Abstract
Hexameric ring-shaped AAA+ molecular motors have a key function of active translocation of a macromolecular chain through the central pore. By performing multiscale molecular dynamics (MD) simulations, we revealed that HslU, a AAA+ motor in a bacterial homologue of eukaryotic proteasome, translocates its substrate polypeptide via paddling mechanism during ATP-driven cyclic conformational changes. First, fully atomistic MD simulations showed that the HslU pore grips the threaded signal peptide by the highly conserved Tyr-91 and Val-92 firmly in the closed form and loosely in the open form of the HslU. The grip depended on the substrate sequence. These features were fed into a coarse-grained MD, and conformational transitions of HslU upon ATP cycles were simulated. The simulations exhibited stochastic unidirectional translocation of a polypeptide. This unidirectional translocation is attributed to paddling motions of Tyr-91s between the open and the closed forms: downward motions of Tyr-91s with gripping the substrate and upward motions with slipping on it. The paddling motions were caused by the difference between the characteristic time scales of the pore-radius change and the up-down displacements of Tyr-91s. Computational experiments on mutations at the pore and the substrate were in accord with several experiments.
Keywords: coarse-grained model, fully atomistic simulation, HslU, molecular dynamics, multiple basin model
Adenosine 5′-triphosphate (ATP)-driven molecular motors that translocate substrate macromolecules are ubiquitous in all cells. Many of them function as ring-shaped hexamers, where the substrates are threaded into the central pore of the ring. They include translocators of duplex DNAs, those of single-stranded nucleic acids, and those that move polypeptide chains (1, 2). Despite their diverse cellular functions, they have similar structures and biophysical functions, suggesting some underlying common working mechanisms. Indeed, many of these motors are members of the AAA+ superfamily sharing some sequence/structure features (3). Here, we studied the structure-based mechanisms of HslU (heat shock locus gene products U), one of the hexameric AAA+ ATPases that translocate substrate polypeptides.
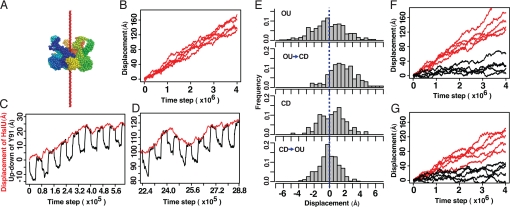
HslU is part of the bacterial energy-dependent proteolytic complex HslUV, a homologue of eukaryotic proteasome (4–6), which degrades the cell division inhibitor SulA (7, 8) and the heat shock sigma factor σ32 (9). HslUV functions as the cylinder-shaped complex that one or two (HslU)6 bind to (HslV)12 (Fig. 1A). Many of the in vitro experiments revealed that HslU unfolds substrate proteins at its narrow pore that only a single stretched peptide can pass through and then HslU translocates the unfolded substrates into the degradation chamber of HslV (Fig. 1 A–C) (10–15). In addition, the structure of HslUV is characterized; indeed, HslUV is the only one energy-dependent protease for which the entire structure is known (10–12, 16–19). These together make HslU an ideal target for studying detailed mechanisms of translocation by molecular simulations. To date, however, no simulation study of HslU has yet been reported. Here, we performed multiscale molecular dynamics (MD) simulations of HslU to understand how substrates are translocated unidirectionally.
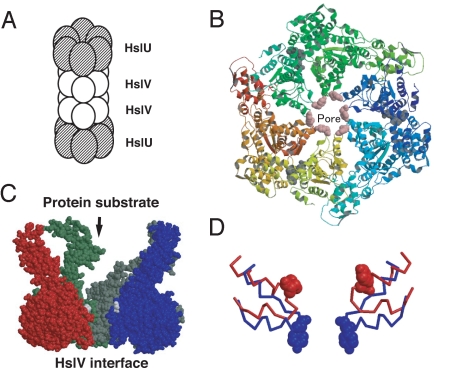
Fig. 1.
Structures of HslU. (A) Diagram of the (HslU)6(HslV)12(HslU)6 complex. HslUV is functional in both (HslU)6(HslV)12(HslU)6 and (HslU)6(HslV)12 forms. (B) The OU form of (HslU)6 viewed from the substrate entrance side. Tyr-91s (pink) form the pore. (C) The side view of HslU. For clarity, only four subunits are shown. The substrates enter the HslU pore from the top and are translocated to the bottom, the HslV side. (D) Structures around the GYVG motif in the OU (red) and CD (blue) forms are superimposed by fitting the 214th residues that are at the binding interface to HslV. For clarity, only two subunits are shown. Residues 80–100 are drawn in the backbone mode, except that Tyr-91s are in the spacefill mode.
The substrates are translocated through the narrow pore of HslU, which consists of the highly conserved GYVG (Gly-90–Tyr-91–Val-92–Gly-93) motif. Some mutations at the pore had fatal effects on the protease activity of HslUV (15, 16). For a homologue, ClpX, the GYVG pore was shown to grip its substrate (20). The GYVG pore undergoes a large-amplitude motion among the conformations of HslU determined in several nucleotide states, which are linked to each other by a hinge motion (17, 18). The GYVG pore is the most open and the central Tyr-91s take the “up” conformations (up means the most distant from the HslV side) when three of six subunits bind nucleotides (the red structure in Fig. 1D) (11). Thus, we call this structure the “open-up” (OU) form. The other extreme, the “closed-down” (CD) form, is the one where all six subunits bind nucleotides, and it has the closed pore and the down conformations of Tyr-91s (the blue structure in Fig. 1D) (see Materials and Methods and SI Appendix for details) (18). These apparently suggest that, during the motions from the OU to the CD forms, Tyr-91s grip a substrate and move it downward (18). Yet, two questions arise.
The first question involves the interactions between the GYVG pore of HslU and the substrate. How does the GYVG pore of HslU grip the substrate? Because the interactions are sequence-specific, the atomistic analysis is indispensable. We conducted fully atomistic simulations investigating details of the interactions between HslU and the N-terminal segment of Arc repressor, a known substrate for HslUV (13). The simulations showed that the GYVG pore is too narrow for the substrate to slip in both the OU and the CD forms of HslU. Tyr-91s and Val-92s were indicated to grip the substrate, and their grip strength was conformation dependent: The grip was firm in the CD form and loose in the OU form.
If HslU grips the substrates in both the OU and CD forms and exerts the up-down motions, how does HslU translocate the substrates unidirectionally, instead of the up-down motions? This is the second question. Because, ATP hydrolysis cycle is far beyond in time scale of the current reach of atomistic simulations, the insights obtained by the above atomistic simulations were transferred into a coarse-grained (CG) model. CG models have been used for simulating large-amplitude motions of proteins and molecular motors (21–24). Using the CG model of HslU with a substrate polypeptide, we simulated multiple ATP cycles, leading to the unidirectional translocation of the substrate. The unidirectional translocation was produced by the paddling motions of Tyr-91s: the downward stroke of Tyr-91s with the gripped substrate during the OU → CD transition and the upward return with the substrate slippage during the CD → OU transition. These mechanisms may be shared by other protease-associated ATPases that have the GYVG motif.
Results
Interactions Between HslU Pore and Substrate Revealed by Fully Atomistic Simulation.
A hexameric HslU with a substrate, which was embedded into explicit water solvent, was simulated by the fully atomistic MD (see Materials and Methods and SI Appendix for details). Simulations were conducted for both the OU and CD conformations of HslU. In each case, the entire chains of HslU were used, where the HslU structure except for the pore region and its neighbor was constrained to the corresponding X-ray structure throughout the simulations (HslU was aligned so that the symmetric axis was parallel to z axis and the HslV side corresponded to the negative direction). As a substrate, the N-terminal 12-residue fragment of Arc repressor (13) was prepared in an extended form. Then, as the initial settings of the simulations, the substrate was forcibly threaded into the HslU pore, where the N terminus of the substrate was directed to the HslV side (HslV was not included in the simulations) and the N-terminal three residues were out of the HslU pore (Fig. 2A). Performing three simulations of 4-ns long for both the OU and CD forms, we investigated the mobility of the substrate in both forms. The mobility was monitored by the z coordinate of the N terminus of the substrate, which was set to 0 at the start.
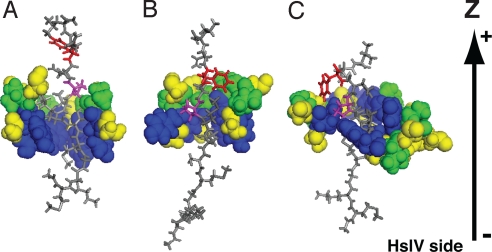
Fig. 2.
Side views of the (HslU)6 pore and the substrate interactions in fully atomistic models. Pore region (GYVG) of HslU is shown by the spacefill mode, and the substrate peptide is shown by the stick mode. (A) The initial structure in the simulations of the CD form of HslU. (B) A snapshot structure after 4-ns simulation in the CD-form HslU with the pulling force (corresponding to the red arrow in Fig. 3B). (C) A snapshot structure paused at near z = −10 Å in the OU-form HslU with the pulling force (the green arrow in Fig. 3B). In the HslU pore, Tyr-91s are in blue, Val-92s in green, and Gly-90s and Gly-93s in yellow. In the substrate, Pro-8 is in magenta and Phe-10 in red. The bottom is the HslV side. Some residues of the pore region are omitted.
In the simulations, the substrate did not move significantly in both the OU and the CD forms of HslU (green and red lines in Fig. 3A). As a control, the same substrate was simulated in bulk water without HslU for 4 ns (blue lines in Fig. 3A). Not surprisingly, the substrate in bulk water largely fluctuated and moved diffusively. Thus, at least in the 4-ns scale, motions of the substrate in HslU were highly constrained by the pore in both the OU and CD forms.
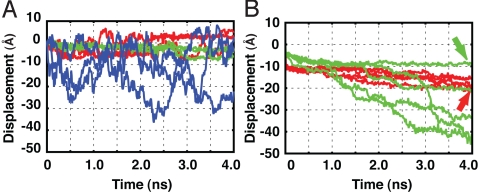
Fig. 3.
Substrate displacements in fully atomistic simulations. The displacements were monitored by the N terminus of the substrate. (A) Simulations without the pulling force. The three trajectories in the CD form of HslU are shown in red, those in the OU form are in green, and those without HslU are in blue. (B) Simulations with the substrate pulled from the HslV side. The five trajectories in the CD form of HslU are in red, and those in the OU form in green. The red and the green arrows correspond to the snapshots in Fig. 2 B and C, respectively.
Next, to investigate how the constraint differs between the OU and CD forms, we weakly pulled the N terminus of the substrate toward the direction of HslV (the −z direction) by using the same setup as for the above simulations. In the CD form of HslU, the substrate moved slightly and stopped for all five trajectories (red lines in Fig. 3B). Conversely, in the OU form, the substrate passed through the pore for three of five trajectories (green lines in Fig. 3B; the complete exit corresponds to z ≈ −30 Å in Fig. 3B).
We inspected the structures in the trajectories (see Movie S1 and Movie S2). In the CD form (Movie S1), at z = −10 ∼ −15Å, the backbone kink at Pro-8 (magenta) of the substrate collided with Val-92 (green) and Tyr-91 (blue) of the HslU pore and the long side chains of Lys-6, Met-7, and Gln-9 clogged the GYVG pore. At z = −15 ∼ −20 Å (a representative structure in Fig. 2B), the backbone kink at Pro-8 (magenta) contacted with Tyr-91(blue), and Phe-10(red) in the substrate started to stick to the GYVG pore. In the OU form (Movie S2), the substrate paused at particular few positions in most trajectories. The most marked one was at z ∼ − 10 Å, where the pore region contacted with the kink of Pro-8, the long side chains of Lys-6 and Met-7, and the large side chain of Phe-10 (Fig. 2C).
These simulation results show that the GYVG pore is too narrow for the substrate to slip in both the OU and CD forms. Tyr-91s and Val-92s in the GYVG motif play important roles for the grip, which is in harmony with the recent experiment on ClpX (20). The strength of the grip depends on the HslU conformations. The GYVG pore grips the substrate firmly in the CD form and loosely in the OU form. In addition, the grip depends on the local structure of the substrate: The backbone kink, long side chains, and hydrophobic side chains markedly contributed to the grip. These results remind us that substrates containing glycine–alanine repeat, substrates with contiguous smaller side chains, impaired their digestion by proteasome (25).
Conformational Transitions of HslU Simulated by CG Model Exhibited Large-Amplitude Motions of Tyr-91s.
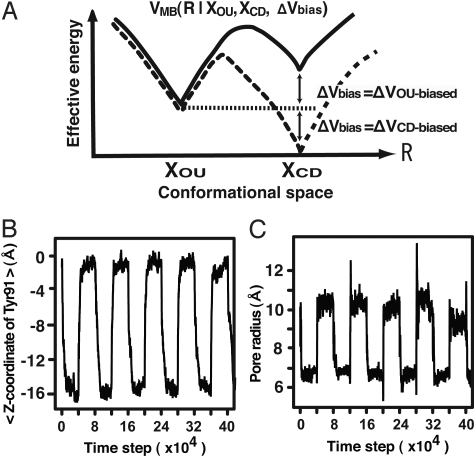
We conducted CG MD simulations of the conformational transitions between the OU and CD forms of the hexameric HslU. Previously, we simulated the rotary motion of F1-ATPase by using a CG model (21). Here, in contrast to the previous model, we used the multiple-basin (MB) CG model (22) that enables us to describe thermally activated conformational transitions of proteins. The model concisely represents the energy landscape of a protein that is overall funnel shape and the bottom of the funnel possesses multiple energy basins (26, 27). In the model, each amino acid of HslU is represented by a single sphere located on the position of the Cβ atom, and the energy function VMB (R | XOU, XCD, ΔVbias) of a simulated structure, R, possesses two energy basins, which correspond to the OU structure, XOU and the CD structure, XCD (Fig. 4A). The relative stability between two basins can be controlled by the parameter ΔVbias (see Materials and Methods and SI Appendix).
Fig. 4.
Conformational change cycles of (HslU)6 in CG simulations. (A) A schematic view of the MB model of HslU. The solid (dashed) curve corresponds to the positive (negative) value of ΔVbias where the landscape is biased to the OU (CD) form. (B) Time series of the average of z coordinates of Tyr-91s, which monitors the up-down motions. (C) The pore radius in a simulation trajectory of the conformational transition cycles.
The cycle of conformational transitions of HslU powered by the ATP hydrolysis is realized by changing the bias parameter ΔVbias. We started the simulation from the structure in the OU form with ΔVbias = ΔVCD-biased, by which the CD state was more stable and the structure changed to the CD form at the very beginning of the simulation. At the 4 × 104 time step, we switched the bias ΔVbias to ΔVOU-biased. This switching stabilized the OU state, inducing the conformational transition to the OU state. By the switching, we inputted energy into the simulation system. The reverse conformational transition was induced at the 8 × 104 time step by returning the bias from ΔVOU-biased to ΔVCD-biased. We regard a cycle of conformational transitions between the OU and CD forms in 8 × 104 time steps as a single cycle of ATP hydrolysis of HslU. The 214th residues of HslU at the binding interface to HslV were fixed, and HslU was aligned so that the symmetric axis was parallel to the z axis and the HslV side corresponded to the negative direction. Upon the conformational changes of HslU, Tyr-91s underwent large-amplitude (≈14 Å) up-down motions monitored by the average of their z coordinates (Fig. 4B; z = 0 corresponds to the OU form), which were achieved not only by the flip-flops of Tyr-91's side-chains but by the also backbone movements of the GYVG pore. At the same time, the conformational changes of Tyr-91s induced the pore-radius changes from 6.7 to 10 Å (Fig. 4C). The pore radius was measured at Tyr-91s (see SI Appendix). These results show that the CG model can recover the conformational transitions between the OU and CD forms.
Unidirectional Translocation Realized by Conformational Transitions of HslU.
Next, a substrate, represented by single sphere per amino acid, was included to the simulation system, and the same cycles of the HslU conformational transitions as above were simulated. In the simulations, we fixed the substrate molecule on the z axis in an extended form, which played the role of a rail, and the substrate rail was encircled by the HslU ring. HslU was placed at the middle of the rail (z = 0) at time 0 (see Fig. 5A and Materials and Methods). By this setting, we could eliminate the effect of the substrate chain entropy on the translocation, which made the problem simpler. The interactions between HslU and the substrate rail were steric repulsion and hydrophobic attraction (Tyr-91s and Val-92s of HslU and all of the substrate residues were regarded as hydrophobic). Because the fully atomistic MD suggested that the GYVG pore grips the threaded substrate in both the OU and CD forms, we set an appropriate attractive range of hydrophobic interactions of Tyr-91s (Val-92s) as 10 Å (8 Å) from the Cβ and the radius of steric repulsion of Tyr-91s (Val-92s) as 8 Å (6 Å), which enabled a grip similar to that in the fully atomistic MD.
Fig. 5.
Unidirectional translocations in CG simulations of HslU and a substrate. (A) The simulation setting is illustrated. The hexameric HslU encircled the substrate that was stretched out and pinned along the z axis to form a rail (red). HslU was initially placed at z = 0. (B) When HslU changing its conformation upon ATP cycles how far HslU moved along the rail was observed. The five independent trajectories were drawn. (C and D) The displacements of HslU (red) and the up-down motions of Tyr-91 (black) in narrow time windows of one trajectory in B. (E) Histograms of the displacements of HslU along the substrate in each of four periods: (i) the OU dwell, (ii) the OU → CD period, (iii) the CD dwell, and (iv) the CD → OU period. (F) The five trajectories of the HslU mutant where the six Tyr-91s were mutated to smaller and nonhydrophobic residues (black), and those of the other mutant where the six Val-92s were mutated to nonhydrophobic residues (red). (G) The five trajectories of the HslU mutant where two subunits of hexameric HslU were defunct and four subunits were functional (red). The five trajectories of HslU along a thinner rail substrate (black) are shown.
During the multiple cycles of the conformational transitions, we monitored the HslU movement along the rail, finding that the unidirectional translocation of HslU in all independent simulations (Fig. 5B and Movie S3). This unidirectional translocation was surprising because the iterative up-down motions of Tyr-91s and their grips on the rail apparently lead HslU to move up and down only at near the initial position. In each cycle of conformational transitions (black lines in Fig. 5 C and D), the movements of HslU were stochastic (red lines in Fig. 5 C and D). However, as longtime behavior, HslU moved unidirectionally in all trajectories with the average rate of ≈3 Å per cycle. We emphasize that no force pulling HslU unidirectionally by design was applied. To decipher which processes in the cycle play what roles, we divided a cycle into four periods: (i) the period dwelling in the OU form (the OU dwell), (ii) the conformational change period from the OU to the CD forms (the OU → CD period), (iii) the CD dwell, and (iv) the CD → OU period. We investigated the HslU movements in each of the four periods (Fig. 5E; the histograms of the HslU movements in z direction in each period). First, both dwells did not show significant bias in the histograms (Fig. 5E). The fluctuation in the OU form was slightly larger than that in the CD form, which is consistent with the grip strength observed in fully atomistic MD results. The OU → CD period produced the forward bias (Fig. 5E), suggesting that the power stroke was exerted in this period by the GYVG pore. Conversely, the CD → OU period did not produce significant bias (Fig. 5E). Namely, no backward bias was produced, which suggests that HslU slipped on the rail substrate at this period. The difference in the bias between the OU → CD and the CD → OU periods resulted in the unidirectional translocation of HslU.
Next, to elucidate the roles of Tyr-91 and Val-92 in the GYVG pore for the unidirectional displacement, two mutational simulations were carried out. (i) The six Tyr-91s were mutated to smaller and nonhydrophobic amino acids (like Ser), which reduced the steric hindrance and deleted the attractive interactions with the substrate. The mutant showed little, if any, ability of the translocation (black lines in Fig. 5F). (ii) The six Val-92s were changed to nonhydrophobic ones with the same residue size (red lines in Fig. 5F). As the result, the mutant at Val-92s moved unidirectionally with the same rate as the wild type. Experimentally, the degradation activity for SulA was impaired for the mutant Tyr-91–Ser of HslUV, but was mostly retained for the Val-92–Ser mutant (15).
We then performed the simulations with a rail substrate thinner than the rail used in the previous simulations (see Materials and Methods). The thin rail tried to mimic the substrates made of small amino acids, such as Gly and Ala. As the result, the unidirectional movements of HslU were still observed in all five trajectories, but it became much slower with the average rate ≈0.7Å per cycle (black lines in Fig. 5G). These simulation results remind us that the substrates containing Gly–Ala repeats impair their digestion by proteasome (25).
The simulations described so far assumed that all six HslU subunits made conformational transitions between the OU and CD forms simultaneously, but this assumption is not imperative. In fact, when we allowed only four HslU subunits to change the conformations between the OU and CD forms, the translocation occurred with the average rate of 2.2 Å per cycle (red lines in Fig. 5G). The rate per cycle was smaller than that of the wild type, but notably, the rate per the number of consumed ATPs was almost identical to that of the wild type. The same tendency was observed in the experiment of ClpX (28).
Finally, we also performed the translocation simulations by the opposite setup. The hexameric HslU was spatially fixed at its interface to HslV, whereas the substrate threaded into the HslU pore was free to move. Except this difference, using the same simulation protocols as above, we monitored the translocations of the substrate (see Fig. S1). The substrate translocations to the HslV side were observed over long simulations, although the translocations were more stochastic and less efficient because of the chain entropy effect of the substrate.
Paddling Motions of Tyr-91s Produce Translocation.
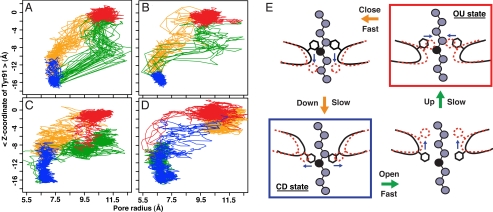
Toward the elucidation of molecular mechanisms of the translocation, the immediate question is why Tyr-91s grip the substrate firmly in the OU → CD transition but slip on the substrate in the opposite transition. To address this question, for the above simulation where the wild-type HslU was translocated unidirectionally along the rail substrate (Fig. 5B), we plotted the trajectories of Tyr-91 on the radius-z plane: the pore radius on the horizontal axis and the up-down displacement (z) of Tyr-91 on the vertical axis (Fig. 6A). As before, each cycle was divided into four periods. In Fig. 6, the movements in each period are drawn in a different color: the OU dwell in red, the OU → CD period in yellow, the CD dwell in blue, and the CD → OU period in green. We found that Tyr-91s followed the different pathways of the conformational changes between the OU → CD and the CD → OU transitions. On the OU → CD transitions, Tyr-91s approached the substrate closer than those on the CD → OU. Namely, in both transitions, the pore-radius change proceeded three to six times faster than the up-down motions of Tyr-91s (see Fig. S2), which resulted in the anticlockwise rotations of the trajectories on the radius-z plane. During these rotary motions, Tyr-91s exert the downward stroke for the substrate translocation and return upward with slipping on the substrate, which we call the paddling motions of Tyr-91s. The paddling motions clearly explain the unidirectional translocation.
Fig. 6.
Trajectories of Tyr-91s in ATP cycles and the paddling motions. (A and B)Trajectories of Tyr-91s during the ATP cycles with (A) and without (B) the substrate were drawn in the radius-z plane: the average of z coordinates of Tyr-91s and the pore radius of Tyr-91s. Each cycle was divided into four periods: (i) the OU dwell (red), (ii) the OU → CD period (yellow), (iii) the CD dwell (blue), and (iv) the CD → OU period (green). (C and D) Trajectories of Tyr-91s when the changes in the intrasubunit and intersubunit interactions were induced separately and sequentially in the following order: for C (i) the change in the intrainteractions for the OU → CD transitions (yellow), (ii) the change in the interinteractions for the OU → CD (blue), (iii) the change in the intrainteractions for the CD → OU (green), and (iv) the change in the interinteractions for the CD → OU (red); for D (i) the change in the interinteractions for the OU → CD (yellow), (ii) the change in the intrainteractions for the OU → CD transitions (blue), (iii) the change in the interinteractions for the CD → OU (green), and (iv) the change in the intrainteractions for the CD → OU (red). (E) The paddling motion of Tyr-91 between the OU and CD states.
The anticlockwise rotary motions of Tyr-91s were also observed in the simulations of conformational changes of HslU without the substrate (Fig. 6B), suggesting the paddling motions are inherent features of HslU.
Origin of the Paddling Motions: The Time-Scale Difference Between Intrasubunit and Intersubunit Motions.
Seeking origin of the paddling motions, we investigated why the pore-radius change proceeded faster than the up-down motions of Tyr-91s. For the purpose, we separated the interactions in hexameric HslU into two parts: intrasubunit and intersubunit interactions. We conducted two sets of simulations of HslU without the substrate. In the first set of simulations, we induced the OU → CD transition in two sequential steps: (i) the intrasubunit conformational change (yellow in Fig. 6C), and then (ii) the change of interface between subunits (blue in Fig. 6C), and then induced the CD → OU transition in the two sequential steps: (iii) the intrasubunit conformational change (green), and finally (iv) the change of interface between subunits (red). This first set of simulations led to the anticlockwise rotations on the radius-z plane (Fig. 6C). In the second set of simulations, we reversed the order of the two sequential steps in both the OU → CD and CD → OU transitions; the transitions were proceeded in the order of ii, i, iv, and iii, which showed Tyr-91s trajectories rotated clockwise on the radius-z plane (Fig. 6D). These results suggest that the intrasubunit (intersubunit) changes are correlated with the pore-radius change (the up-down motions). In reality, both the intrasubunit and intersubunit changes are initiated simultaneously upon the nucleotide state change. If the intrasubunit motions are faster than the intersubunit motions, Tyr-91s rotate anticlockwise on the radius-z plane, which is consistent with our simulations (Fig. 6B). Thus, the origin of the paddling motions of Tyr-91s is attributed to the time-scale difference between the intrasubunit and intersubunit motions.
Discussion
The simulations elucidated that the unidirectional translocation of the substrate was realized by the paddling motions of Tyr-91s in the pore of HslU (Fig. 6E). Starting from the OU form (Fig. 6E Upper Right), Tyr-91s, the paddles, first grip the substrate by closing (Fig. 6E Upper Left), which is followed by the downward stroke to push the substrate (Fig. 6E Lower Left). Upon the return, Tyr-91s first open to lose the firm grip (Fig. 6E Lower Right), and then move upward to the OU form without pushing back the substrate. This asymmetry of the Tyr-91 motions (Fig. 6 A and B) is an essence of the function derived from the difference between the time scale of the pore-radius change and that of the up-down displacements of Tyr-91s. The former (the latter) is linked to the intrasubunit (intersubunit) motions and is faster (slower) (Fig. 6 C and D).
Next, we discuss the ATP hydrolysis cycles and the structure changes in HslU, which are currently rather unclear (14). In most of our simulations, we assumed the concerted model: all subunits in hexameric HslU change their conformations simultaneously during ATP cycles. However, in the middle of our study, Sauer's group (14) reported that HslU binds three or four ATP molecules at saturation, supporting the probabilistic model: each subunit independently hydrolyzes ATP and changes its structure. The result also suggested that the structure binding 4 nucleotides [Protein Data Bank (PDB) ID code 1DO0] (11) is a candidate structure to be used in simulations. Inspired by their work, we tried to test whether the paddling mechanism is applicable to the probabilistic model. The structure 1DO0, however, has the completely filled pore, making it difficult for the substrate to be threaded into the pore in simulations. Instead, still using the OU and CD forms as references, we conducted the simulations where only four subunits make conformational transitions, which resulted in the unidirectional translocation (red lines in Fig. 5G). This result indicates that the paddling mechanism should be applicable to the probabilistic model and is in harmony with the implication that ClpX works under asymmetric ATP cycles (28). Yet, further simulations should be performed based on the availability of other structures.
Finally, the current CG simulations predicted that the unidirectional translocation is realized by the paddling motion, which needs to be verified experimentally.
Materials and Methods
Reference Structures.
We used the two reference structures: the OU form with PDB ID code 1DO2 (11) and the CD form with PDB ID code 1G4b (18). The 1DO2 structure binds 3 nucleotides, and the 1G4b most likely binds 6 nucleotides. Proteins were drawn with PyMOL (DeLano Scientific).
Fully Atomistic MD Simulations.
As a substrate, the capped N-terminal 12 residues of Arc repressor, Ac-MKGMSKMPQFNL-NH2, were used because they are known to serve as the signal for HslUV (13). Initially, the substrate in an extended form was threaded into the HslU pore so that the N-terminal three residues, MKG, were out of the pore at the HslV side. The fully atomistic simulations were performed at T = 350 K by GROMACS 3.3.1 (29, 30) with the OPLS/AA force field (31) for proteins and the TIP4P model (32) for water molecules. The total charge in the simulations was neutralized by Na+ or Cl−. The Cα atoms of HslU, except for residues 78–100, were constrained by harmonic springs to their initial positions. For both the CD and OU forms, after equilibration, 4-ns-long simulations were conducted three times without the pulling force. As a control, the same substrate was simulated in bulk water without HslU under the same condition. Next, the N terminus of the substrate was weakly pulled from the HslV side, and for both the CD and OU forms 4-ns-long simulations were conducted five times. Details are in SI Appendix.
CG MD Simulations.
For the CG simulations, the details are in SI Appendix. In the CG simulations, HslU was represented by beads located at the Cβ atoms, instead of the Cα atoms used in previous works (21, 33). A stretched substrate used as the rail, which was also represented by one bead per residue, was prepared by connecting 100 residues by virtual bonds with the length 3.8 Å and the dihedral angle around the bond 230°. The angle between two adjacent bonds was set to 130° (150° for the thinner substrate).
The total energy function consists of five terms: (i) VMBHslU is the MB energy function of (HslU)6, where the bias parameter ΔVbias was set to ΔVCD-biased = −3,000 (ΔVOU-biased = +3,000) when the CD (OU) state was more stable. (ii) Vsubstrate is the energy function of the substrate. (iii) The hydrophobic interaction VHPsubstrate YV(HslU) acts between substrate residues and Tyr-91s and Val-92s of HslU (34). (iv) Vexvolsubstrate-HslU is the repulsive interaction between HslU and the substrate. (v) Vfix is for fixing the substrate or HslU on their initial positions. MD simulations were conducted by the underdamped Langevin dynamics (35) at the temperature T = 0.16, with the mass of all residues 10, the friction constant 0.01, and the time step 0.2. The pore radius is defined by the averaged distance between six Tyr-91s and the center of mass of Tyr-91s. One cycle of ATP hydrolysis contains 8 × 104 MD steps, and ΔVbias was changed every 4 × 104 steps.
Supplementary Material
Acknowledgments.
We thank Rie Tatsumi-Koga for critical reading of the manuscript. This work was supported by Grant-in-Aid for Scientific Research and Research and Development of the Next-Generation Integrated Simulation of Living Matter of the Ministry of Education, Culture, Sports, Science, and Technology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904756106/DCSupplemental.
References
- 1.Hopfner KP, Michaelis J. Mechanisms of nucleic acid translocases: Lessons from structural biology and single-molecule biophysics. Curr Opin Struct Biol. 2007;17:87–95. doi: 10.1016/j.sbi.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Tucker PA, Sallai L. The AAA+ superfamily: A myriad of motions. Curr Opin Struct Biol. 2007;17:641–652. doi: 10.1016/j.sbi.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 3.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA plus ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Rohrwild M, et al. HslV–HslU: A novel ATP-dependent protease complex in Escherichia coli related to the eukaryotic proteasome. Proc Natl Acad Sci USA. 1996;93:5808–5813. doi: 10.1073/pnas.93.12.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoo SJ, et al. Purification and characterization of the heat shock proteins HslV and HslU that form a new ATP-dependent protease in Escherichia coli. J Biol Chem. 1996;271:14035–14040. doi: 10.1074/jbc.271.24.14035. [DOI] [PubMed] [Google Scholar]
- 6.Missiakas D, Schwager F, Betton JM, Georgopoulos C, Raina S. Identification and characterization of HsIV HsIU (ClpQ ClpY) proteins involved in overall proteolysis of misfolded proteins in Escherichia coli. EMBO J. 1996;15:6899–6909. [PMC free article] [PubMed] [Google Scholar]
- 7.Seong IS, Oh JY, Yoo SJ, Seol JH, Chung CH. ATP-dependent degradation of SulA, a cell division inhibitor, by the HslVU protease in Escherichia coli. FEBS Lett. 1999;456:211–214. doi: 10.1016/s0014-5793(99)00935-7. [DOI] [PubMed] [Google Scholar]
- 8.Kanemori M, Yanagi H, Yura T. The ATP-dependent HslVU/ClpQY protease participates in turnover of cell division inhibitor SulA in Escherichia coli. J Bacteriol. 1999;181:3674–3680. doi: 10.1128/jb.181.12.3674-3680.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanemori M, Nishihara K, Yanagi H, Yura T. Synergistic roles of HslVU and other ATP-dependent proteases in controlling in vivo turnover of σ32 and abnormal proteins in Escherichia coli. J Bacteriol. 1997;179:7219–7225. doi: 10.1128/jb.179.23.7219-7225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sousa MC, et al. Crystal and solution structures of an HslUV protease–chaperone complex. Cell. 2000;103:633–643. doi: 10.1016/s0092-8674(00)00166-5. [DOI] [PubMed] [Google Scholar]
- 11.Bochtler M, et al. The structures of HsIU and the ATP-dependent protease HsIU–HsIV. Nature. 2000;403:800–805. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- 12.Sousa MC, Kessler BM, Overkleeft HS, McKay DB. Crystal structure of HslUV complexed with a vinyl sulfone inhibitor: Corroboration of a proposed mechanism of allosteric activation of HslV by HslU. J Mol Biol. 2002;318:779–785. doi: 10.1016/S0022-2836(02)00145-6. [DOI] [PubMed] [Google Scholar]
- 13.Burton RE, Baker TA, Sauer RT. Nucleotide-dependent substrate recognition by the AAA+ HslUV protease. Nat Struct Mol Biol. 2005;12:245–251. doi: 10.1038/nsmb898. [DOI] [PubMed] [Google Scholar]
- 14.Yakamavich JA, Baker TA, Sauer RT. Asymmetric nucleotide transactions of the HslUV protease. J Mol Biol. 2008;380:946–957. doi: 10.1016/j.jmb.2008.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park E, et al. Role of the GYVG pore motif of HslU ATPase in protein unfolding and translocation for degradation by HslV peptidase. J Biol Chem. 2005;280:22892–22898. doi: 10.1074/jbc.M500035200. [DOI] [PubMed] [Google Scholar]
- 16.Song HK, et al. Mutational studies on HslU and its docking mode with HslV. Proc Natl Acad Sci USA. 2000;97:14103–14108. doi: 10.1073/pnas.250491797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, et al. Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU. Structure (London) 2001;9:1107–1116. doi: 10.1016/s0969-2126(01)00670-0. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, et al. Crystal structures of the HslVU peptidase–ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure (London) 2001;9:177–184. doi: 10.1016/s0969-2126(01)00570-6. [DOI] [PubMed] [Google Scholar]
- 19.Wang J. A corrected quaternary arrangement of the peptidase HslV and atpase HslU in a cocrystal structure. J Struct Biol. 2001;134:15–24. doi: 10.1006/jsbi.2001.4347. [DOI] [PubMed] [Google Scholar]
- 20.Martin A, Baker TA, Sauer RT. Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat Struct Mol Biol. 2008;15:1147–1151. doi: 10.1038/nsmb.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koga N, Takada S. Folding-based molecular simulations reveal mechanisms of the rotary motor F1-ATPase. Proc Natl Acad Sci USA. 2006;103:5367–5372. doi: 10.1073/pnas.0509642103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okazaki K, Koga N, Takada S, Onuchic JN, Wolynes PG. Multiple-basin energy landscapes for large-amplitude conformational motions of proteins: Structure-based molecular dynamics simulations. Proc Natl Acad Sci USA. 2006;103:11844–11849. doi: 10.1073/pnas.0604375103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyeon C, Onuchic JN. Mechanical control of the directional stepping dynamics of the kinesin motor. Proc Natl Acad Sci USA. 2007;104:17382–17387. doi: 10.1073/pnas.0708828104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hyeon C, Lorimer GH, Thirumalai D. Dynamics of allosteric transitions in GroEL. Proc Natl Acad Sci USA. 2006;103:18939–18944. doi: 10.1073/pnas.0608759103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoyt MA, et al. Glycine-alanine repeats impair proper substrate unfolding by the proteasome. EMBO J. 2006;25:1720–1729. doi: 10.1038/sj.emboj.7601058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Onuchic JN, Luthey-Schulten Z, Wolynes PG. Theory of protein folding: The energy landscape perspective. Annu Rev Phys Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 27.Ma B, Kumar S, Tsai CJ, Nussinov R. Folding funnels and binding mechanisms. Protein Eng. 1999;12:713–720. doi: 10.1093/protein/12.9.713. [DOI] [PubMed] [Google Scholar]
- 28.Martin A, Baker TA, Sauer RT. Rebuilt AAA + motors reveal operating principles for ATP-fueled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- 29.Berendsen HJC, Vanderspoel D, Vandrunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;91:43–56. [Google Scholar]
- 30.Lindahl E, Hess B, van der Spoel D. GROMACS 3.0: A package for molecular simulation and trajectory analysis. J Mol Mod. 2001;7:306–317. [Google Scholar]
- 31.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem B. 2001;105:6474–6487. [Google Scholar]
- 32.Jorgensen WL, Madura JD. Temperature and size dependence for Monte Carlo simulations of TIP4P water. Mol Phys. 1985;56:1381–1392. [Google Scholar]
- 33.Clementi C, Nymeyer H, Onuchic JN. Topological and energetic factors: What determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? An investigation for small globular proteins. J Mol Biol. 2000;298:937–953. doi: 10.1006/jmbi.2000.3693. [DOI] [PubMed] [Google Scholar]
- 34.Fujitsuka Y, Takada S, Luthey-Schulten ZA, Wolynes PG. Optimizing physical energy functions for protein folding. Proteins. 2004;54:88–103. doi: 10.1002/prot.10429. [DOI] [PubMed] [Google Scholar]
- 35.Honeycutt JD, Thirumalai D. The nature of folded states of globular proteins. Biopolymers. 1992;32:695–709. doi: 10.1002/bip.360320610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.