later experiments measuring overstretching as a function of salt, pH, and temperature quantitatively verified this prediction and showed that solution conditions that inhibit DNA annealing also induced strong hysteresis in the DNA stretching curves, as expected for melting (7). Later studies on DNA stretched in the presence of binding ligands were also consistent with this model (8).
Despite the thermodynamic evidence in favor of force-induced melting, several authors either suggested that the structure of overstretched DNA was still an open question (9, 10) or that aspects of DNA overstretching could not be explained by a melting model (11, 12). Because there was no clear definition of S-DNA, fits of the experimental data to some free parameters were often used as a definition of S-DNA (13, 14). However, there were no quantitative predictions made by an S-DNA model that were verified experimentally. One additional experiment that appeared to favor S-DNA was one in which DNA was attached to beads by both strands using magnetic tweezers (15). Instead of overstretching at 65 pN, another transition at 110 pN, which occurred over the same extension range as the overstretching transition (Fig. 1), was observed. Although this result is thermodynamically consistent with force-induced melting, the transition was instead interpreted to consist of a portion of S-DNA and a portion of overwound, melted P-DNA.
Because S-DNA was defined thermodynamically by the observed transition force, structural evidence of DNA melting was needed to fully establish overstretching as force-induced melting. The only prediction that could be attributed to an S-DNA model was that the DNA base pairs were not exposed during the transition. To test this prediction, Shokri et al. (16) used glyoxal modification of the DNA base pairs exposed during overstretching to show that the base pairs were indeed exposed to solution during overstretching. However, even this structural evidence was questioned as allegedly occurring because of nicks in the DNA backbone (17). Thus, despite many years of detailed study, DNA stretching experiments lacked the smoking gun to directly demonstrate what happens as the DNA is overstretched, until now.
To directly visualize the form of DNA undergoing force-induced structural transitions, van Mameren et al. (1) first stretched DNA through the transition in the absence of ligand and then moved the DNA molecule into a region of their flow cell that contained an intercalating dye (binding only to dsDNA) or a fluorescently labeled ssDNA binding (SSB) protein. This procedure captures and visualizes the double-stranded and single-stranded regions of the stretched DNA that occur in the absence of the ligand. Moreover, both stretched and relaxed single DNA strands can be visualized by using two different types of fluorescent SSB proteins, adding the capability to visually follow the ssDNA state.
van Mameren et al. (1) first stretched torsionally unconstrained DNA to various extensions within the low-force transition and then exposed the molecule to an intercalating dye, revealing the fraction of dsDNA at that extension. This dsDNA fraction corresponded directly to the fraction of the transition remaining to be stretched, and the dsDNA disappeared only from the ends of the molecule or from a nick if one existed. They repeated this experiment with labeled mitochondrial SSB (mtSSB) protein, which wraps ssDNA. The strands were wrapped at each end, forming a fluorescent spot that revealed the location of the moving front of melted DNA (Fig. 1A). To further illustrate this effect, they exposed overstretched DNA to the SSB protein RPA, which does not wrap ssDNA. By flowing liquid perpendicular to the axis of the DNA molecule, they showed that ssDNA was indeed peeling from the ends of the molecule, and the peeled fluorescently labeled ssDNA stuck out in the direction of the flow (Fig. 1B). Finally, they performed the same experiments using DNA that was attached to the beads by both strands, revealing the torsionally constrained transition at 110 pN. In this case, they observed multiple melted domains in the middle of the DNA molecule, with no evidence of separate S-DNA and P-DNA phases (Fig. 1C).
Although these experiments clearly demonstrate that dsDNA preferentially melts from its free ends, this situation can be altered in the presence of DNA binding ligands. For example, whereas SSB proteins will most likely favor end-melting, intercalators and other binding ligands that stabilize dsDNA will lead to DNA melting in the middle, similar to that observed for torsionally constrained DNA in this study. These details will be important for studies of the kinetics of protein–DNA interactions (8, 18).
Interestingly, van Mameren et al. (1) showed that at the very end of the overstretching transition at forces far above the melting plateau only a single short region of dsDNA remains. They attempted to explain the anomalously high force required to continue stretching dsDNA by the presence of this small fraction of dsDNA. However, this region of the DNA stretching curve depends on the DNA pulling rate (19, 20), so a complete understanding of this phenomenon will require further experimental and theoretical studies describing the kinetics of force-induced melting.
Because the authors first stretched DNA into the transition and then observed fluorescence due to ssDNA or dsDNA binding, the results present a picture of the transition in the absence of ligand. We must consider the possibility that the presence of the binding ligands quickly alters the equilibrium from an S-DNA state towards ssDNA, thereby changing the initial structure before it can be observed. However, the rapid appearance of the fluorescence pattern just seconds after the DNA was exposed to the intercalator, which remains unchanged upon further intercalator binding, makes conversion of S-DNA to ssDNA on this timescale extremely unlikely. Moreover, S-DNA would nucleate preferentially in the middle of the B-form DNA, while the melting occurs from its ends. Similarly, the transition force is independent of the presence of mtSSB and the patterns of ssDNA and dsDNA obtained in the presence of two different intercalators and SSB proteins were identical, making an active role of these ligands in converting S-DNA to ssDNA unlikely. Finally, the primary argument made to support an intermediate S-DNA state is the existence of the second high force transition after the overstretching transition. However, this second transition exists even when the DNA has been exposed to binding ligands, which would not be expected if the ligands converted S-DNA into ssDNA.
It is also possible that RPA might bind to P-DNA when torsionally constrained DNA is stretched, and this could not be distinguished from ssDNA. However, at 135 pN the authors see uniform coating of the molecule by RPA. Because P-DNA is a specific form of melted DNA that is overwound, the maximum fraction of P-DNA that could occur, assuming equilibrium with S-DNA, is 1/5 (15). The data appears to preclude such an interpretation.
By establishing force-induced DNA melting as the mechanism by which DNA undergoes conformational transitions under force, van Mameren et al. (1) have significantly advanced the field of single-molecule DNA stretching. Based on these results, DNA stretching studies with or without DNA binding ligands must be interpreted in terms of DNA melting. Therefore, it is untenable to simply attribute poorly understood features of DNA structural transitions to some unknown structure such as S-DNA. Instead, these features should be explained by quantitative and predictive models that can be related to measurable biophysical properties of DNA.
Footnotes
The authors declare no conflict of interest.
See companion article on page 18231.
References
- 1.van Mameren J, et al. Unraveling the structure of DNA during overstretching by using multicolor, single-molecule fluorescence imaging. Proc Natl Acad Sci USA. 2009;106:18231–18236. doi: 10.1073/pnas.0904322106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith SB, Cui Y, Bustamante C. Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 3.Cluzel P, et al. DNA: An extensible molecule. Science. 1996;271:792–794. doi: 10.1126/science.271.5250.792. [DOI] [PubMed] [Google Scholar]
- 4.Lebrun A, Lavery R. Modeling extreme stretching of DNA. Nucleic Acids Res. 1996;24:2260–2267. doi: 10.1093/nar/24.12.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rouzina I, Bloomfield VA. Force-induced melting of the DNA double helix 1. Thermodynamic analysis. Biophys J. 2001;80:882–893. doi: 10.1016/S0006-3495(01)76067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rouzina I, Bloomfield VA. Force-induced melting of the DNA double helix. 2. Effect of solution conditions. Biophys J. 2001;80:894–900. doi: 10.1016/S0006-3495(01)76068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams MC, Rouzina I, Bloomfield VA. Thermodynamics of DNA interactions from single-molecule stretching experiments. Acc Chem Res. 2002;35:159–166. doi: 10.1021/ar010045k. [DOI] [PubMed] [Google Scholar]
- 8.McCauley MJ, Williams MC. Optical tweezers experiments resolve distinct modes of DNA-protein binding. Biopolymers. 2009;91:265–282. doi: 10.1002/bip.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bustamante C, Bryant Z, Smith SB. Ten years of tension: Single-molecule DNA mechanics. Nature. 2003;421:423–427. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- 10.Strick TR, et al. Stretching of macromolecules and proteins. Rep Prog Phys. 2003;66:1–45. [Google Scholar]
- 11.Whitelam S, Pronk S, Geissler PL. There and (slowly) back again: Entropy-driven hysteresis in a model of DNA overstretching. Biophys J. 2008;94:2452–2469. doi: 10.1529/biophysj.107.117036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitelam S, Pronk S, Geissler PL. Stretching chimeric DNA: A test for the putative S-form. J Chem Phys. 2008;129:205101. doi: 10.1063/1.3009266. [DOI] [PubMed] [Google Scholar]
- 13.Cocco S, Yan J, Leger JF, Chatenay D, Marko JF. Overstretching and force-driven strand separation of double-helix DNA. Phys Rev E. 2004;70 doi: 10.1103/PhysRevE.70.011910. 011910. [DOI] [PubMed] [Google Scholar]
- 14.Storm C, Nelson PC. Theory of high-force DNA stretching and overstretching. Phys Rev E. 2003;67 doi: 10.1103/PhysRevE.67.051906. 051906. [DOI] [PubMed] [Google Scholar]
- 15.Léger JF, et al. Structural transitions of a twisted and stretched DNA molecule. Phys Rev Lett. 1999;83:1066–1069. [Google Scholar]
- 16.Shokri L, McCauley MJ, Rouzina I, Williams MC. DNA overstretching in the presence of glyoxal: Structural evidence of force-induced DNA melting. Biophys J. 2008;95:1248–1255. doi: 10.1529/biophysj.108.132688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danilowicz C, et al. The structure of DNA overstretched from the 5′5′ ends differs from the structure of DNA overstretched from the 3′3′ ends. Proc Natl Acad Sci USA. 2009;106:13196–13201. doi: 10.1073/pnas.0904729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart-Maynard KM, et al. Retroviral nucleocapsid proteins display nonequivalent levels of nucleic acid chaperone activity. J Virol. 2008;82:10129–10142. doi: 10.1128/JVI.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rief M, Clausen-Schaumann H, Gaub HE. Sequence-dependent mechanics of single DNA molecules. Nat Struct Biol. 1999;6:346–349. doi: 10.1038/7582. [DOI] [PubMed] [Google Scholar]
- 20.Clausen-Schaumann H, Rief M, Tolksdorf C, Gaub HE. Mechanical stability of single DNA molecules. Biophys J. 2000;78:1997–2007. doi: 10.1016/S0006-3495(00)76747-6. [DOI] [PMC free article] [PubMed] [Google Scholar]