Abstract
Reactive oxygen species (ROS) interact with DNA, frequently generating highly mutagenic 7,8-dihydro-8-oxoguanine (8-oxo-G) lesions. Replicative DNA polymerases (pols) often misincorporate adenine opposite 8-oxo-G. The subsequent repair mechanism allowing the removal of adenine and formation of C:8-oxo-G base pair is essential to prevent C:G to A:T transversion mutations. Here, we show by immunofluorescence experiments, in cells exposed to ROS, the involvement of MutY glycosylase homologue (MUTYH) and DNA pol λ in the repair of A:8-oxo-G mispairs. We observe specific recruitment of MUTYH, DNA pol λ, proliferating cell nuclear antigen (PCNA), flap endonuclease 1 (FEN1) and DNA ligases I and III from human cell extracts to A:8-oxo-G DNA, but not to undamaged DNA. Using purified human proteins and a DNA template, we reconstitute the full pathway for the faithful repair of A:8-oxo-G mispairs involving MUTYH, DNA pol λ, FEN1, and DNA ligase I. These results reveal a cellular response pathway to ROS, important to sustain genomic stability and modulate carcinogenesis.
Keywords: base excision repair, DNA ligase I, DNA ligase III, oxidation damage
One of the often generated oxidative DNA lesions, upon exposure of the cells to reactive oxygen species (ROS) is 7,8-dihydro-8-oxoguanine (8-oxo-G). The estimated steady-state level of 8-oxo-G lesions is ≈103 per cell/per day in normal tissues and up to 105 lesions per cell/per day in cancer tissues (1). The presence of 8-oxo-G on the replicating strand leads to frequent (10–75%) misincorporations of adenine opposite a lesion (formation of A:8-oxo-G mispairs), subsequently resulting in C:G to A:T transversion mutations (2). These mutations are one of the most predominant somatic mutations in lung, breast, ovarian, gastric, and colorectal cancers (3). Thus, human cells require a base excision repair (BER) pathway ensuring correct and efficient repair of A:8-oxo-G mismatches to reduce the mutational burden of ROS. MutY glycosylase homologue (MUTYH) is initiating this repair by recognizing A:8-oxo-G mispair and removing the A (4, 5). During subsequent BER, a specialized repair DNA polymerase (pol) that will catalyze with high preference the accurate (incorporation of dCTP) bypass of 8-oxo-G is needed. In vitro studies have indicated that several DNA pols may be implicated in BER (6–8). DNA pol β, a member of DNA pol family X, was shown to be the major enzyme involved in gap filling (9–11), thus playing a central role in BER (12, 13). Another member of the DNA pol family X, DNA pol λ (14) has been implicated in BER (6, 15), nonhomologues end joining (16, 17) and translesion synthesis (18–20). We have recently shown (18, 21) that DNA pol λ is very efficient in the accurate bypass of 8-oxo-G lesion both on primed and 1-nucleotide (nt) gapped DNA templates. In the presence of the auxiliary proteins, replication protein A (RP-A) and proliferating cell nuclear antigen (PCNA), DNA pol λ incorporates dATP opposite 8-oxo-G with a very low frequency (10−3). At the same time, these two auxiliary proteins prevent binding of DNA pol β to 1-nt gapped 8-oxo-G template. Overall the presence of RP-A and PCNA results in a 145-fold more efficient DNA pol λ than DNA pol β incorporation of dCTP opposite 8-oxo-G on 1-nt gaps (21). Although these data implicate that DNA pol λ could play an important role, the mechanism ensuring accurate and efficient repair of A:8-oxo-G mismatches in human cells stays to be elucidated.
In the present work we show a key role of MUTYH and DNA pol λ in the repair of 8-oxo-G. Additionally we identify the critical repair components, by specific recruitment of MUTYH, DNA pol λ, PCNA, flap endonuclease 1 (FEN1), and DNA ligases I and III from human whole cell extracts (WCEs) to A:8-oxo-G DNA. Using purified human proteins and an 8-oxo-G specificity assay, we prove that DNA pol δ preferentially forms A:8-oxo-G mispair during replication and that MUTYH recognizes this mispair thereby generating an apurinic (AP) site, subsequently processed by apurinic endonuclease 1 (APE1). We show that on the newly formed 1-nt gapped template, in the presence of auxiliary proteins RP-A and PCNA, DNA pol λ preferentially incorporates dCTP opposite 8-oxo-G and additionally elongates by adding 1 nt, thereby creating a short 1-nt flap. Interestingly, no elongation occurs when in the rare cases DNA pol λ synthesizes an A:8-oxo-G mispair. Finally, we find that DNA ligase I in the presence of flap endonuclease 1 (FEN1) ligates 2-fold better a correct C:8-oxo-G product of DNA pol λ repair synthesis than an incorrect A:8-oxo-G product. In summary, our findings provide a mechanism for the accurate repair of highly mutagenic A:8-oxo-G mispairs, important to prevent mutagenesis and sustain genomic stability.
Results
Exposure to ROS Leads to the Colocalization of MUTYH and DNA Polymerase λ with the Sites of Oxidative DNA Damage and to the Increase in Their Protein Levels.
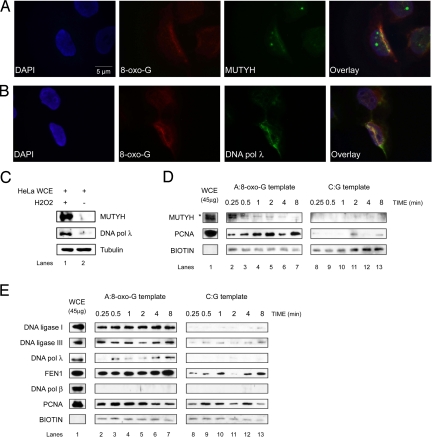
We introduced oxidative DNA damage in synchronized HeLa cells (Fig. S1A), by UVA laser microirradiation (Fig. 1 A and B) or H2O2 treatment (Fig. 1C and Fig. S1), and tested whether MUTYH and DNA pol λ accumulate at the sites of damage. Immunofluorescence analysis revealed that both MUTYH (Fig. 1A and Fig. S1C) and DNA pol λ (Fig. 1B and Fig. S1E) efficiently colocalize with the sites of oxidative DNA damage, such as 8-oxo-G lesion sites. Recently recruitment of FEN1, X-ray repair cross-complementing 1 protein (XRCC1) and PCNA was reported in a similar experimental setup (22). Additionally, H2O2 treatment of HeLa cells caused a dramatic increase in the protein levels of MUTYH and DNA pol λ (Fig. 1C). These data clearly suggest activation of MUTYH/DNA pol λ-dependent repair pathway upon induction of oxidative DNA damage.
Fig. 1.
Recruitment of BER proteins to the sites of oxidative DNA damage. The experiments were done under the conditions as specified in Materials and Methods. (A and B) Fluorescent microscope images of 355-nm laser-irradiated HeLa cells synchronized in G1/S phase and stained with antibodies against 8-oxo-G and MUTYH (A) or DNA pol λ (B). Colocalization was observed in 50% of the analyzed cells. (C) Immunoblot analysis of MUTYH and DNA pol λ protein level, 5 h upon recovery, in HeLa whole cell extract (WCE) treated with and without H2O2. (D and E) Protein recruitment assay using A:8-oxo-G (Left) or C:G (Right) biotinylated hairpin substrate and HeLa WCE in the absence (D) or presence (E) of Mg2+ for the times indicated (*, the higher molecular weight band corresponds to MUTYH localized in mitochondria). Only proteins that bind to hairpin substrate can be cross-linked and visualized by immunoblotting.
MUTYH, DNA Polymerase λ, PCNA, FEN1, and DNA Ligases I and III Are Specifically Recruited to DNA Containing A:8-oxo-G Mispair.
To identify BER proteins directly involved in the repair of A:8-oxo-G mispairs, we used a reversible cross-linking approach (23). The 3′-biotinylated oligonucleotides containing a hairpin loop and a single A:8-oxo-G mispair or C:G base pair (Fig. S2) were incubated with HeLa WCE in the presence of a cross-linking agent. Bound proteins were released, separated and detected by immunoblotting. During incubation of the A:8-oxo-G substrate with WCE, MUTYH, removes A from the substrate DNA, and APE1 incises the AP site, creating a 1-nt gap. This can be further processed by DNA pols β or λ and DNA ligase IIIα/XRCC1 or FEN1 and DNA ligase I. Because the recruitment of glycosylases to DNA is considered to be very fast, we first omitted Mg2+ from the reactions and observed a rapid recruitment (within 0.25 min of incubation) of MUTYH and a slower, but very robust recruitment of PCNA (Fig. 1D). Cross-linking of hairpin DNA substrates with WCE in the presence of Mg2+ resulted in a strong damage-specific recruitment of DNA ligases I and III, FEN1, as well as DNA pol λ (Fig. 1E). Interestingly, under the same conditions we were not able to detect the recruitment of DNA pol β to either A:8-oxo-G or to a control undamaged DNA substrate, although the protein was present in the WCE (Fig. 1E). Recruitment of PCNA in the presence of Mg2+ was damage-unspecific, which was expected because PCNA plays a role in variety of cellular processes besides DNA repair (24, 25). These results indicated a direct role of MUTYH, DNA pol λ, PCNA, FEN1, and DNA ligases I and III in the repair of A:8-oxo-G mispairs.
The Replicative DNA Polymerase δ Preferentially Incorporates dATP Opposite 8-oxo-G and This Mispair Is Specifically Cut by MUTYH.
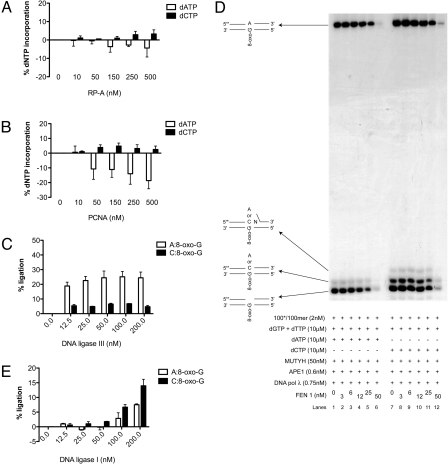
Next, we addressed the mechanism of A:8-oxo-G repair by developing an 8-oxo-G specificity assay (Fig. S3A). This assay enables accurate determination of DNA pols fidelity, based on a specific DNA sequence of the template strand (Fig. S3B). During DNA polymerization reaction on such template, dATP or dCTP can be incorporated only opposite a single 8-oxo-G lesion present on the template. Thus, a direct correlation can be made between the amount of dATP or dCTP incorporated opposite 8-oxo-G and the quantified signal intensity of the polymerization reaction products separated on a polyacrylamide gel. Compared with the generally used single nucleotide incorporation assays, the 8-oxo-G specificity assay more accurately reflects the physiological situation where replicative DNA pols synthesize longer stretches of DNA. Because in the 8-oxo-G specificity assay direct competition between dATP and dCTP within the same reaction is not possible, we first tested the fidelity of DNA pol δ under identical conditions in two experimental setups: (i) containing unlabeled primer/template and all 4 dNTPs when either dATP (Fig. S4A, lanes 2–5) or dCTP (Fig. S4A, lanes 6–9) were radioactively labeled; and (ii) containing labeled primer/template, dGTP, dTTP, and dATP (Fig. S4B, lanes 2–5) or dCTP (Fig. S4B, lanes 6–9). Independently whether in the reactions all four or only three dNTPs were present, DNA pol δ preferentially and with similar efficiencies incorporated dATP opposite 8-oxo-G lesion. The same effect was observed when PCNA was titrated in the reaction (Fig. S4C). Next we checked whether the product of the DNA pol δ polymerization reaction containing A:8-oxo-G mispair could be recognized as a substrate by MUTYH. For this MUTYH was titrated into the reactions in the presence of DNA pol δ and APE1. MUTYH efficiently removed A from A:8-oxo-G product synthesized by DNA pol δ, and together with APE1 created a 1-nt gap opposite the lesion (Fig. 2A, lanes 5 and 6). As expected, MUTYH was not able to act on C:8-oxo-G product of DNA pol δ (Fig. 2A, lanes 11 and 12).
Fig. 2.
DNA polymerase λ physically interacts with MUTYH and preferentially incorporates the correct dCTP opposite an 8-oxo-G after inaccurate replication by DNA polymerase δ. The experiments were done under the conditions as specified in Materials and Methods. (A) MUTYH titration in the presence of the indicated amounts of DNA pol δ, APE1, dGTP, dTTP and dATP (lanes 1–6) or dCTP (lanes 7–12). (B) Interaction of MUTYH and DNA pol λ using a GST pull-down assay. Incubation of APE1, DNA pols δ, and λ with GST-MUTYH (lanes 1–3) and GST (lanes 4–6). Lanes 7–9: input purified recombinant APE1, DNA pols δ, and λ, respectively. (C) dGTP, dTTP, and dATP (lanes 1–6) or dCTP (lanes 7–12) incorporation by DNA pol λ with MUTYH and APE1. (D) Summary of DNA pol λ activity in the presence of dATP (●) or dCTP (■) from three different experiments as the one documented in C; error bars, ± SD values.
DNA Polymerase λ Physically Interacts with MUTYH and Preferentially Incorporates the Correct dCTP Opposite 8-oxo-G on a 1-nt Gapped Substrate.
It has been shown that MUTYH interacts with the components of long patch (LP) BER, such as APE1, PCNA, and RP-A, but does not interact with DNA pols δ and β (26). Therefore, we tested whether MUTYH interacts with DNA pol λ. GST pull-down experiments clearly showed that MUTYH specifically interacted with APE1 and DNA pol λ (Fig. 2B, lanes 1 and 3), but not DNA pol δ (Fig. 2B, lane 2). Based on this observation, we checked whether DNA pol λ could faithfully fill the gap opposite 8-oxo-G lesion, created by MUTYH and APE1. The double-stranded (ds) DNA template containing an A:8-oxo-G mispair was first incubated with MUTYH and APE1, followed by the addition of DNA pol λ into the reaction. DNA pol λ showed a 3-fold preference for dCTP versus dATP incorporation opposite 8-oxo-G lesion (Fig. 2 C and D).
PCNA and RP-A Promote the Accurate Incorporation Opposite 8-oxo-G by DNA Polymerase λ.
We have shown that the auxiliary proteins RP-A and PCNA positively influence the accurate bypass of 8-oxo-G by DNA pol λ (18, 21). To test this effect in the 8-oxo-G specificity assay, RP-A or PCNA were titrated in the presence of MUTYH, APE1 and DNA pol λ. RP-A promoted accurate bypass of 8-oxo-G by DNA pol λ, by repressing the formation of A:8-oxo-G mispair and stimulating the formation of C:8-oxo-G base pair (Fig. 3A). As shown in Fig. 3B, PCNA inhibited the incorporation of dATP opposite 8-oxo-G by DNA pol λ thereby stimulating the accurate bypass. Thus, in a MUTYH and APE1 initiated reaction, the auxiliary proteins RP-A and PCNA can additionally promote the accurate bypass of an 8-oxo-G lesion by DNA pol λ.
Fig. 3.
RP-A and PCNA stimulate accurate incorporation by DNA polymerase λ and this product is specifically processed by FEN1 and DNA ligase I. The experiments were done under the conditions specified in Materials and Methods. (A and B) Incorporation of dGTP, dTTP, and dATP (white bars) or dCTP (black bars) by DNA pol λ in the presence of RP-A (A) or PCNA (B). Error bars, ± SD values of three independent experiments. (C) Products of the DNA pol λ repair synthesis containing A:8-oxo-G mispair (white bars) or C:8-oxo-G base pair (black bars) ligated by DNA ligase III in the presence of MUTYH and APE1. Error bars, ± SD values of three independent experiments. (D) dGTP, dTTP, and dATP (lanes 1–6) or dCTP (lanes 7–12) incorporation by DNA pol λ with MUTYH, APE1, and FEN1. (E) Products of the DNA pol λ repair synthesis containing A:8-oxo-G mispair (white bars) or C:8-oxo-G base pair (black bars) ligated by DNA ligase I in the presence of MUTYH, APE1, and FEN1. Error bars, ± SD values of three independent experiments.
FEN1 and DNA Ligase I Are Required for Accurate Repair of A:8-oxo-G Mispairs.
When DNA pol λ incorporated dCTP opposite an 8-oxo-G, addition of + 2nt was observed (Fig. 2C, lanes 11 and 12). However, this was not the case when dATP was incorporated, indicating that accurate repair could be mediated via the LP-BER (2–12-nt patch), and inaccurate (DNA containing A:8-oxo-G mispair) via the short patch (SP) BER (1-nt patch). To address this, we titrated the SP-BER protein, DNA ligase III, into the 8-oxo-G assay containing MUTYH, APE1, and DNA pol λ. DNA ligase III ligated 5-fold better products of repair synthesis containing A:8-oxo-G mispair, than C:8-oxo-G base pair (Fig. 3C). Next we tested the effect of the LP-BER protein, FEN1 on the repair fidelity. Addition of FEN1 in the presence of MUTYH and APE1 had no effect on DNA pol λ fidelity or polymerization activity (Fig. 3D). At higher amounts 5′->3′ exonuclease activity of FEN1 was observed (Fig. 3D, lanes 5, 6, 11, and 12). To test, whether another LP-BER protein, DNA ligase I, promotes inaccurate or accurate repair, we titrated it in the presence of constant amount of FEN1. Interestingly, DNA ligase I ligated 2-fold better a C:8-oxo-G than an A:8-oxo-G product of the DNA pol λ reaction (Fig. 3E), thereby mediating the accurate repair. In summary, the specific recruitment of MUTYH, DNA pol λ, PCNA, FEN1, and DNA ligases I and III from the WCE to the A:8-oxo-G lesion (Fig. 1 D and E) is in strong correlation with our in vitro observations that MUTYH initiated repair of 8-oxo-G can proceed either through an inaccurate SP-BER or an accurate LP-BER and that DNA pol λ acts as the key enzyme to promote the accurate pathway.
Discussion
The repair of A:8-oxo-G mispair is suggested to follow LP-BER involving DNA pols δ or ε (26–28). However, both of these DNA pols are significantly inaccurate during 8-oxo-G bypass, by incorporating dATP in 30–50% of the cases (18). In addition, both human DNA ligase III, acting in SP-BER and DNA ligase I, involved in LP-BER join 3′dA-terminated primer paired to 8-oxo-G much more efficiently than 3′dC-terminated primer (29). Thus, there is no available evidence suggesting a preferential role of either SP-BER or LP-BER in the repair of A:8-oxo-G mispairs. In this work we propose the accurate LP-BER pathway of A:8-oxo-G mispairs involving MUTYH, APE1, DNA pol λ, FEN1, and DNA ligase I. This repair pathway is in particular coordinated by the activities of MUTYH and DNA pol λ. MUTYH acts as a sensor for the A:8-oxo-G mispairs and initiates the repair by removing the A whereas DNA pol λ preferential incorporates dCTP opposite 8-oxo-G and directly promotes the accurate repair. In addition we show that upon exposure of the cells to ROS protein level of MUTYH and DNA pol λ is increased and both proteins directly localize to the site of oxidative DNA damage. Role of DNA pol λ, in protection of the cells against ROS, is supported by the previous finding that DNA pol λ-null cells are highly hypersensitive to oxidative DNA damaging agents (30). In addition DNA pol λ-null cell extracts exhibit lower level of dCTP incorporation opposite 8-oxo-G lesion compared with wild-type cell extracts (18). MUTYH-null mouse embryonic stem (ES) cells, deficient in the repair of A:8-oxo-G mispairs, have a spontaneous mutation rate in Hprt locus 2-fold higher than the wild-type cells (31). The expression of wild-type mouse MUTYH restores the increased spontaneous mutation rate of the MUTYH-null (ES) cells to the wild-type level (31). Thus, the accurate A:8-oxo-G repair pathway is from a great importance in suppressing C:G to A:T transversion mutations and maintaining genomic stability.
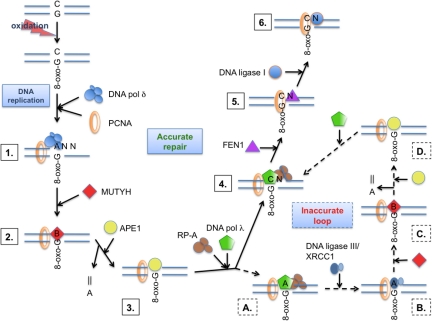
Based on our data and known protein–protein interactions, we propose a model for the accurate MUTYH/DNA pol λ-coordinated repair of A:8-oxo-G mispairs (Fig. 4). Replicative DNA pols, such as DNA pol δ, during DNA replication, bypass 8-oxo-G in inaccurate manner. The MUTYH is through the interaction with PCNA recruited to the A:8-oxo-G mispair and removes the A. To restore a 3′-OH moiety and create gapped intermediate, APE1, another PCNA-interacting protein (32), is recruited. The created gapped substrate can be further recognized, by several factors, through competition for PCNA and RP-A binding. These auxiliary proteins will further promote the accurate bypass of 8-oxo-G and additional + 1-nt elongation reaction by DNA pol λ. After the lesion bypass, RP-A and DNA pol λ dissociate, allowing FEN1 to be recruited onto the resulting 1-nt flap intermediate through interaction with PCNA (33, 34). In the subsequent step DNA ligase I interacts with PCNA (35, 36), binds to the created nicked intermediate and ligates it. In this way accurate repair is ensured at the lesion bypass step, by the combined action of PCNA, RP-A, and DNA pol λ. Alternatively when DNA pol λ incorporates dATP opposite 8-oxo-G an inaccurate short patch loop will be initiated. After the lesion bypass, RP-A and DNA pol λ dissociate from the nicked intermediate enabling binding of DNA ligase III/XRCC1 (37) through interaction with PCNA, and subsequent ligation. Thus, the resulting product contains an A:8-oxo-G mispair that can be further recognized and processed by MUTYH and APE1, thereby creating another chance for an accurate repair to occur. Overall, our data suggest that the repair of A:8-oxo-G mispairs is an interplay between accurate long-patch BER and inaccurate short patch BER, in which DNA pol λ acts as the key enzyme to regulate the error rate.
Fig. 4.
Model for the MUTYH initiated long patch BER of 8-oxo-G, after misincorporation by the replication machinery. 1) DNA replication over an 8-oxo-G by DNA pol δ. 2) recognition of an A:8-oxo-G mispair by MUTYH, removal of the A and formation of an AP site (designated as B). 3) recruitment of APE1 mediated by MUTYH/PCNA and generation of 5′-P, 3′-OH gapped intermediate. 4) protection of the 1-nt gap by RP-A and PCNA mediated recruitment of DNA pol λ, with accurate gap filling (dCTP incorporation). 5) PCNA mediated recruitment of FEN1 and removal of 1-nt flap. 6) ligation of the nick by recruited DNA ligase I and further faithful OGG1 initiated DNA pol β mediated SP-BER of C:8-oxo-G product. Alternatively an inaccurate loop is initiated: (A) DNA pol λ catalyzes inaccurate gap filling; (B) recruitment of DNA ligase III/XRCC1-mediated by PCNA and ligation of the nick; (C) recognition of A:8-oxo-G mispair by MUTYH, removal of A and generation of AP site (designated as B); and (D) recruitment of APE1 mediated by MUTYH/PCNA, generation of 5′-P, 3′-OH gapped intermediate. This creates an opportunity for DNA pol λ to catalyze accurate LP-BER. For further details, see text.
Materials and Methods
Chemicals.
Deoxynucleotides were purchased from Sigma. Labeled γ[32P] ATP, α[32P] dATP and α[32P] dCTP were purchased from Hartmann Analytic. All of the other reagents were of analytic grade and purchased from Fluka, Sigma, or Merck. The 39-mer and 100-mer were purchased from Microsynth and the 100-mer containing 8-oxo-G and 3′-biotinylated 58-mer were from Purimex. Streptavidin-coupled magnetic beads were purchased from Invitrogen.
DNA Substrates.
All oligonucleotides were purified from polyacrylamide denaturing gels (see SI Text for sequences, purification, and template preparation details). Annealing of 100-mer containing 8-oxo-G lesion with the unlabeled or 5′-labeled 39-mer primer or 100-mer created primer/template or ds substrate containing A:8-oxo-G mispair, respectively. Hairpin oligonucleotide substrates were created from 3′-biotinylated 58-mer.
Cells and Extracts.
Human HeLa cells were purchased from American Type Culture Collection and grown according to standard protocols (see SI Text for details). HeLa cell pellets were purchased from Computer Cell Culture Center. WCEs were prepared as described in ref. 38 and stored at −80 °C.
Antibodies and Proteins.
Antibodies against GST, DNA pols λ, and δ (polyclonal rabbit) were from our laboratory. Antibodies against APE1, MUTYH, and PCNA were purchased from Santa Cruz. Antibodies against 8-oxo-G were purchased from Millipore, against DNA ligase I from Genetex, and against DNA ligase III from Abcam. Recombinant human DNA pol λ, RP-A, PCNA, DNA pol δ, FEN1, DNA ligase I, and APE1 were expressed and purified as described (15, 18, 35, 39–44). The bacterial expression vector for human DNA ligase III was provided by G.L. Dianov. DNA ligase III was purified on Ni-NTA agarose (Invitrogen) as recommended by the manufacturer. The human hMUTYH gene was isolated from pGEV1-hMYH (gift from A.L. Lu) as a XhoI-NheI-digested fragment and transferred into pET41a (Novagene) to obtain the pET41a-MUTYH expression construct. GST-tagged MUTYH fusion protein was bound to the glutathione Sepharose (GE Healthcare) and purified as recommended by the manufacturer (see SI Text for details). The bacterial expression vector GST was expressed and purified as described in ref. 44.
Cell Treatment.
Double thymidine block was achieved by seeding the HeLa cells in six-well plates 8 h before incubation in medium with 2 mM thymidine for 16 h. The cells were washed and incubated in fresh medium for 8 h. A second 16-h incubation in 2 mM thymidine was carried out before releasing the block. Upon synchronization at G1/S boundary, cells were treated with 5 mM H2O2 for 40 min at 37 °C and then recovered in fresh medium for 5 h. For analysis of the cell synchronization at G1/S boundary (Fig. S1A), the cells were fixed in 70% ethanol, washed and incubated with 200 μg/mL RNase A (Roche Diagnostics) in PBS for 30 min at 37 °C. Propidium iodide (20 μg/mL; Sigma) was added and the DNA content was analyzed by flow cytometric analysis using a Cytomics FC 500 (Beckman Coulter).
Microscopy and UVA Laser Microirradiation.
HeLa cells were grown on glass coverslips, synchronized by double thymidine block, and laser microirradiated (Fig. 1) or treated with 5 mM H2O2 (Fig. S1 B–E) as described above. Upon fixation, cells were incubated with primary 8-oxo-G, MUTYH, or DNA pol λ antibody. Next, cells were washed three times with PBS and incubated with secondary FITC or Texas Red antibody (Jackson ImmunoResearch) for 30 min at room temperature. Images were captured with an Olympus BX51 fluorescent microscope and acquired with CCD camera (Orca AG) using CellR software (Olympus). At least 100 nuclei were analyzed in each experiment. For additional information see SI Text.
Cross-Linking Assay.
The cross-linking assay with hairpin oligonucleotide substrates attached to streptavidin magnetic beads was performed as described in ref. 23. For direct comparison, cross-linked proteins from different substrates were analyzed in parallel on the two immunoblots treated simultaneously, under the same conditions and with the same exposure time.
GST Pull-Down Assay.
Purified recombinant GST tagged MUTYH (3 μg) was coupled to glutathione Sepharose beads and used in a pull-down assay with 800 ng of purified recombinant human APE1, DNA pols δ, and λ. As an input control, 100 ng of purified recombinant human APE1, DNA pols δ, and λ was applied. Pull-down assay was performed as described in SI Text.
8-oxo-G Specificity Assays.
Primer extension assay.
For denaturing gel analysis of DNA synthesis products, the reaction mixture (10 μL) contained 50 mM Tris·HCl (pH 7.5), 20 mM KCl, 2 mM DTT, and 10 mM Mg2+. Concentrations of DNA pol δ, MUTYH, APE1, PCNA, RP-A, dNTPs, the 5′ 32P-labeled, and unlabelled primer/template were as indicated in the Figs. and Fig. Legends. Reaction products were analyzed on 7M urea/10% polyacrylamide gel (see SI Text for details).
Repair assay.
For denaturing gel analysis of DNA repair products the reaction mixture (10 μL) contained 50 mM Tris·HCl (pH 7.5), 20 mM KCl, and 2 mM DTT. Concentrations of MUTYH, APE1, DNA pol λ, FEN1, DNA ligases I and III, PCNA, RP-A, dNTPs, and the 5′ 32P-labeled double-stranded substrate containing A:8-oxo-G mispair were as indicated in the Figs. and Fig. Legends. Reaction products were separated on a 7 M urea/15% polyacrylamide gel. For reaction and assay details see SI Text.
Steady-State Kinetic Analysis.
Reactions were performed as described above. Quantification was done by densitometry with a PhosphorImager (Typhoon Trio, GE Healthcare). The initial velocities of the reaction were calculated from the values of integrated gel band intensities with the programs ImageQuant and GraphPad Prism 5.0 (see SI Text).
Supplementary Material
Acknowledgments.
We thank G.L. Dianov, J.L. Parsons, I. Shevelev, and A.L. Lu for providing expression constructs; E. Ferrari and R. Imhof for purifying DNA pol λ, FEN1, and DNA ligase III; and G. Villani, G. Maga, K. Ramadan, and M. Stucki for critically reading of the manuscript and their suggestions. This work is supported by the Swiss National Science Foundation Grant 3100–109312/2 and the University of Zürich.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907280106/DCSupplemental.
References
- 1.Collins AR. Oxidative DNA damage, antioxidants, and cancer. Bioessays. 1999;21:238–246. doi: 10.1002/(SICI)1521-1878(199903)21:3<238::AID-BIES8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 2.Avkin S, Livneh Z. Efficiency, specificity and DNA polymerase-dependence of translesion replication across the oxidative DNA lesion 8-oxoguanine in human cells. Mutat Res. 2002;510:81–90. doi: 10.1016/s0027-5107(02)00254-3. [DOI] [PubMed] [Google Scholar]
- 3.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slupska MM, Luther WM, Chiang JH, Yang H, Miller JH. Functional expression of hMYH, a human homolog of the Escherichia coli MutY protein. J Bacteriol. 1999;181:6210–6213. doi: 10.1128/jb.181.19.6210-6213.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takao M, Zhang QM, Yonei S, Yasui A. Differential subcellular localization of human MutY homolog (hMYH) and the functional activity of adenine:8-oxoguanine DNA glycosylase. Nucleic Acids Res. 1999;27:3638–3644. doi: 10.1093/nar/27.18.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braithwaite EK, et al. DNA polymerase lambda mediates a back-up base excision repair activity in extracts of mouse embryonic fibroblasts. J Biol Chem. 2005;280:18469–18475. doi: 10.1074/jbc.M411864200. [DOI] [PubMed] [Google Scholar]
- 7.Fortini P, et al. Different DNA polymerases are involved in the short- and long-patch base excision repair in mammalian cells. Biochemistry. 1998;37:3575–3580. doi: 10.1021/bi972999h. [DOI] [PubMed] [Google Scholar]
- 8.Stucki M, et al. Mammalian base excision repair by DNA polymerases delta and epsilon. Oncogene. 1998;17:835–843. doi: 10.1038/sj.onc.1202001. [DOI] [PubMed] [Google Scholar]
- 9.Allinson SL, Dianova II, Dianov GL. DNA polymerase beta is the major dRP lyase involved in repair of oxidative base lesions in DNA by mammalian cell extracts. EMBO J. 2001;20:6919–6926. doi: 10.1093/emboj/20.23.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dianov GL, Prasad R, Wilson SH, Bohr VA. Role of DNA polymerase beta in the excision step of long patch mammalian base excision repair. J Biol Chem. 1999;274:13741–13743. doi: 10.1074/jbc.274.20.13741. [DOI] [PubMed] [Google Scholar]
- 11.Podlutsky AJ, Dianova II, Podust VN, Bohr VA, Dianov GL. Human DNA polymerase beta initiates DNA synthesis during long-patch repair of reduced AP sites in DNA. EMBO J. 2001;20:1477–1482. doi: 10.1093/emboj/20.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sobol RW, et al. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 13.Sobol RW, et al. The lyase activity of the DNA repair protein beta-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000;405:807–810. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 14.Ramadan K, Shevelev I, Hubscher U. The DNA-polymerase-X family: Controllers of DNA quality? Nat Rev Mol Cell Biol. 2004;5:1038–1043. doi: 10.1038/nrm1530. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Diaz M, Bebenek K, Kunkel TA, Blanco L. Identification of an intrinsic 5′-deoxyribose-5-phosphate lyase activity in human DNA polymerase lambda: A possible role in base excision repair. J Biol Chem. 2001;276:34659–34663. doi: 10.1074/jbc.M106336200. [DOI] [PubMed] [Google Scholar]
- 16.Lee JW, et al. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J Biol Chem. 2004;279:805–811. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- 17.Nick McElhinny SA, et al. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 18.Maga G, et al. 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 2007;447:606–608. doi: 10.1038/nature05843. [DOI] [PubMed] [Google Scholar]
- 19.Maga G, et al. Human DNA polymerase lambda functionally and physically interacts with proliferating cell nuclear antigen in normal and translesion DNA synthesis. J Biol Chem. 2002;277:48434–48440. doi: 10.1074/jbc.M206889200. [DOI] [PubMed] [Google Scholar]
- 20.Picher AJ, Blanco L. Human DNA polymerase lambda is a proficient extender of primer ends paired to 7,8-dihydro-8-oxoguanine. DNA Repair (Amst) 2007;6:1749–1756. doi: 10.1016/j.dnarep.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Maga G, et al. Replication protein A and proliferating cell nuclear antigen coordinate DNA polymerase selection in 8-oxo-guanine repair. Proc Natl Acad Sci USA. 2008;105:20689–20694. doi: 10.1073/pnas.0811241106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong X, et al. Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells. Nucleic Acids Res. 2009;37:e68. doi: 10.1093/nar/gkp221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parsons JL, Dianov GL. Monitoring base excision repair proteins on damaged DNA using human cell extracts. Biochem Soc Trans. 2004;32:962–963. doi: 10.1042/BST0320962. [DOI] [PubMed] [Google Scholar]
- 24.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): A dancer with many partners. J Cell Sci. 2003;116:3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 26.Parker A, et al. Human homolog of the MutY repair protein (hMYH) physically interacts with proteins involved in long patch DNA base excision repair. J Biol Chem. 2001;276:5547–5555. doi: 10.1074/jbc.M008463200. [DOI] [PubMed] [Google Scholar]
- 27.Fortini P, et al. 8-Oxoguanine DNA damage: At the crossroad of alternative repair pathways. Mutat Res. 2003;531:127–139. doi: 10.1016/j.mrfmmm.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Parlanti E, Fortini P, Macpherson P, Laval J, Dogliotti E. Base excision repair of adenine/8-oxoguanine mispairs by an aphidicolin-sensitive DNA polymerase in human cell extracts. Oncogene. 2002;21:5204–5212. doi: 10.1038/sj.onc.1205561. [DOI] [PubMed] [Google Scholar]
- 29.Hashimoto K, Tominaga Y, Nakabeppu Y, Moriya M. Futile short-patch DNA base excision repair of adenine:8-oxoguanine mispair. Nucleic Acids Res. 2004;32:5928–5934. doi: 10.1093/nar/gkh909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braithwaite EK, et al. DNA polymerase lambda protects mouse fibroblasts against oxidative DNA damage and is recruited to sites of DNA damage/repair. J Biol Chem. 2005;280:31641–31647. doi: 10.1074/jbc.C500256200. [DOI] [PubMed] [Google Scholar]
- 31.Hirano S, et al. Mutator phenotype of MUTYH-null mouse embryonic stem cells. J Biol Chem. 2003;278:38121–38124. doi: 10.1074/jbc.C300316200. [DOI] [PubMed] [Google Scholar]
- 32.Dianova II, Bohr VA, Dianov GL. Interaction of human AP endonuclease 1 with flap endonuclease 1 and proliferating cell nuclear antigen involved in long-patch base excision repair. Biochemistry. 2001;40:12639–12644. doi: 10.1021/bi011117i. [DOI] [PubMed] [Google Scholar]
- 33.Warbrick E, Lane DP, Glover DM, Cox LS. Homologous regions of Fen1 and p21Cip1 compete for binding to the same site on PCNA: A potential mechanism to co-ordinate DNA replication and repair. Oncogene. 1997;14:2313–2321. doi: 10.1038/sj.onc.1201072. [DOI] [PubMed] [Google Scholar]
- 34.Li X, Li J, Harrington J, Lieber MR, Burgers PM. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J Biol Chem. 1995;270:22109–22112. doi: 10.1074/jbc.270.38.22109. [DOI] [PubMed] [Google Scholar]
- 35.Jonsson ZO, Hindges R, Hubscher U. Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J. 1998;17:2412–2425. doi: 10.1093/emboj/17.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levin DS, Bai W, Yao N, O'Donnell M, Tomkinson AE. An interaction between DNA ligase I and proliferating cell nuclear antigen: Implications for Okazaki fragment synthesis and joining. Proc Natl Acad Sci USA. 1997;94:12863–12868. doi: 10.1073/pnas.94.24.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan J, Otterlei M, Wong HK, Tomkinson AE, Wilson DM., 3rd XRCC1 co-localizes and physically interacts with PCNA. Nucleic Acids Res. 2004;32:2193–2201. doi: 10.1093/nar/gkh556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manley JL, Fire A, Samuels M, Sharp PA. In vitro transcription: Whole-cell extract. Methods Enzymol. 1983;101:568–582. doi: 10.1016/0076-6879(83)01038-1. [DOI] [PubMed] [Google Scholar]
- 39.Podust VN, Chang LS, Ott R, Dianov GL, Fanning E. Reconstitution of human DNA polymerase delta using recombinant baculoviruses: The p12 subunit potentiates DNA polymerizing activity of the four-subunit enzyme. J Biol Chem. 2002;277:3894–3901. doi: 10.1074/jbc.M109684200. [DOI] [PubMed] [Google Scholar]
- 40.van Loon B, Ferarri E, Hubscher U. In: DNA Replication, Methods and Protocols. Vengrova S, Dalgaard JZ, editors. New York: Humana Press – Springer; 2009. pp. 345–359. [Google Scholar]
- 41.Ramadan K, et al. Human DNA polymerase lambda possesses terminal deoxyribonucleotidyl transferase activity and can elongate RNA primers: Implications for novel functions. J Mol Biol. 2003;328:63–72. doi: 10.1016/s0022-2836(03)00265-1. [DOI] [PubMed] [Google Scholar]
- 42.Stucki M, Jonsson ZO, Hubscher U. In eukaryotic flap endonuclease 1, the C terminus is essential for substrate binding. J Biol Chem. 2001;276:7843–7849. doi: 10.1074/jbc.M008829200. [DOI] [PubMed] [Google Scholar]
- 43.Saparbaev M, et al. 1,N(2)-ethenoguanine, a mutagenic DNA adduct, is a primary substrate of Escherichia coli mismatch-specific uracil-DNA glycosylase and human alkylpurine-DNA-N-glycosylase. J Biol Chem. 2002;277:26987–26993. doi: 10.1074/jbc.M111100200. [DOI] [PubMed] [Google Scholar]
- 44.Toueille M, et al. The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase beta and increases its DNA substrate utilisation utilization efficiency: Implications for DNA repair. Nucleic Acids Res. 2004;32:3316–3324. doi: 10.1093/nar/gkh652. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.