Abstract
A rigorous numerical test of a hypothetical mechanism of a molecular motor should model explicitly the diffusive motion of the motor's degrees of freedom as well as the transition rates between the motor's chemical states. We present such a Brownian dynamics, mechanochemcial model of the coarse-grain structure of the dimeric, linear motor myosin V. Compared with run-length data, our model provides strong support for a proposed strain-controlled gating mechanism that enhances processivity. We demonstrate that the diffusion rate of a detached motor head during motor stepping is self-consistent with known kinetic rate constants and can explain the motor's key performance features, such as speed and stall force. We present illustrative and realistic animations of motor stepping in the presence of thermal noise. The quantitative success and illustrative power of this type of model suggest that it will be useful in testing our understanding of a range of biological and synthetic motors.
Keywords: Brownian dynamics, molecular motor, strain-dependent gating
Myosin V moves cargo through eukaryotic cells by walking in a hand-over-hand fashion along actin filaments (1, 2). Recent high-resolution, single-molecule experiments have provided an unprecedented level of insight into the physical mechanism underlying the coordinated steps of this biological motor. A growing level of support is beginning to form around a hypothetical stepping mechanism in which a step takes place through biased tethered diffusion of a detached head (3–5), and detachment of the two heads is coordinated through intramolecular strain, facilitating hand-over-hand forward stepping and avoiding motor detachment (6–9).
One can expect that the realization of such a stepping mechanism should require delicate mutual fine-tuning of the involved chemical rates, the motor's structural and mechanical properties, and the diffusive motion of the motor's degrees of freedom. For example, the rate of tethered diffusion (a function of motor size and stiffness) must be matched to the binding and unbinding rates of the motor head. Furthermore, the effectiveness of a strain-controlled gating mechanism depends on the detailed diffusive motion of the motor's internal degrees of freedom, because intramolecular strain is a quickly varying function of the instantaneous motor conformation.
To test the feasibility of a given stepping mechanism of dimeric molecular motors, it is therefore necessary to model the random thermal motion of the molecule's degrees of freedom as well as its mechanical and the kinetic properties. Most suited for this task are so-called “mechanochemical” models, which include a discrete set of chemical states and some coarse-grained mechanical features. Their advantage is that they are more detailed than discrete kinetic models that treat the motor as one or two “points” that discretely hop along a track and yet allow full simulation of hand-over-hand stepping, in contrast to computationally expensive full atomistic models that also require large numbers of parameters (10).
Two recent studies have applied mechanochemical approaches to myosin V (11, 12). Both of these models treat the neck domains as elastic filaments and predict transition rates between quasiequilibrium mechanical and chemical states based on the elastic potential energy of the dimer in each state. They correctly predict the predominant mechanochemical cycles as well as some characteristics of the load dependence of myosin V. However, neither of these models reports sensitivity to the choice of mechanical parameters, and each requires five or more free parameters. More importantly, these previous models either do not include conformation-dependent transition rates (12) or do not allow motor dissociation from actin (11) and, thus, do not test the role of strain-dependent gating in achieving motor processivity.
Here we present a mechanochemical model of myosin V that takes the key step forward to explicitly model the thermally activated motion and conformational changes of the molecule throughout all chemical and mechanical transitions. This approach enables us to make several contributions: First, we are able to test a hypothesized strain-dependent coordination mechanism in which head detachment is gated by neck conformation, which depends on molecular fluctuations and intramolecular strain (6–8). We confirm the feasibility of such a mechanism and explore how its effectiveness depends on the motor's mechanical properties. Second, the present model produces computational data amenable to direct comparison with recent single-molecule experimental data on myosin-V walking with unprecedented spatial and temporal resolution, including during the fleeting one-head-bound portion of the step (3, 5). Prior models have not been able to produce such data because they modeled reattachment by a discrete rate constant rather than simulating the motion of the detached head. Finally, but not least important, by modeling the motor's realistic thermal motion we are able to produce unique, highly illustrative animations [see supporting information (SI) Movies S1–S3] of motor stepping, which we believe will be very useful in raising the level of intuitive understanding of the physical conditions under which molecular motors operate.
An additional strength of our model is that its mechanical assumptions (detailed in the next section) are well-supported by the most recent experiments. Most input parameters of our model are taken directly from measurements, and we vary the small number of parameters that are not experimentally constrained to explore how they affect the performance of the motor. Our minimal model reproduces key performance features of myosin V, including the run length, the velocity, the relaxation (“working stroke”) distance, and the stall force. We are able to make predictions of parameters that are yet to be measured, including details of the molecule's flexibility, and to establish experimentally accessible performance characteristics (such as the relative probability for trailing head to leading head detachment) that can be used to test these predictions. We propose specific experiments that could test and fully constrain our model.
Model
Model Assumptions.
To construct a minimal model, we use the following assumptions:
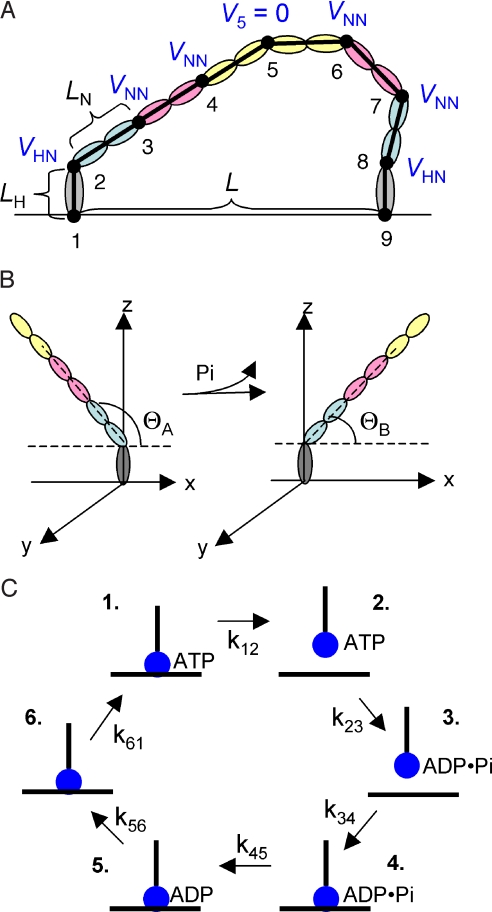
Semiflexible neck domains with three rigid segments. The neck domain of myosin V incorporates six tandem elements called IQ motifs (13, 14). A model of the neck domain based on the crystal structure, combined with FRET measurements of distances between IQ complexes, suggests that the neck domain can be thought of as three pairs of strongly interacting IQ motifs, with minimal interaction between adjacent pairs, allowing bending primarily at these junctures (Fig. 1A) (14).
State-dependent equilibrium angles of neck domains. We treat the juncture between each head and the adjacent neck domain (points 2 and 8 in Fig. 1A) as a semiflexible joint with an equilibrium angle that depends on the chemical state of the head [an assumption that is supported by EM images (15, 16)]. We define the equilibrium angle with respect to the direction of motility along the filament, which we denote as the +x direction. We assume that a forward rotation in the equilibrium angle (from θA to θB) between head and neck is tightly coupled to phosphate release from the head (Fig. 1B).
Free rotation about neck domain juncture. EM images (15) and observations of the motion of the detached neck domain by tracking an attached gold nanoparticle (3) or a microtubule filament (4) support a model in which a detached head and its adjacent neck domain undergo tethered diffusion about the juncture between neck domains, with free rotation about this joint (point 5 in Fig. 1A).
Binding to track. We assume that if a detached head has undergone ATP hydrolysis and diffuses close enough to a binding site to interact with it electrostatically (R < Rscreen; see Table 2), it will attach to this site. We make the simplification of treating the actin filament as a one-dimensional array of evenly spaced binding sites with separation L = 36 nm. We do not include nonspecific binding to actin or other electrostatic interactions with the filament away from the binding sites.
Fig. 1.
Mechanochemical model for myosin V. (A) The neck domains are modeled as chains of three rigid segments, each representing a pair of IQ motifs. Semiflexible joints between segments are characterized by a neck–neck potential VNN (see text), and the semiflexible joints to the head domains (gray) are characterized by VHN (points 2 and 8). The juncture between neck domains (point 5) is treated as a freely rotating joint (V5 = 0). (B) The equilibrium angle of the neck–head junctures (points 2 and 8 in A) is assumed to rotate forward (from θA to θB) upon phosphate release from the head. (C) In our simulations, each head cycles stochastically through the relatively long-lasting states in the observed monomer cycle (see Table 1 for transition rates).
Table 2.
Mechanical input parameters, as defined in Fig. 1
| Parameter | Value | Source |
|---|---|---|
| VNN | Tunable | Estimated range: see SI Text |
| VHN | Tunable | |
| θB | Tunable | |
| θA | 0.72π | Ref. 32 |
| V5 | 0 kT | Refs. 3 and 4 |
| LH | 5 nm | Ref. 33 |
| LN | 10 nm | Refs. 6 and 34 |
| L | 36 nm | Refs. 1, 16, 20, and 22 |
| γH | 6.2 × 10−8 pN/nm | Calculated estimate |
| γN | 4.1 × 10−8 pN/nm | Calculated estimate |
| Rscreen | 0.6 nm | Calculated estimate |
The drag coefficient of a head domain, γH, is calculated using Stoke's Law, and that of a neck domain segment, γN, is approximated by the transverse drag coefficient of a cylinder with length LN = 10 nm and diameter 2 nm. A detached head attaches to an actin binding site if the distance between the head and the site is less than Rscreen = 0.6 nm, equal to the Debye length for electrostatic screening in a fluid under physiological conditions. See SI Text for an estimated range of experimentally supported values of VNN.
Brownian Dynamics (BD) Simulations.
In our model, the myosin-V molecule is represented by a chain of eight rigid segments (two head domains and three segments in each of the two neck domains) (Fig. 1A). Each segment endpoint (i = 1–9) is a mathematical point with a spatial coordinate and an associated drag coefficient, corresponding to the estimated size and shape of each segment of the molecule (see Table 2). To simulate the motion of this semiflexible filament model, we use BD methods based on the overdamped Langevin equation:
where −γiṙi represents the viscous drag force on point i, −∇U(ri) represents the internal forces on point i due to bending or stretching the semielastic filament, and ξi(t) is a Gaussian-distributed white noise term that represents the thermal noise of the environment. The total potential energy of the semiflexible filament is given by
 |
where the first term represents the elastic energy associated with bending at the joints and the second term is a harmonic potential energy included to impose a fixed separation distance between adjacent points. This phenomenological expression for the bending energy, where φi is the angle between filament segments that meet at point i and φi0 is the equilibrium angle, reduces in the limit of small δ = φi − φi0 to an expression that is quadratic in δ, consistent with the form used in Allen and Tildsley (17). The equilibrium angle between adjacent neck domain segments is φi0 = 0, such that the elastic energy of a neck domain is minimized when the neck domain is straight. The stiffness parameter Vi characterizes the elastic stiffness of a joint (for example, if Vi = 100 kT, an elastic energy of U = Vi/2 = 50 kT is required to bend the joint 90° away from its preferred angle). The input parameters VNN and VHN represent the elastic stiffness of the neck domain joints and the head–neck junctures, respectively (Fig. 1A). In the second term of Eq. 2, K is the harmonic stiffness, r0 is the rest length, and rij is the separation distance between adjacent particles i and j, where j = i + 1. We used K = 105 kT and program time step dt = 1.175 × 10−10 s, which keeps fluctuations in interparticle distance small (〈∣rij − r0∣〉 < 0.01r0).
In each time step, we evaluate the positional change of each point in response to internal forces and external stochastic thermal forces. We allow each head to cycle stochastically through the relatively long-lasting states in the predominant observed cycle of a monomer (18, 19), as summarized in Fig. 1C. To limit the number of parameters in our minimal model, we include forward chemical transitions with experimentally measured transition rates (Table 1) and do not allow backward chemical transitions.
Table 1.
Chemical transition rates of a myosin-V monomer
| Rate | Value | Source |
|---|---|---|
| k12 | dt−1 | Ref. 18 |
| k23 | 700 s−1 | Ref. 18 |
| k34 | Diffusion limited | Model output |
| k45 | 200 s−1 | Refs. 19 and 21 |
| k56 | 15 s−1 | Refs. 8 and 21 |
| k61 | 1.6 μM−1·s−1, [ATP] = 1 mM | Refs. 18 and 31 |
These transition rates were used to determine the probability for a transition to occur in each simulation time step (dt = 1.175 × 10−4 s). For example, the probability for a transition from chemical state I to state II in a given time step is equal to k12dt.
Based on measurements and hypotheses in Veigel et al. (6) and Purcell et al. (8), we test a coordination mechanism in which the ADP release rate from a head is reduced if the adjacent neck domain has a rearward rotation with respect to the equilibrium angle, θB. Such a mechanism could enhance processivity, because intramolecular strain after phosphate release from the leading head would make the leading head unlikely to detach prematurely. We incorporate this gating mechanism by using the following expression for the ADP release rate, k56 (Fig. 1C):
 |
where θi is the instantaneous angle with respect to the filament of the neck domain at point “i”, where i = 2 or 8 (Fig. 1A). Here, the parameter ξ (in units of kT) quantifies the sensitivity of k56 to angular displacement from the preferred head-neck juncture angle, θB. Using a procedure described in SI Text, we calibrate the parameter ξ to match experimental measurements (Fig. S1). To adjust to the physical properties of the molecule, a different value for ξ is used for each set of values of VNN and VHN.
We use measured values for almost all of the mechanical and chemical input parameters (see Tables 1 and 2). Three mechanical parameters, namely θB, VNN, and VHN, are not well-known. Estimates of the persistence length, lp, of myosin-V neck domains in the literature varies over a wide range (lp = 100–400 nm) (12). Within the framework of our computational model, this range suggests that VNN is on the order of 10 kT < VNN < 100 kT [using VNN ∼ 2lp kT/LN, where LN is the length of a rigid segment in the neck domain (17)]. To further constrain these remaining three free parameters and to evaluate the validity of our model, we have at our disposal as many as five measured performance features of myosin V (Table 3).
Table 3.
Experimentally measured performance features against which we test our model
| Feature | Value | Source |
|---|---|---|
| Velocity | 550 (± 40) nm/s | Ref. 21 |
| Run length | 20–60 steps | Ref. 6 |
| Stall force | 2–3 pN | Refs. 22 and 23 |
| Relaxation distance, Rx | ≈25 (± 2) nm | Refs. 3 and 5–7 |
| Relaxation distance, Rz | ≈6 (± 4) nm | Ref. 5 |
The experiments cited used saturating ATP concentrations of at least 1 mM.
Results and Discussion
Dominant Dimeric Cycle.
We find that the model reproduces hand-over-hand transport of myosin V at a speed consistent with experimental measurements (for an illustration, see Movie S1) without the need for any fine-tuning of the mechanical parameters, as long as the neck domains have a forward-leaning preferred angle after phosphate release (0 < θB < 0.5π). Using the chemical cycle of the monomer as an input (Fig. 1C), we obtain as a model output that the dimer typically undergoes a mechanochemical cycle consistent with the kinetic model proposed in Rief et al. (20) (Fig. 2A). The mechanochemical states in this cycle can be represented, within the framework of our model, in terms of the three distinct mechanical states that are characterized by the equilibrium angles (θA or θB) of the leading and trailing heads (labeled I–III and illustrated in Fig. 2B). In Fig. 2C, we show a sample of the time-dependent motion of each head and of the neck domain juncture (point 5 in Fig. 1A) during hand-over-hand stepping. It is important to note that the model allows the possibility of backward steps, which can occur through leading head detachment followed by reattachment at a binding site in the −x direction.
Fig. 2.
The typical mechanochemical cycle resulting from our model is consistent with the kinetic model proposed in Rief et al. (20). (A and B) As each head undergoes kinetic state transitions (A), the myosin V dimer typically cycles through three distinct mechanical states, I–III (B). Color-coded boxes indicate the correspondence between mechanical states and kinetic states. (C) Data representative of hand-over-hand stepping as the motor undergoes the mechanochemical transitions illustrated in A and B, showing the time-dependent motion of each head domain (red and blue) and the neck domain juncture (black).
Relaxation Distance.
Several recent experiments demonstrate that the step average of myosin V (i.e., the center-of-mass motion averaged over a large number of synchronized steps) has two characteristic phases in the direction of transport (x direction): a fast 25-nm phase followed by a slower, 11-nm motion to complete the step (3, 5–7). The first phase can be interpreted as the average x motion corresponding to the relaxation of the attached neck domain, suggesting a relaxation distance Rx ≈ 25 nm (sometimes referred to as the “working stroke”). Also, Cappello et al. (5) measure an average motion (Rz ≈ 6 nm) away from the filament aligned in their step averages with the onset of the Rx ≈ 25 nm motion. In our model, these distances correspond to the change in the average position of the neck domain juncture (point 5 in Fig. 1A) that takes place after the motor transitions from mechanical state II to state III upon trailing head detachment (Fig. 3A).
Fig. 3.
Relaxation distance calculations. (A) Schematic of the x and z components of the relaxation distance, Rx and Rz. (B) Simulated values of Rx as a function of VNN/VHN, with the following values of θB from top to bottom: 0.2π, 0.3π, 0.4π, and 0.5π. (C) Rz from the same simulations as in B, with the same symbols to denote the value of θB.
For comparison between our simulations and the above experimental measurements, we assume a stiff coupling between cargo and motor, such that the motion of the juncture between neck domains (point 5 in Fig. 1A) corresponds approximately to the motion of the cargo measured experimentally. This approximation is reasonable for comparison to Cappello et al. (5), because they report that the relaxation time of this linkage in their experiment is very fast compared with the time scale of the step phases. By simulating the average conformation of the motor in mechanical states II and III, we find that the model approximately reproduces Rx = 25 nm as long as θB ≈ 0.4π for all tested values of VNN and VHN, although the optimal value of θB depends somewhat on the stiffness parameters (Fig. 3B). We also obtain Rz > 0, in agreement with experiment, when θB = 0.4π for all tested values of VNN and VHN (Fig. 3C). For the remainder of this study, we use θB = 0.4π unless otherwise noted.
Tethered Diffusion and Stall Force.
To characterize the tethered diffusional search during mechanical state III, we simulate the average time, τ, between when the trailing head detaches and when it reaches the next binding site through tethered diffusion. For a broad range of stiffness parameters, the tethered diffusion rate, 1/τ, under zero load is about an order of magnitude higher than the rate of ATP hydrolysis (k23 = 700 s−1) (Fig. 4A). This computational result provides a consistency test by showing that reattachment is limited by ATP hydrolysis rather than tethered diffusion, consistent with experiment. Our result also suggests that the effectiveness of the tethered diffusion stepping mechanism is not highly dependent on mechanical parameters, such as the stiffness, and that the processivity of the motor is not hampered by the “search” process. Because the tethered diffusion rate is fast compared to the relevant chemical transitions, the simulated motor speed is relatively independent of the mechanical input parameters and depends primarily on the chemical transition rates. For this reason, the speed of the motor is consistent with experimental measurements at saturating ATP concentrations (v ≈ 500–600 nm/s) (21) over the range of mechanical parameters tested.
Fig. 4.
Tethered diffusion and stall force. (A) Simulated values of the tethered diffusion rate, 1/τ, as a function of load force applied to the neck domain juncture (point 5 in Fig. 1A), for VNN = VHN = 100 kT (red triangles); and VNN = 100 kT, VHN = 105 kT (black squares). (B) Stall force, fstall, as a function of VNN for VHN = 100 kT (circles) and VHN = 10,000 kT (squares). Values are based on an exponential fit to the tethered diffusion rate versus load data exemplified in A. (C) fstall as a function of VHN for VNN = 100 kT.
For load forces applied to the cargo domain, several single-molecule experiments find stall forces in the range fstall = 2–3 pN, based on the distance from the center of a stationary optical trap at which the motor stalls and dissociates from actin (5, 22, 23). To enable comparison of our model to these data, we define fstall in a consistent way, namely as the detachment force at which the tethered diffusion rate is equal to the ADP release rate (on the order of k560 = 15 s−1, represented by the dashed line in Fig. 4A), because for large load forces, when 1/τ < k56, the motor is more likely to dissociate than to complete a step. (Note that this approach is likely to yield a conservative estimate of the stall force; if rearward load decreases the ADP release rate, the motor has a better chance to complete a step without premature dissociation and could thus take consecutive steps under higher loads than we estimate in Fig. 4.)
We find that fstall increases with stiffness parameters VNN and VHN, leveling off for high values of either parameter, as shown in Fig. 4 B and C. A range of VNN and VHN combinations can produce stall forces in the experimentally observed range fstall = 2–3 pN. However, if we assume that VNN is in the range 10–100 kT (based on estimated persistence lengths; see SI Text), then we need VNN in the upper end of this range, with VHN ≫ VNN, to produce a stall force of at least 2 pN (Fig. 4C).
Processivity and Gating.
In Fig. 5A we show data on the motor's dynamics with and without the stepping coordination introduced in Eq. 3 (for illustrative animations, see Movies S2 and S3). When ADP release is taken independent of conformation (no gating, k56 = k560), the leading head and trailing head detach with equal likelihood. Trailing head detachments are often followed by forward steps, whereas leading head detachments often result in futile cycles with no net progress (Fig. 5A). The mean number of steps the motor takes along actin before detaching is ≈11 steps (Fig. 5B), which falls short of the experimentally observed run lengths of 20–60 steps (7). When conformation-dependent gating of ADP release is included (Eq. 3), the mean run length increases significantly (Fig. 5C) and is within the margin of error of experimental estimates over the range of VHN and VNN we tested.
Fig. 5.
Run length and processivity simulations (θB = 0.4π, and VNN = VHN = 100 kT, unless noted). (A) Sample data of the x position (along the filament) of the neck domain juncture (point 5 in Fig. 1A), with (black) and without (red) conformation-dependent coordination between heads (Eq. 3). The small “spikes” apparent in the simulations without head–head coordination correspond to futile cycles of the motor in which a head detaches from actin and reattaches in its original location. (B) Histogram of run length (number of steps before detachment) for ≈250 individual walking simulations with uncoordinated steps (ξ = 0, Eq. 3). (C) Run length histogram with coordinated stepping (ξ = 4.0 kT, based on calibration to experiment for the mechanical parameters used here). (D) Mean run length as a function of VHN, averaged over ≈250 individual runs with conformation-dependent, head–head coordination (data points), and without head–head coordination (dashed line). (E) Average kinetic asymmetry, 〈λ〉, as a function of VNN, with the following values of VHN from bottom to top: 102, 103, 104, and 105 kT.
To continue to quantitatively assess the effectiveness of this angle-dependent gating mechanism (Eq. 3), we define kinetic asymmetry, λ, as the probability for ADP dissociation from the trailing head, relative to the total probability of ADP dissociation from either head:
 |
The kinetic asymmetry, λ(t), must be modeled numerically, because it depends on the instantaneous conformation of the motor (λ is highest when the leading neck domain is rotated in the rearward direction with respect to the preferred angle, θB). Because ADP release is the rate-limiting process for detachment of a myosin head from actin, λ is an approximate measure of the likelihood for trailing head detachment, relative to the overall probability for either head to detach.
BD simulations of the motor in mechanical state II demonstrate that the average kinetic asymmetry, 〈λ〉, increases with increasing VNN, because the rearward rotation of the leading neck domain increases as the neck domains become stiffer (Fig. 5E), making the gating mechanism more effective. In the limit of large VHN (which produces the best stall force performance in our model), the value of 〈λ〉 depends very little on VHN and is primarily a function of VNN. If future experiments show that the head–neck juncture can be treated as a very stiff joint (VHN > 103 kT), then comparing a measurement of the ratio of leading to trailing head detachment with the theoretical prediction of 〈λ〉 could indicate whether the neck domains of myosin V are relatively flexible (VNN ≈ 10 kT) or relatively stiff (VNN ≈ 100 kT).
Concluding Remarks.
In conclusion, the known kinetic, mechanical, and structural parameters of myosin V (Tables 1 and 2) interplay with thermal noise in such a way that the feasibility of the hypothetical stepping mechanism for myosin V was demonstrated: Individual hand-over-hand steps occur through a process of biased tethered diffusion of a detached head, and processive hand-over-hand stepping is facilitated by a conformation-dependent intramolecular coordination mechanism that reduces the likelihood of leading head detachment. Furthermore, we find that our model reproduces the measured relaxation distance and stall force of myosin V particularly well if we choose the values θB = 0.4π, VNN ≈ 100 kT, and VHN ≫ VNN.
By varying the few unfixed parameters, we explore the sensitivity to these parameters and gain a fuller understanding of the range of possible motor performance. For example, a previous model assumes completely rigid state-dependent attachment angles and concludes as a result of this assumption that energetic constraints prevent the leading-head working stroke from occurring before trailing head detachment (12). In contrast, our model treats the head–neck junctures as elastic joints of variable stiffness and thus allows the motor to transition into a mechanochemical state characterized by a “telemark” configuration, similar to those that have been observed in some EM images (16).
The following set of experiments could completely constrain this model: (i) If the average run length and the relevant transition rates under consistent solution conditions were measured more precisely, it would be possible to further constrain the head–neck stiffness, VHN (see Fig. 5D). (ii) In our model, if the head–neck juncture is very stiff, the trailing/leading head detachment ratio depends only on VNN (Fig. 5E), which means that a measurement of this performance feature would amount to a measurement of VNN. To experimentally determine the trailing/leading head detachment ratio, high temporal resolution is needed to detect the fleeting events in which the leading head detaches and reattaches in its original place. Recent high-resolution experimental studies such as Cappello et al. (5) show evidence of such events and provide a promising route toward more stringent performance tests of this model. Detailed comparison between our computational data and the experimental data of Cappello et al. (5) is necessary.
The model developed here is particularly powerful because it combines a limited set of kinetic and mechanical parameters and explicitly models diffusion and conformational changes due to thermal noise. The quantitative conclusions enabled by such a well-constrained model and the illustrative nature of resulting animations (see Movies S1–S3 and Table S1) will be useful for many other motor systems. For example, a similar approach could be taken to study the strain-dependent gating of kinesin-1 (24) and to help interpret recent high-resolution observations of the unbound head of kinesin-1 during steps (25). The model could be extended to include reverse chemical rate transitions to study the physical mechanism of the recently observed processive backward stepping of myosin V under superstall load forces (26). This type of model could also be used to model the dynamics of molecular motors at filament intersections (27, 28), to study the more complex cooperative gating of multiple motors attached to a single cargo (29), or in the design of synthetic molecular motors (30).
Supplementary Material
Acknowledgments.
We thank Giovanni Cappello, Alex Dunn, Steven Lade, Alf Månsson, Michael Plischke, and Martin Zuckermann for helpful and interesting discussions. This work was supported by National Science Foundation CAREER Grant 0239764 and Integrative Graduate Education and Research Traineeship Grant DGE-0549503 and by the use of the Bugaboo computing facility and the Research in the Mathematical and Computational Sciences Centre of Simon Fraser University, which are funded in part by the Canada Foundation for Innovation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908192106/DCSupplemental.
References
- 1.Yildiz A, et al. Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 2.Warshaw DM, et al. Differential labeling of myosin-V heads with quantum dots allows direct visualization of hand-over-hand processivity. Biophys J. 2005;88:L30–L32. doi: 10.1529/biophysj.105.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunn AR, Spudich JA. Dynamics of the unbound head during myosin-V processive translocation. Nat Struct Mol Biol. 2007;14:246–248. doi: 10.1038/nsmb1206. [DOI] [PubMed] [Google Scholar]
- 4.Shiroguchi K, Kinosita K. Myosin V walks by lever action and Brownian motion. Science. 2007;316:1208–1212. doi: 10.1126/science.1140468. [DOI] [PubMed] [Google Scholar]
- 5.Cappello G, et al. Myosin V stepping mechanism. Proc Natl Acad Sci USA. 2008;104:15328–15333. doi: 10.1073/pnas.0706653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veigel C, Wang F, Bartoo ML, Sellers JR, Molloy JE. The gated gait of the processive molecular motor, myosin V. Nat Cell Biol. 2002;4:59–65. doi: 10.1038/ncb732. [DOI] [PubMed] [Google Scholar]
- 7.Veigel C, Schmitz S, Wang F, Sellers JR. Load-dependent kinetics of myosin V can explain its high processivity. Nat Cell Biol. 2005;7:861–869. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- 8.Purcell TJ, Sweeney HL, Spudich JA. A force-dependent state controls the coordination of processive myosin V. Proc Natl Acad Sci USA. 2005;102:13873–13878. doi: 10.1073/pnas.0506441102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trybus KM. No strain, no gain. Nat Cell Biol. 2005;7:861–869. doi: 10.1038/ncb0905-854. [DOI] [PubMed] [Google Scholar]
- 10.Vilfan A. Five models for myosin V. Frontiers Biosci. 2009;14:2269–2284. doi: 10.2741/3378. [DOI] [PubMed] [Google Scholar]
- 11.Lan G, Sun SX. Dynamics of myosin-V processivity. Biophys J. 2005;88:999–1008. doi: 10.1529/biophysj.104.047662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vilfan A. Elastic lever-arm model for myosin V. Biophys J. 2005;88:3792–3805. doi: 10.1529/biophysj.104.046763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheney RE, et al. Brain myosin V is a two-headed unconventional myosin with motor activity. Cell. 1993;75:13–23. doi: 10.1016/S0092-8674(05)80080-7. [DOI] [PubMed] [Google Scholar]
- 14.Terrak M, Rebowski G, Lu RC, Grabarek Z, Dominguez R. Proc Natl Acad Sci USA. 2005;102:12718–12723. doi: 10.1073/pnas.0503899102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burgess S, et al. The prepower stroke conformation of myosin V. J Cell Biol. 2002;159:983–991. doi: 10.1083/jcb.200208172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker ML, et al. Two-headed binding of a processive myosin to F-actin. Nature. 2000;405:804–807. doi: 10.1038/35015592. [DOI] [PubMed] [Google Scholar]
- 17.Allen MP, Tildsley DJ. Computer Simulations of Liquids. New York: Oxford Univ Press; 1987. [Google Scholar]
- 18.De La Cruz EM, Wells AL, Rosenfeld SS, Ostap EM, Sweeney HL. The kinetic mechanism of myosin V. Proc Natl Acad Sci USA. 1999;96:13726–13731. doi: 10.1073/pnas.96.24.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De La Cruz EM, Wells AL, Sweeney HL, Ostap EM. Actin and light chain isoform dependence of myosin-V kinetics. Biochemistry. 2008;39:14196–14202. doi: 10.1021/bi001701b. [DOI] [PubMed] [Google Scholar]
- 20.Rief M, et al. Myosin-V stepping kinetics: A molecular model for processivity. Proc Natl Acad Sci USA. 2002;97:9482–9486. doi: 10.1073/pnas.97.17.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baker JE, et al. Myosin V processivity: Multiple kinetic pathways for head-to-head coordination. Proc Natl Acad Sci USA. 2004;101:5542–5546. doi: 10.1073/pnas.0307247101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehta AD, et al. Myosin-V is a processive actin-based motor. Nature. 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 23.Uemura S, Higuchi H, Olivares AO, De La Cruz EM, Ishiwata S. Mechanochemical coupling of two substeps in a single myosin-V motor. Nature Struct Mol Biol. 2004;11:877–883. doi: 10.1038/nsmb806. [DOI] [PubMed] [Google Scholar]
- 24.Rosenfeld SS, Fordyce PM, Jefferson GM, King PH, Block SM. Stepping and stretching. How kinesin uses internal strain to walk processively. J Biol Chem. 2003;278:18550–18556. doi: 10.1074/jbc.M300849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guydosh NR, Block SM. Direct observation of individual kinesin head motions. Biophys J. 2009;96:215. (abstr) [Google Scholar]
- 26.Gebhardt JCM, Clemen AE-M, Jaud J, Rief M. Myosin-V is a mechanical ratchet. Proc Natl Acad Sci USA. 2006;103:8680–8685. doi: 10.1073/pnas.0510191103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross JL, Ali MY, Warshaw DM. Cargo transport: Molecular motors navigate a complex cytoskeleton. Curr Opin Cell Biol. 2008;20:41–47. doi: 10.1016/j.ceb.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snider J, et al. Intracellular actin-based transport: How far you go depends on how often you switch. Proc Natl Acad Sci USA. 2004;101:13204–13209. doi: 10.1073/pnas.0403092101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ali MY, Lu H, Bookwater CS, Warshaw DM, Trybus KM. Myosin V and kinesin act as tethers to enhance each others' processivity. Proc Natl Acad Sci USA. 2008;105:4691–4696. doi: 10.1073/pnas.0711531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bromley EHC, et al. The tumbleweed: Toward a synthetic protein motor. HFSP J. 2009;3:204–212. doi: 10.2976/1.3111282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yengo CM, De La Cruz EM, Safer D, Ostap EM, Sweeney HL. Kinetic characterization of the weak binding states of Myosin V. Biochemistry. 2002;41:8508–8517. doi: 10.1021/bi015969u. [DOI] [PubMed] [Google Scholar]
- 32.Forkey JN, Quinlan ME, Shaw MA, Corrie JET, Goldman YE. Three-dimensional structural dynamics of myosin V by single-molecule fluorescence polarization. Nature. 2003;422:399–404. doi: 10.1038/nature01529. [DOI] [PubMed] [Google Scholar]
- 33.Liu J, Taylor DW, Krementsova EB, Trybus KM, Taylor KA. Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography. Nature. 2006;442:208–211. doi: 10.1038/nature04719. [DOI] [PubMed] [Google Scholar]
- 34.Moore JR, Krementsova EB, Trybus KM, Warshaw DM. Myosin V exhibits a high duty cycle and large unitary displacement. J Cell Biol. 2001;155:625–635. doi: 10.1083/jcb.200103128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.