Abstract
Periglacial soils are one of the least studied ecosystems on Earth, yet they are widespread and are increasing in area due to retreat of glaciers worldwide. Soils in these environments are cold and during the brief summer are exposed to high levels of UV radiation and dramatic fluctuations in moisture and temperature. Recent research suggests that these environments harbor immense microbial diversity. Here we use sequencing of environmental DNA, culturing of isolates, and analysis of environmental variables to show that members of the Chytridiomycota (chytrids) dominate fungal biodiversity and perhaps decomposition processes in plant-free, high-elevation soils from the highest mountain ranges on Earth. The zoosporic reproduction of chytrids requires free water, yet we found that chytrids constituted over 70% of the ribosomal gene sequences of clone libraries from barren soils of the Himalayas and Rockies; by contrast, they are rare in other soil environments. Very few chytrids have been cultured, although we were successful at culturing chytrids from high-elevation sites throughout the world. In a more focused study of our sites in Colorado, we show that carbon sources that support chytrid growth (eolian deposited pollen and microbial phototrophs) are abundant and that soils are saturated with water for several months under the snow, thus creating ideal conditions for the development of a chytrid-dominated ecosystem. Our work broadens the known biodiversity of the Chytridomycota, and describes previously unsuspected links between aquatic and terrestrial ecosystems in alpine regions.
Keywords: alpine ecology, glacial retreat, nival zone, psychrophiles
Vast regions of the terrestrial biosphere, including deserts, polar regions, and the slopes of the Earth's highest mountains, are devoid of plant life. Research on life in these extreme soil environments has increased in recent years, especially in unvegetated areas of the high Arctic and Antarctica (1–3). Less well studied are high-elevation soils that may contain large reservoirs of undocumented biodiversity (4, 5). High-elevation soils are unique and may be challenging for life, mainly due to their oligotrophic nature (5) and dramatic freeze-thaw cycles, even in summer (6).
We know very little about how life is supported in high-elevation soils in the absence of primary production from plants. Swan and others (7, 8) hypothesized that life in such soils must be supported by eolian transport of organic matter from lower elevations; whereas Freeman et al. (5) presented preliminary evidence of an algal and cyanobacterial supported food web in “barren” high-elevation soils. Previous work suggests that fungi are important decomposers in these soils, but the fungal community has never been characterized (9).
A majority of molecular surveys of fungal community composition have shown that Basidiomycota and Ascomycota are the dominant fungi in vegetated soils (10, 11). In contrast, Zygomycota are often rare, although recent studies suggest they can be abundant in subtropical soils (12) and may be abundant in transiently favorable habitats like beneath late-winter snowpacks in coniferous forests (13). While the relative abundance of fungi varies among ecosystems, some fungi are consistently rare in all previous surveys of fungal diversity in soil (e.g., the Mucoromycotina and Chytridiomycota). Consequentlty, these groups, particularly the Chytridiomycota, have been regarded as having little importance to soil ecosystem function (14).
This present study demonstrates the importance of understanding the unique organisms that inhabit high-elevation soils. These areas are changing rapidly due to global warming (15, 16) and many high-elevation ecosystems that depend on snowmelt and long periods of snow cover may disappear in the future, leading to the extinction of many species before they are even known to science. Here we report the surprising diversity of chytrids and the unexpectedly high carbon inputs to unvegetated, high-elevation soils of the Rocky, Andes, and Himalayan mountains. Our results show that, unlike all other soil systems studied to date, these soils are dominated by members of the Chytridiomycota (chytrids); and that these fungi with aquatically dispersed (flagellated) reproductive stages depend on soil moisture from melting snowbanks, eolian inputs of organic matter (mostly pollen), and especially on endogenous primary production by microbial phototrophs.
Results and Discussion
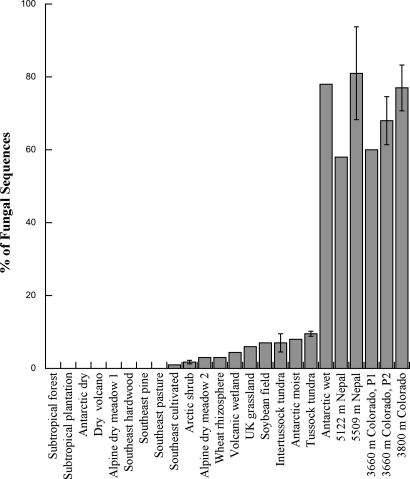
Multiple surveys of 18S rRNA genes from the Rockies and Himalayas and extensive culturing of new isolates from throughout the world revealed an unexpected diversity and abundance of Chytridiomycota in high-elevation, unvegetated soils. In Colorado soils, chytrids comprised over 30% of the eukaryotic and 70% of the fungal sequences (Fig. 1). Consistent with our results from Colorado, chytrids constituted 20% of the eukaryotic sequences and 75% of all fungal sequences in soils collected from six sites (all above 5,100 m elevation) in the Himalayas of Nepal (Fig. 1). These results suggest that chytrids are the dominant members of the fungal community and perhaps even the eukaryotic community in high-elevation, unvegetated soils. In contrast, in most other general surveys of fungal abundance in soils, chytrids were either undetectable or constituted less than 10% of the fungal sequences obtained (for references see Fig. 1). The one exception to this pattern is a site in Antarctica (Antarctic wet in Fig. 1) that receives large inputs of seasonal snowmelt, much like our sites in Nepal and Colorado.
Fig. 1.
Percent relative abundance of chytrid phylotypes in fungal clone libraries from soils collected across a range of environments. More than 60% of phylotypes from our Nepal and Colorado unvegetated, high-elevation soils were chytrids, whereas in all other environments chytrids comprised less than 10% of fungi. P1 and P2 represent different primer sets used at our sites in Colorado. Error bars are standard error of the mean of spatially separated replicates from a given environment. Data for other sites were obtained from the literature: subtropical forest and plantation (12), Antarctic dry (38), dry volcano (4), alpine dry meadow (39), four land-use types in Southeast USA (10), Arctic shrub (26), alpine dry meadow 2 (11), wheat rhizosphere (14), volcanic wetland (4), U.K. grassland (17), soybean field (40), intertussock and tussock tundra (26), Antarctic moist and wet (38). For our data, fungi comprised 45% and 36% of all eukaryotic sequences obtained from Colorado and Nepal, respectively. In contrast, in the only other study that reported similar data for soils, fungi comprised only 9% of the total eukaryotes (4).
It is possible that the low relative abundance of chytrids across most previously studied environments reflects bias associated with PCR amplification; however, there is no evidence for such bias across a range of different primer pairs (14, 17) and in this study two different primer pairs yielded the same high relative abundance of chytrids in the soil (Fig. 1). It is also possible that rDNA copy number and nuclei density per cell can bias estimates of the abundance of fungal groups in studies such as this one. However, based on preliminary results from whole genome shotgun reads of two chytrids (Batrachochytrium dendrobatidis and Spizellomyces sp.) by Christina Cuomo of the Broad Institute (www.broad.harvard.edu/genome_bio/bios/bio-cuomo.html), 18S copy number ranges from approximately 50–80 in chytrids, comparable to that found in other fungi. In addition, based on their phylogenetic position (see below) most of the chytrids identified in the present study are monocentric, meaning that individuals have only one nucleus per cell for much of their life cycle. Therefore, although we cannot completely rule out variability in rDNA copies as a partial explanation for the dominance of chytrid sequences in high elevation soils, at present there is no evidence that copy number is higher in chytrids in general or that chytrids from high elevations would have higher copy number than chytrids from lower elevations.
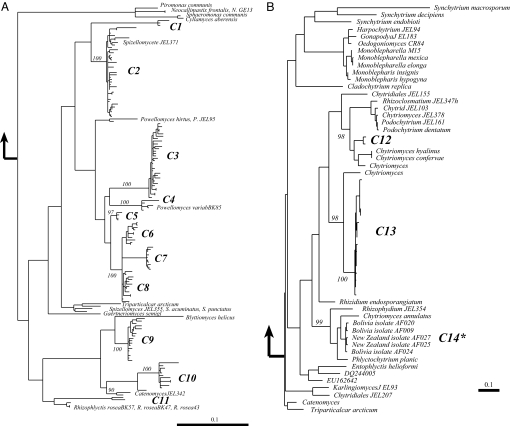
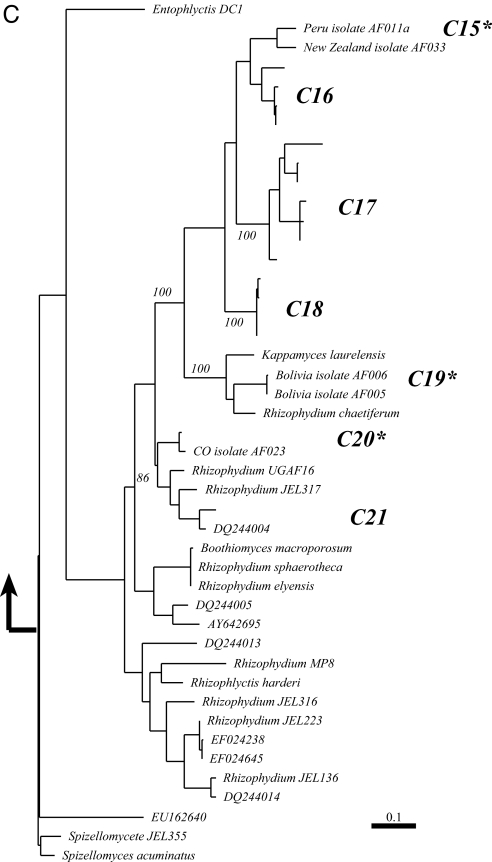
Phylogenetic analysis of our isolates and environmental sequences revealed that high-elevation, unvegetated soil supports a diverse assemblage of many Chytridiomycota (Fig. 2). There were 27 distinct clades of chytrids, 21 of which were analyzed in detail (Fig. 2). Only three of the clades found in this study included sequences that were nearly identical to known taxa, Spizellomyces JEL371, Powellomyces variablis, and Catenomyces JEL342. All other sequences grouped separately and distinctly from taxa in the GenBank and AFTOL databases, suggesting that unvegetated, high-elevation soils harbor a largely unstudied and previously unknown chytrid community.
Fig. 2.
Inferred phylogenetic relationships of sequences obtained from cultures and clone libraries in the present study compared to the known diversity of major groups of chytrids. (A) shows clades C1–C11; sequences in these clades are most closely related to the orders Spizellomycetales and Rhizophlyctidales. (B) clades C12–C14, sequences in these clades are most closely related to the order Chytridiales. (C) clades C15–C21; sequences in these clades are most closely related to the order Rhizophydiales. Black arrows indicate where on the tree the other groups of chytrids attach. An overview of how the major groups of chytrids link to one another is presented in Fig. S1. Bootstrap values are indicated at nodes above 90%; numbered clades had bootstrap support of at least 90% and/or branch lengths of subtending clades long enough to warrant separate discussion (>1% sequence evolution). An asterisk indicates those clades that include a cultured isolate reported in this study.
The majority of our environmental sequences grouped in a deeply divergent clade containing the Spizellomycetales and its sister order the Rhizophlyctidales (18, 19) (Fig. 2A), both of which are notable in their ability to tolerate freezing and dessication (20). Members of the Spizellomycetales are generally saprotrophic, and can use pollen as a substrate (21). Powellomyces species grow inside pollen grains and thus may have increased protection from changes in temperature and water availability (22). Some of our Rhizophlyctidales phylotypes (Fig. 2A) grouped with Blyttiomyces helicus (C9), a known pollen degrader (18).
Two clades of environmental sequences grouped within the order Chytridiales (18) (Fig. 2B). Sequences in clades C12 and C13 are related to the freshwater species Chytriomyces hyalinus and C. confervae, organisms that grow on pollen and algae, respectively (24).
A number of divergent sequences grouped within the Rhizophydiales (18), including several isolates that we have cultured from unvegetated soils in Peru, Bolivia, New Zealand, and Colorado (clades C15, C19, C20) (Fig. 2C). Fifteen sequences from this study and two cultured chytrids from high-elevation, Peruvian soil comprised a clade that has a sister taxa relationship with Kappamyces laurelensis and R. chaetiferum (Fig. 2C). Based on this tree, there may be as many as seven unique taxa within this clade. Clades C20 and C21 are related to Aquamyces chlorogonii, an organism isolated from a unicellular green algae (25). Members of the Rhizophydiales (Fig. 3C) can use both algae and pollen as carbon sources for growth (25). Furthermore, some Rhizophydiales microscopic animals as substrates, suggesting the the Alveolata, Cercozoa, and Metazoa found in our soils could be an additional food source.
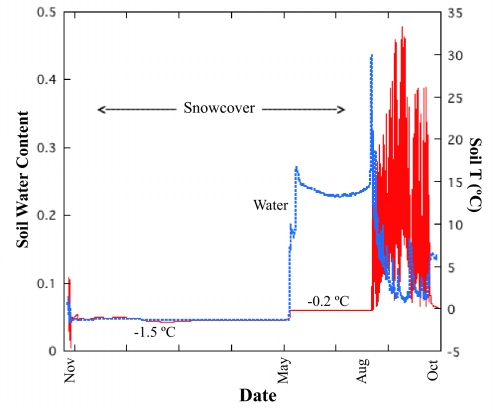
Fig. 3.
Soil temperature and moisture data (4 cm depth) from a typical year at our Colorado sites. Soils are saturated for several months under the melting snowpack, ideal conditions for the growth of chytrids. In contrast, the short summer period was dry with large, diurnal temperature fluctuations. The spike in soil water at the end of the snow-covered period was due to a large rainfall event that washed away the remaining snowpack (8).
Finally, several of the environmental sequences grouped basally on the tree, suggesting high-alpine soil environments, may harbor deeply divergent lineages. These sequences were not similar to any known taxa, but grouped with other environmental sequences from diverse sites around the globe, including Swiss snowmelt water (AJ867631), anoxic marine sediment (AY180024), lake sediment in Japan (AB252775), a lake in Greece (FJ157332), and soil of the Netherlands (AM114806).
Do High-Elevation Environments Favor Chytrids?
Given the abundance and diversity of chytrids in high-elevation soils we examined the environmental variables that could help explain their abundance in these extreme soils. We installed data loggers at our sites to record year-round soil temperature and moisture data. At our sites in Colorado, deep snowpack ameliorated the extreme conditions, leading to a period when soils under snowpack were saturated with water for several months and remained at temperatures conducive to microbial activity (Fig. 3). This explains why fungal biomass and decomposition processes in these soils reach peak levels during and just after the late snow-covered period (9). We hypothesize that chytrids can dominate these sites partially because the soils are essentially aquatic habitats during the period of saturation under the snow. Tussock tundra soils also undergo periods of water saturation in the late spring and early summer (26) and this may also help explain why tussock tundra sites have higher chytrid abundances than most other soils studied to date (Fig. 1).
Next we examined the potential carbon inputs to high-elevation soils to determine if substrates known to support chytrids were present at our sites. Chytrids are known to degrade pollen and parasitize and/or decompose dead algae and cyanobacteria (see above). Despite the complete lack of plants at our sites, there were high levels of pollen in these soils (Table 1). At our Colorado sites, we quantified pollen at two depths from four locations, for three dates during the snow-free period of 2007 (Fig. S2). Large amounts of pollen were observed at both depths on all dates indicating that there was pollen to support pollen-degrading chytrids in these soils. Based on size and obvious morphological features, most of the pollen observed in these soils was from Ponderosa and Lodge Pole Pines. These trees are very common at lower elevation throughout Colorado and prevailing west-to-east winds probably carry pollen across the Continental Divide (the ridge above our highest sites) and deposit it at our sites. To compare pollen inputs with other possible carbon sources for chytrids (see below), we estimated the carbon content of pollen (Table 1). It should be pointed out that pollen grains can persist in soil long after they have been stripped of soluble nutrients, thus we were probably over-estimating C inputs from pollen compared to C inputs from microbial photosynthesis. In addition, surveys of soils at our Nepalese sites found much lower pollen densities (Fig. S2), indicating that chytrids may depend more on microbial phototrophs (see below) than on degrading pollen in high elevation soils.
Table 1.
Carbon pools and inputs to our high-elevation sites in Colorado (2007)
| Date | Pollen C, grams of C per square meter | DOC, grams of C per square meter | Microb. Biomass, grams of C per square meter | Microb. Photosyn., grams of C per square meter per day |
|---|---|---|---|---|
| 8/13 | 14 (5) | 4.7 (1.0) | 1.0 (0.4) | 0.8 (0.1) |
| 8/27 | 14 (1) | 3.8 (0.6) | 4.3 (2.0) | 0.5 (0.2) |
| 9/21 | 27 (8) | 6.5 (1.4) | 1.7 (0.3) | 0.4 (0.1) |
| Mean | 18 (5) | 5.0 (0.6) | 2.3 (0.7) | 0.6 (0.1) |
| N | 12 | 18 | 18 | 18 |
Pollen, DOC and microbial biomass were standing concentrations in soil on each date, whereas microbial photosynthesis is the rate of C input per day as estimated from CO2 flux measurements. From these CO2 flux estimates and measurements from multiple dates in 2002 and 2007 (5), approximately 24 g of C enters the soils from microbial photosynthesis per year at these sites, making microbial C fixation the largest C input to this system.
Finally, we also have carried out molecular surveys, direct epifluorescence counts, and light-driven, CO2-uptake measurements to determine if algae and cyanobacteria could be helping to maintain the high diversity and abundance of chytrids in these soils. Cyanobacteria and algae are extremely diverse and abundant in these soils, as previously shown at our sites in the Rockies (5), Andes (6, 27, 28), and Himalayas. In addition, light-driven CO2 uptake occurred at high rates on all dates at our Colorado sites indicating that there was ample primary production to support a robust food web in these seemingly barren soils (Table 1). Based on the data presented here and multiple years of data from Freeman et al. (5), we estimate a yearly influx of carbon of 24 g C m−2 yr−1 enters these soils during a typical year, as the result of microbial photosynthesis. This number is much higher than the observed total standing crop of pollen in these soils, perhaps indicating that chytrids in these soils depend more on parasitism and/or decomposition of algae and cyanobacteria than on pollen degradation.
Conclusion
Through the use of sequencing and culturing approaches, we have discovered 27 distinct clades of Chytridiomycota in soils from high-elevation sites in Nepal, Colorado, Hawaii, Bolivia, Peru, and New Zealand. Many of these phylotypes are unique among known chytrid sequences indicating that high-elevation soils may harbor a large reservoir of previously unstudied chytrids that utilize both pollen and microbial phototrophs as sources of C.
Methods
Study Sites and Sample Collection.
Our main sampling sites were in the Annapurna Conservation Area (ACA) of the Himalaya Mountains (Nepal) and the Front Range of the Colorado Rocky Mountains (USA). At both locations we sampled sites just above the highest plants in each region and sites, as high as were attainable due to prevailing conditions (e.g., presence of ice and snow) at the time of sampling. The low-elevation sites in the ACA are unvegetated, southeast facing slopes (28°43′N, 83°55′E) at 5,122 m above sea level (m.a.s.l.) and approximately 8 km east of the northern tip of Tilicho Lake. The high-elevation sites in the ACA are unvegetated southeast facing slopes from 5,503–5,516 m.a.s.l., 1 km north of Thorong Pass (28°48′N, 83°56′E). Thorong Pass crosses the great divide separating the Marsyangdi River to the east and the Kali Gandaki River to the west. The low and high-elevation sites in Colorado are unvegetated southeast facing slopes at 3,660 and 3,800 m.a.s.l., respectively in the Green Lakes Valley (40°03′N, 105°35′W); an area that is off-limits to the general public. The high-elevation site is 0.1 km north of the Arikaree glacier and 0.1 km east of the Continental Divide and the low elevation site is 2 km east of the Arikaree glacier and the Continental Divide. Full descriptions of our research sites can be found elsewhere (5, 29–31). Soils were collected from all sites in Colorado and Nepal from late July to early October. Samples were collected from at least three spatially separated points at each site, with a minimum of 5 m distance between each sample point. In previous work this sampling scheme was shown to adequately represent the variation of microbial parameters within a sampling area (29). Collected soils were frozen until DNA was extracted.
DNA Extraction, Clone Libraries, and Sequence Analysis.
DNA was extracted using the MO BIO UltraClean Mega Prep Soil DNA kit (MO BIO Laboratories, Inc.). Two sets of eukaryotic-specific 18S primers were used to amplify the soil DNA. In our initial extractions (Colorado soils only) we used primer set 1 (P1 in Fig. 1). P1 consisted of Eukaryotic-specific forward primer EukA (32) (5′-AACCTGGTTGATCCTGCCAGT-3′) and reverse primer 1195RE (5′-GGGCATCACAGACCTG-3′). All other amplifications (Colorado and Nepal) were done with primer set 2 (P2 in Fig. 1). P2 consisted of forward primer 4Fa (5′-TCCGGTTGATCCTGCCRG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′). Previous studies using minor variants of these primers have shown no bias for or against chytrids (14, 17, 33). For detailed methods of PCR amplification and cloning see Freeman et al. (5).
Species of Chytridiomycota analyzed in James et al. (18), as well as additional sequences that closely matched those sampled in this study based on BLAST surveys, were aligned with the sequences obtained from the high-elevation, unvegetated sites using MUSCLE (http://www.ebi.ac.uk/Tools/muscle/index.html). We used this large tree (Fig. S1) as a means of narrowing the scope of analysis to particular groups. In particular, we selected subsets of species and repeated the alignment and phylogenetic analysis for a more robust assessment of relationships. Trees were constructed assuming an HKY model of sequence evolution and the neighbor joining clustering algorithm using PAUP. Nonparametric bootstrap analysis using 1,000 replicates was used to assess support for inferring the relationship of taxa sampled from the alpine to known species or previously published environmental sequences. Additionally, we labeled distinct clades on the trees (Fig. 2) that included sequences from the high-elevation, unvegetated sites based on the combined assessment of bootstrap scores (90% or greater) and/or if the branch lengths subtending clades were sufficiently long to warrant recognition (>1% sequence evolution). Of the 21 labeled clades, three included previously described isolates Spizellomyces JEL371, Powellomyces variablis, and Catenomyces JEL342. Importantly, the trees are not meant as robust phylogenetic hypotheses for chytrids, but rather serve as a means to describe chytrid diversity and assess whether the sequences and cultured isolates are similar to previously described samples or lineages. Robust phylogenetic inference will require the analysis of multiple genes.
Soil Carbon Pools and Fluxes.
Soil dissolved organic carbon (DOC) and microbial biomass C were measured using standard methods as previously reported (29). Briefly, soil samples were extracted with 0.5 M K2SO4 with and without chloroform fumigation. Samples extracted without fumigation are reported as DOC, and microbial biomass C was obtained by taking the difference between fumigated and unfumigated samples as described elsewhere (29). Soil pollen concentrations were determined by vortexing 1 g soil in 2 mL of water for one minute. Solutions were allowed to settle for 5 min and 50 μL of slurry was air-dried on a Reich counting slide (Bellco Glass Inc.). Pollen grains were counted using a wide-field fluorescence microscope (Leica Microsystems DMLB) with a CY3 filter cube (Leica Microsystems), green excitation filter (bandpass 535/50 nm) and an orange-red suppression filter (bandpass 610/70 nm). Counts were then converted to pollen grains per g of dry soil. To estimate C contributed to the soil by pollen, we used the average estimate of 1.8 million grains corn pollen per g (34–36). The volumes (assuming a spherical shape) of corn pollen (di = 76–106 μm) (34) and pine pollen (di = 45 μm from our sites) were determined resulting in an estimate of 13.5 million grains of pine pollen per g. Pine pollen contains approximately 500 mg of C per g (37). We estimate an average of 0.31 mg pollen C per g soil in the top 4 cm of soil. This estimate was converted to g of pollen C per m2 (Table 1) using the soil bulk density and assuming an average soil depth of 4 cm.
Field rates of soil CO2 fixation were measured using a PP Systems EGM-4 infrared gas analyzer (PP Systems). Measurements were done with a clear 1.2 L chamber to allow measurement of both light and dark CO2 flux. Measurements of light-driven CO2 flux were done with the chamber uncovered, while dark measurements were done with the chamber covered with a dark cloth to block ambient light. The chamber was flushed with ambient CO2 before each soil flux measurement and the autozero function was active throughout the measurement process. Light measurements include both CO2 uptake and CO2 respiration, while dark measurements are primarily a measure of respiration. Therefore, light-driven CO2 uptake rates were calculated by subtracting CO2 efflux rates measured in the light from those measured in the dark. The mean of a minimum of three measurement locations at each site were used to calculate mean flux as described elsewhere (5).
Supplementary Material
Acknowledgments.
We thank Laszlo Nagy, Nima Sherpa, Karma Gurung, and Andrew King for assistance with field work and Christina Cuomo for genomic analyses. This project was supported by National Science Foundation Microbial Observatories Program Grant MCB-0455606. Logistical support in Colorado was provided by the Niwot Ridge Long-Term Ecological Research site. Travel to foreign sites was funded by the National Geographic Committee for Research and Exploration and University of Colorado Faculty Fellowships (to S.K.S. and A.P.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. GQ995246–GQ995438 and GQ995439–GQ995448).
This article contains supporting information online at www.pnas.org/cgi/content/full/0907303106/DCSupplemental.
References
- 1.Aislabie JM, et al. Dominant bacteria in soils of Marble Point and Wright Valley, Victoria Land, Antarctica. Soil Biol Biochem. 2006;38:3041–3056. [Google Scholar]
- 2.Cowan DA, et al. Antarctic Dry Valley mineral soils contain unexpectedly high levels of microbial biomass. Extremophiles. 2002;6:431–436. doi: 10.1007/s00792-002-0276-5. [DOI] [PubMed] [Google Scholar]
- 3.Parsons AN, et al. Soil carbon dioxide flux in Antarctic Dry Valley ecosystems. Ecosystems. 2004;7:286–295. [Google Scholar]
- 4.Costello EK, et al. Fumarole-supported islands of biodiversity within a hyperarid, high-elevation landscape on Socompa Volcano, Puna de Atacama, Andes. Appl Environ Microbiol. 2009;75:735–747. doi: 10.1128/AEM.01469-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freeman KR, et al. Soil CO2 flux and photoautotrophic community composition in high-elevation, ‘barren’ soil. Environ Microbiol. 2009;11:674–686. doi: 10.1111/j.1462-2920.2008.01844.x. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt SK, et al. Microbial activity and diversity during extreme freeze-thaw cycles in periglacial soils, 5,400 m Elevation, Cordillera Vilcanota, Perú. Extremophiles. 2009 doi: 10.1007/s00792-009-0268-9. doi 10.1007/s00792-009-0268-9. [DOI] [PubMed] [Google Scholar]
- 7.Swan LW. The aeolian biome. Bioscience. 1992;42:262–270. [Google Scholar]
- 8.Ley RE, Williams MW, Schmidt SK. Microbial population dynamics in an extreme environment: Controlling factors in talus soils at 3,750 m in the Colorado Rocky Mountains. Biogeochem. 2004;68:313–335. [Google Scholar]
- 9.Ley RE, Schmidt SK. Fungal and bacterial responses to phenolic compounds and amino acids in high altitude barren soils. Soil Biol Biochem. 2002;34:989–995. [Google Scholar]
- 10.Lauber CL, et al. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol Biochem. 2008;40:2407–2415. [Google Scholar]
- 11.Nemergut DR, et al. The effects of chronic nitrogen fertilization on alpine tundra soil microbial communities: Implications for carbon and nitrogen cycling. Environ Microbiol. 2008;10:3093–3105. doi: 10.1111/j.1462-2920.2008.01735.x. [DOI] [PubMed] [Google Scholar]
- 12.He JZ, Xu ZH, Hughes J. Analyses of soil fungal communities in adjacent natural forest and hoop pine plantation ecosystems of subtropical Australia using molecular approaches based on 18S rRNA genes. FEMS Microb Lett. 2005;247:91–100. doi: 10.1016/j.femsle.2005.04.033. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt SK, et al. Exponential growth of “snow molds” at sub-zero temperatures: An explanation for high beneath-snow respiration rates and Q10 values. Biogeochemistry. 2009;95:13–21. [Google Scholar]
- 14.Smit E, et al. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl Environ Microbiol. 1999;65:2614–2621. doi: 10.1128/aem.65.6.2614-2621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barnett TP, Adam JC, Lettenmaier DP. Potential impacts of a warming climate on water availability in snow-dominated regions. Nature. 2005;438:303–309. doi: 10.1038/nature04141. [DOI] [PubMed] [Google Scholar]
- 16.Seimon TA, et al. Upward range extension of Andean anurans and chytridiomycosis to extreme elevations in response to deglaciation. Global Change Biol. 2007;13:288–299. [Google Scholar]
- 17.Anderson IC, Campbell CD, Prosser JI. Potential bias of fungal 18S rDNA and internal transcribed spacer polymerase chain reaction primers for estimating fungal biodiversity in soil. Environ Microbiol. 2003;5:36–47. doi: 10.1046/j.1462-2920.2003.00383.x. [DOI] [PubMed] [Google Scholar]
- 18.James TY, et al. A molecular phylogeny of the flagellated fungi (Chytridiomycota) and description of a new phylum (Blastocladiomycota) Mycologia. 2006;98:860–871. doi: 10.3852/mycologia.98.6.860. [DOI] [PubMed] [Google Scholar]
- 19.Letcher PM, et al. Rhizophlyctidales-a new order in Chytridiomycota. Mycol Res. 2008;112:1031–1048. doi: 10.1016/j.mycres.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Gleason FH, Letcher PM, McGee PA. Some Chytridiomycota in soil recover from drying and high temperatures. Mycol Res. 2004;108:850–850. doi: 10.1017/s0953756204009736. [DOI] [PubMed] [Google Scholar]
- 21.Lozupone CA, Klein DA. Molecular and cultural assessment of chytrid and Spizellomyces populations in grassland soils. Mycologia. 2002;94:411–420. [PubMed] [Google Scholar]
- 22.Longcore JE, Barr DJS, Desaulniers N. Powellomyces, a new genus in the Spizellomycetales. Can J Bot. 1995;73:1385–1390. [Google Scholar]
- 23.Mozley-Standridge SE, et al. Cladochytriales-a new order in Chytridiomycota. Mycol Res. 2009;113:498–507. doi: 10.1016/j.mycres.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Letcher PM, et al. Ultrastructural and molecular phylogenetic delineation of a new order, the Rhizophydiales (Chytridiomycota) Mycol Res. 2006;110:898–915. doi: 10.1016/j.mycres.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 25.Letcher PM, et al. Ultrastructural and molecular analyses of Rhizophydiales (Chytridiomycota) isolates from North America and Argentina. Mycol Res. 2008;112:759–782. doi: 10.1016/j.mycres.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 26.Wallenstein MD, McMahon S, Schimel J. Bacterial and fungal community structure in Arctic tundra tussock and shrub soils. FEMS Microbiol Ecol. 2007;59:428–435. doi: 10.1111/j.1574-6941.2006.00260.x. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt SK, et al. The earliest stages of ecosystem succession in high-elevation (5,000 meters above sea level), recently deglaciated soils. Proc Roy Soc B. 2008;275:2793–2802. doi: 10.1098/rspb.2008.0808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nemergut DR, et al. Microbial community succession in an unvegetated, recently deglaciated soil. Microb Ecol. 2007;53:110–122. doi: 10.1007/s00248-006-9144-7. [DOI] [PubMed] [Google Scholar]
- 29.King AJ, Meyer AF, Schmidt SK. High levels of microbial biomass and activity in unvegetated tropical and temperate alpine soils. Soil Biol Biochem. 2008;40:2605–2610. [Google Scholar]
- 30.Williams MW, Hood E, Caine N. Role of organic nitrogen in the nitrogen cycle of a high-elevation catchment, Colorado Front Range. Wat Resources Res. 2001;37:2569–2581. [Google Scholar]
- 31.Williams MW, et al. Changes in climate and hydrochemical responses in a high-elevation catchment in the Rocky Mountains, USA. Limnol Oceanog. 1996;41:939–946. [Google Scholar]
- 32.Medlin L, et al. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene. 1988;71:491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- 33.Diez B, et al. Application of denaturing gradient gel electrophoresis (DGGE) to study the diversity of marine picoeukaryotic assemblages and comparison of DGGE with other molecular techniques. Appl Environ Microbiol. 2001;67:2942–2951. doi: 10.1128/AEM.67.7.2942-2951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aylor DE. Settling speed of corn (Zea mays) pollen. J Aerosol Sci. 2002;33:1601–1607. [Google Scholar]
- 35.Hellmich RL, et al. Monarch larvae sensitivity to Bacillus thuringiensis-purified proteins and pollen. Proc Natl Acad Sci USA. 2001;98:11925–11930. doi: 10.1073/pnas.211297698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller J, Georgian T. Estimation of fine particulate transport in streams using pollen as a seston analog. J North Amer Benthol Soc. 1992;11:172–180. [Google Scholar]
- 37.Doskey PV, Ugoagwu BJ. Atmospheric deposition of macronutrients by pollen at a semi-remote site in northern Wisconsin. Atmos Environ. 1989;23:2761–2766. [Google Scholar]
- 38.Bridge PD, Newsham KK. Soil fungal community composition at Mars oasis, a southern maritime Antarctic site, assessed by PCR amplification and cloning. Fungal Ecol. 2009;2:66–74. [Google Scholar]
- 39.Schadt CW, et al. Seasonal dynamics of previously unknown fungal lineages in tundra soils. Science. 2003;301:1359–1361. doi: 10.1126/science.1086940. [DOI] [PubMed] [Google Scholar]
- 40.de Castro AP, et al. Diversity of soil fungal communities of Cerrado and its closely surrounding agriculture fields. Arch Microbiol. 2008;190:129–139. doi: 10.1007/s00203-008-0374-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.