Abstract
We report the X-ray crystal structure of a phosphodiesterase (PDE) that includes both catalytic and regulatory domains. PDE2A (215–900) crystallized as a dimer in which each subunit had an extended organization of regulatory GAF-A and GAF-B and catalytic domains connected by long α-helices. The subunits cross at the GAF-B/catalytic domain linker, and each side of the dimer contains in series the GAF-A and GAF-B of one subunit and the catalytic domain of the other subunit. A dimer interface extends over the entire length of the molecule. The substrate binding pocket of each catalytic domain is occluded by the H-loop. We deduced from comparisons with structures of isolated, ligand-bound catalytic subunits that the H-loop swings out to allow substrate access. However, in dimeric PDE2A (215–900), the H-loops of the two catalytic subunits pack against each other at the dimer interface, necessitating movement of the catalytic subunits to allow for H-loop movement. Comparison of the unliganded GAF-B of PDE2A (215–900) with previous structures of isolated, cGMP-bound GAF domains indicates that cGMP binding induces a significant shift in the GAF-B/catalytic domain linker. We propose that cGMP binding to GAF-B causes movement, through this linker region, of the catalytic domains, such that the H-loops no longer pack at the dimer interface and are, instead, free to swing out to allow substrate access. This increase in substrate access is proposed as the basis for PDE2A activation by cGMP and may be a general mechanism for regulation of all PDEs.
Keywords: cGMP activation, GAF domains, PDE-2A
The cyclic nucleotides cAMP and cGMP are ubiquitous intracellular signaling molecules that mediate a vast array of biological processes throughout the body. The means by which these two molecules participate in diverse functions, in different cell types and within single cells, involves tight regulation of the spatial and temporal residence of their concentrations. The phosphodiesterases (PDEs) are a superfamily of enzymes that metabolically inactivate cAMP and cGMP (1) to play key roles in both these aspects of regulation. The PDEs are modular enzymes characterized by a relatively conserved C-terminal catalytic domain and a more variable N-terminal domain involved in regulation of activity, subcellular localization, and interactions with other proteins. There are 11 PDE gene families, with different families encoded by one to four genes, and further diversity derived from alternative splicing. The PDE families differ broadly in specificity and affinity for cAMP and cGMP. Much has been learned about the molecular bases for these differences from studies of X-ray crystal structures of the catalytic domains of PDE1, -2, -3, -4, -5, -7, -8, -9, and -10 (2–15).
The regulation of catalytic activity is a key element in tuning of cyclic nucleotide signaling by the PDEs. PDE activity is regulated by parallel or feedback signaling pathways mediated largely through the N-terminal domains. PDE1 is activated by calcium/calmodulin binding to a site in the N-terminal regulatory domain. The long isoforms of PDE4 are regulated by protein kinase A (PKA) catalyzed phosphorylation at a site at the N-terminal end of UCR1 (16) that results in increased affinity for Mg2+, a component of the active site obligatory for catalysis (17). PDE5 is also activated by phosphorylation in the N-terminal region (18). However, the most common mechanism of regulation of PDE activity appears to be through N-terminal GAF domains. PDEs 2, 5, 6, 10, and 11 contain GAF domains, and for PDEs 2, 5, and 6, cGMP binding to these GAF domains activates the enzymes. For PDE5, cGMP-induced activation is mediated by binding to the most N-terminal, GAF-A, domain, whereas for PDE2 cGMP interacts with GAF-B, which is more proximal to the catalytic domain. PDE6 is also regulated by cGMP binding to GAF-A, in the unique context of interaction with the Pγ subunit (19, 20). The physiological role of GAF domain regulation for PDE10 and 11 is still to be determined (21).
The molecular mechanism(s) whereby binding of signaling molecules in the N-terminal domains of different PDEs regulates activity of the catalytic domain is not yet understood. Here, we report the X-ray crystal structures of a PDE protein that includes both catalytic and regulatory domains, namely, PDE2 containing the N-terminal GAF domains and the catalytic domain. Analysis of these structures suggests that the N-terminal domain of PDE2 regulates access to the substrate binding pocket as the means of regulating catalytic activity.
Results and Discussion
Native human PDE2A is a protein of 941 amino acids that is organized in four domains [N terminus (1–214), GAF-A (215–372), GAF-B (393–541), and catalytic (579–941)] and that functions as a homodimer (22). We experimented with constructs of different lengths, seeking one that would crystallize, while still maintaining the essential characteristics of the full-length protein, namely, dimerization, and activation by cGMP. The construct used in this analysis spans residues 215–900 and, thus, encompasses three of the four domains (Fig. S1). It is catalytically active, with a Vmax of 32 ± 11 μmol/min/mg for cGMP hydrolysis, which is comparable to values reported previously for the native enzyme purified from bovine heart (22) or for purified recombinant mouse PDE2A holoenzyme (23). Activation by cGMP is also similar: approximately 4-fold at a half-maximal cGMP concentration of 1.4 μM (Fig. S2). Light scattering and size-exclusion chromatography indicate the construct is a dimer in solution (Fig. S3). Thus, PDE2A (215–900) closely mimics the characteristics of the native full-length enzyme. The structure of PDE2A (215–900) was determined by molecular replacement, with data to 3.0 Å resolution and refined to an Rwork/Rfree of 21.0/31.1 (Table S1). This protein crystallized as a dimer in an asymmetric unit.
Description of the PDE2A (215–900) Structure.
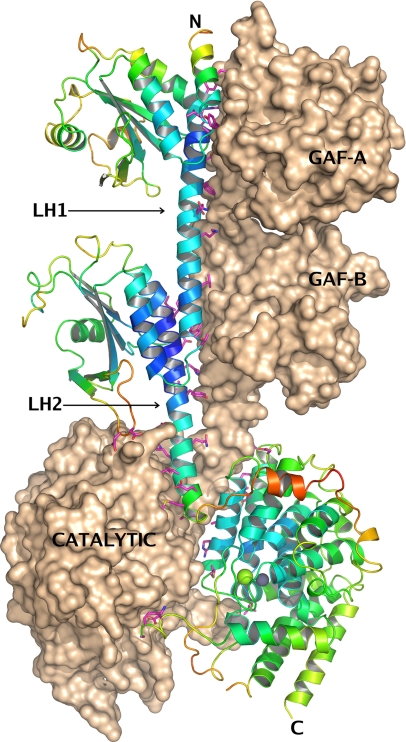
The structure of PDE2A (215–900) shows a linear extended organization of the three domains (GAF-A, GAF-B, catalytic) connected by long α-helices, with a dimer interface that extends over the entire length of the molecule (Fig. 1). The dimer is organized head-to-head with three major interfaces between GAF-A/GAF-A (total area buried at dimer interface: 2,306 Å2), GAF-B/GAF-B (1,911 Å2), and catalytic domain-A/catalytic domain-B (3,065 Å2). The overall dimensions of the dimer are approximately 140 × 70 × 30 Å. The same overall structure, with a linear organization of GAF-A, GAF-B, and catalytic domains, arranged as head-to-head dimers was also evident in cryo-EM images of PDE5 and PDE6, two other GAF-domain-containing proteins (24).
Fig. 1.
Structure of hPDE2A (215–900). The asymmetric unit contains two molecules, A and B, related by a noncrystallographic 2-fold axis of symmetry, which, in this view, is roughly parallel to the plane of the paper in the vertical direction. The three domains are labeled in the figure, as well as the linker helices LH1 and LH2 that connect them. Molecule B is shown in surface representation, and molecule A is shown in a ribbons representation, with the ribbons colored by crystallographic B-factor, blue being low and red being high. Regions of the structure with higher B-factors, such as the linker between the GAF-B domain and the catalytic domain, are expected to be more flexible. All side chains from molecule A that are within 3.5 Å of molecule B are shown as magenta sticks, The dimer interface extends over the surface of the entire molecule. The two catalytic sites in the vicinity of the Zn2+ and Mg2+ ions (shown as gray and green spheres) mutually occlude each other at the dimer interface. All figures showing the structure were generated with PyMOL (www.pymol.org).
GAF-A and GAF-B have similar overall topology that is also similar to those of the published GAF domain structures, namely, the GAF domains from yeast YKG9, mouse PDE2A, Cyanobacterium anabena adenylate cyclase, PDE5 GAF-A, PDE6 GAF-A, and PDE10 GAF-B (25–30). Each domain contains a six-stranded antiparallel β-sheet (β3-β2-β1-β6-β5-β4) sandwiched by a three-helix bundle (α1, α2, and α5) on one side and two short α-helices (α3 and α4) on the other. The C-terminal helix of GAF-A (α5, 360–372) is connected to the N-terminal helix of GAF-B (α1, 393–403) by a linker helix (LH1). Although the entire span covering residues 360–403 is α-helical, it is described here as three connected helices (α5GAF-A, LH1, α1GAF-B) for consistency with the previously established nomenclature of individual GAF domains.
The two GAF-B/GAF-B and GAF-A/GAF-A dimer interfaces are similar, with the majority of the interacting residues coming from α1 and α5 from each subunit. Additionally, the α1-α2 loop packs against the β5′-β6′ loop. The two LH1 helices are roughly parallel to the dimer axis and form extensive hydrophobic contacts along their entire length. The C-terminal helix of GAF-B (α5, 534–541) has a kink at residue 541, followed by a second linker helix (LH2, 542–571) that connects the GAF-B domain to the catalytic domain. LH2 helices of the two subunits cross over the 2-fold axis, so that the catalytic domain of one molecule lies directly below the GAF-B domain of the other molecule of the dimer.
The all α-helical structure of the catalytic domain in PDE2A (215–900) is nearly identical to the previously published structure of the isolated PDE2A catalytic domain (7). The significant exception is in conformational differences in the H- and M-loops that flank the catalytic site, discussed below.
Comparison with Other Structures of Tandem GAF Domains.
PDE2A (215–900) is a structure of a phosphodiesterase containing both the regulatory GAF domains and the catalytic domain. This is also a first GAF domain structure without a bound cyclic nucleotide. The PDE2A (215–900) structure has two significant differences from the structures of tandem GAF domains described previously (26, 27). These are 1) the relative orientation of GAF-A and GAF-B within the dimer and 2) the conformation of GAF-B in the absence of bound ligand.
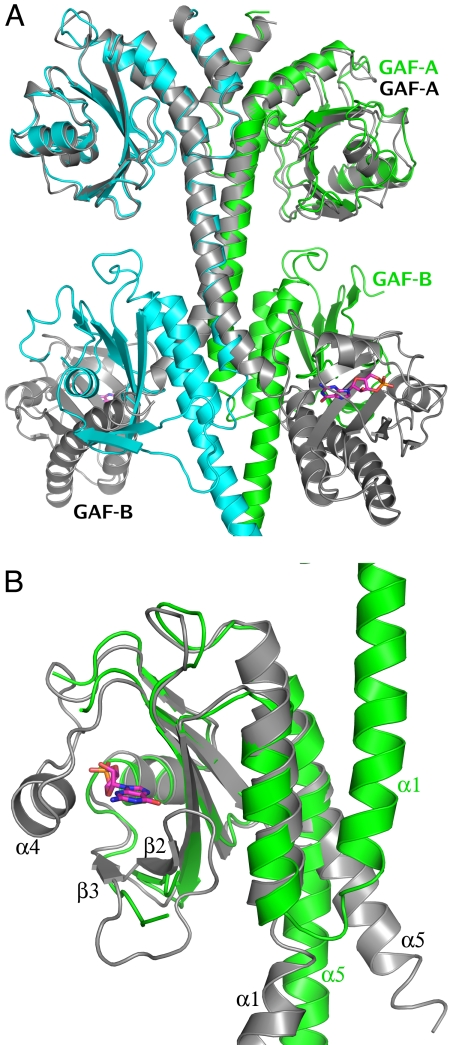
A superposition of the nucleotide-bound structure of the isolated GAF domains from mouse PDE2A (26) and the present structure is shown in Fig. 2A. In both structures, the GAF domains are dimeric, and the GAF-A/GAF-A dimers can be superimposed very closely (r.m.s.d. of 0.8 Å for 130 overlapping Cα atoms). However, with the GAF-A domains superimposed, the GAF-B domains from the two structures do not overlap. In the structure of the isolated GAF domains, the connecting helices cross, which separates the centers of the GAF-B domains by approximately 65 Å. In contrast, in PDE2A (215–900) the connecting helices are roughly parallel, with the GAF-A/GAF-B domains of each molecule in series and the GAF-B domains in contact, forming a tight interface.
Fig. 2.
Comparison of PDE2A (215–900) with the structure of the cGMP-bound GAF domains from mouse PDE2A (PDB ID code 1MC0) (26). (A) Superposition of the two dimers [1mc0, gray; PDE2A (215–900), green and cyan] using just the Cα atoms from the GAF-A domain of one subunit shows the GAF-A dimer interface to be nearly identical in the two structures, while the GAF-B domains do not overlap at all. GAF-A and GAF-B domains from one subunit of each structure have been labeled. (B) Superposition of just the GAF-B domains from the two structures [1mc0, gray; PDE2A (215–900), green] shows the conformational changes between the cGMP-bound and apo GAF-B domain. There are significant differences (A) in the immediate vicinity of the cGMP binding site—α4 helix and β2-β3 loop, which interact with the bound cyclic nucleotide (shown in magenta sticks), are completely disordered in the PDE2A (215–900) structure, and (B) in the orientation of α1 and α5, the two helices that define the N and C terminii of this domain are rotated by nearly 180° and 30°, respectively.
Two major areas of conformational difference are also noted between the unliganded (apo) GAF-B of PDE2A (215–900) and the nucleotide-bound GAF-B from mouse PDE2A published previously (26), clearly seen when the two GAF-B domains are superimposed individually (Fig. 2B). All crystal structures of nucleotide-bound GAF domains show the nucleotide to be completely buried (25–30). Helix α4 and the loop connecting strands β2 and β3 are fully ordered and are “clamped down” on the nucleotide, making multiple specific interactions. In contrast, in the “apo” structure described here, no connected electron density is visible for helix α4, implying that it is flexible and exists in multiple conformations in the crystal. The loop connecting strands β2 and β3 is similarly disordered in molecule B of the dimer and is just barely traceable in molecule A, where it is partially stabilized by a crystal lattice contact. This suggests that pocket opening may be achieved by movement of α4 and the β2-β3 loop, while the rest of the pocket, in particular the helix dipole at the N terminus of α3 that may attract the negatively charged phosphate group of the cyclic nucleotide, remains stationary. A similar observation was reported in the NMR structure of the GAF-A domain of PDE5 (28), where the spectra in the absence of bound nucleotide showed extreme line broadening, suggesting that in the absence of a bound nucleotide, α4 and β2-β3 sample multiple conformations and do not adopt a defined tertiary structure.
The second major conformational difference between the “apo-” and nucleotide-bound GAF-B structures is in the relative orientation of α1 and α5, the first and last helices of GAF-B (Fig. 2B). In the “apo” structure, the helical axes of α1 and α5 are nearly parallel, and the two helices interact at the dimer interface with their dimer-related partners α1′ and α5′. In the cGMP-bound structure, α1 is flipped by nearly 180°, so that it is antiparallel to α5, and there is no four-helix bundle at the dimer interface. The C-terminal helix, α5, which is the connector to the catalytic domain, is rotated by about 30°. It is noted that the magnitude of these differences may be exaggerated by the comparison of the isolated GAF domains with the present structure that has the catalytic domain attached to the end of α5.
The data discussed above indicate that cGMP binds into a highly flexible pocket in GAF-B, which becomes ordered upon ligand binding. Helix α4 and loop β2-β3 clamp down on the nucleotide, engendering multiple specific side-chain interactions that contribute to the observed high affinity binding. Occupation of the binding pocket by cGMP also appears to shift the orientation of the α1 and α5 helices of GAF-B, the latter being part of the linker to the catalytic domain. We believe it reasonable to infer that movement in the linker helix connected to α5 leads to a rearrangement of the dimer interface between the catalytic domains. Insight into how such rearrangement of the catalytic domains could lead to enzyme activation follows from observation of the changes in configuration of the catalytic site between liganded and unliganded state, discussed in the next section.
The Catalytic Domain Has “Open” and “Closed” Conformations.
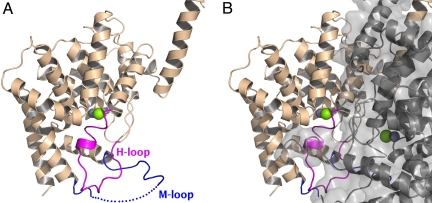
The overall α-helical architecture of the PDE2A (215–900) catalytic domain is similar to that seen in all of the crystal structures of isolated catalytic domains of other PDEs. However, in the vicinity of the substrate binding site, there are two major exceptions never previously observed (Fig. 3A). First, residues 702–723 of the H-loop are in a conformation in which they fold into and completely occlude the substrate-binding site. Second, residues 830–856 (M-loop), between helices 14 and 15 are also in a very different conformation. Helix 14 is shorter by almost three turns at its C-terminal end as compared to other PDE catalytic domains, while helix 15 is longer at its N-terminal end by about two turns. The loop connecting these two helices (M-loop) is folded away from the substrate-binding pocket and participates in dimer contacts. In contrast, the M-loop in other PDE catalytic domain structures is folded in toward the substrate binding pocket. With the H- and M-loops folded in as in PDE2A (215–900), the substrate-binding face of the catalytic domain is packed against the substrate-binding face of the second catalytic domain of the dimer, closing off access to the substrate binding sites (Fig. 3B). Thus, in the PDE2A (215–900) structure, the catalytic domain appears in a “closed” conformation.
Fig. 3.
The two catalytic sites mutually occlude each other at the dimer interface. (A) The catalytic domain of a single subunit of the PDE2A (215–900) dimer is shown in ribbons representation, with the H-loop colored in magenta and the M-loop colored in blue. The active site, whose location can be inferred from the position of the Zn2+ and Mg2+ ions (shown as gray and green spheres) is partially occupied by residues from the H-loop. Residues 840–850 of the M-loop have not been modeled due to disorder and are indicated by a dotted line. (B) The second subunit is shown as a semitransparent gray surface, as well as in ribbons representation, keeping exactly the same orientation as in panel A. The H-loop is blocked from swinging out of the active site by the dimer interface.
For substrate to access the catalytic site of PDE2A (215–900), the H-loop would have to swing out, allowing the catalytic site to adopt an “open” conformation similar to that seen in the structures of other isolated PDE catalytic domains. Our efforts to crystallize PDE2A (215–900) with substrates, substrate analogs, or inhibitors were unsuccessful. To investigate such a conformational change, we crystallized the isolated catalytic domain (residues 579–919) with and without 3-isobutyl-1-methylxanthine (IBMX), a weak, nonselective inhibitor of several phosphodiesterases.
As shown in Fig. 4A, the overall topology of the catalytic domain with bound IBMX is very similar to the previously published structures of other PDEs, including with respect to the H- and M-loops. IBMX (IC50 = 40 μM) makes a pair of hydrogen bonds with the conserved glutamine (Q859), and the purine ring sits in the “hydrophobic clamp” (15), sandwiched between the hydrophobic side chains F862, I826, and F830. The orientation of the conserved glutamine and its interactions with IBMX are identical to those reported in the structures of IBMX bound to the cGMP-specific phosphodiesterases, PDE5 and PDE9 (5, 31), and different from the structures of the IBMX complex with the cAMP-specific phosphodiesterases PDE4 and PDE7 (5, 11) (Fig. S4). In contrast, in the structure obtained in the absence of IBMX, the H-loop (residues 702–728) folds into and occupies this substrate binding site (Fig. 4B). This configuration is similar to what is seen in the PDE2A (215–900) structure, although the actual trajectories of the H-loop differ. This difference appears to be due to the absence of the Mg2+ in the structure of the unliganded catalytic domain, which allows the side chain of Ser-721 to occupy the conserved metal binding site. In contrast, both Zn2+ and Mg2+ ions are retained in PDE2A (215–900). It is possible that the removal of Mg2+ from the active site is an artifact of the conditions needed for crystallization of the catalytic domain, which include 200 mM sodium citrate, a known Mg2+ chelator.
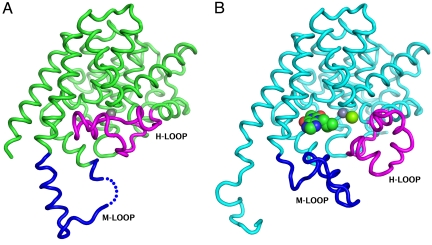
Fig. 4.
Crystal structures of the isolated catalytic domains also show “open” and “closed” conformations of the H-loop. The Cα trace of the isolated catalytic domain (579–919) is shown as green tubes. The H-loop (700–723) and M-loop (830–856) are colored magenta and blue, respectively. (A) Structure of the unliganded catalytic domain that shows the H-loop folded into the catalytic site and displacing the Mg2+ ion. The Zn2+ ion is shown as a gray sphere. (B) Structure of the catalytic domain co-crystallized with IBMX, showing the H-loop swung out. Zn2+ and Mg2+ ions, shown as gray and green spheres, are both visible in this structure.
In summary, the findings with the unliganded catalytic domain of PDE2A essentially parallel those obtained for PDE2A (215–900) and suggest that the substrate binding pocket can be “closed” by the H-loop. The fact that the H-loop is displaced from the binding pocket by ligand is consistent with a hypothesis that this loop swings away to provide access to the binding pocket. Indeed, early biochemical characterization of native PDE2A (32, 33) had turned up the apparently paradoxical observation that low concentrations of inhibitors could stimulate cGMP binding. Thus, displacement of the H-loop is hypothesized to serve as the mechanism of enzyme activation. How this is accomplished by cGMP binding to GAF-B is described in the final section.
The GAF-B/Catalytic Domain Interface and the Basis for Enzyme Activation.
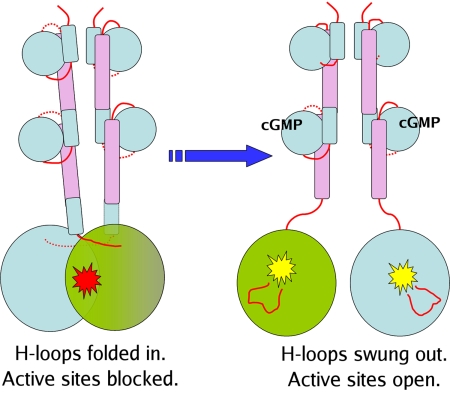
The dimer interface between the catalytic domains in PDE2A (215–900) can only be maintained if the H-loop is in the “closed” conformation. Thus, access to the substrate-binding site is available only if the two catalytic domains move apart to let the H-loop swing into the “open” conformation. Our model for activation of PDE2A is based on these considerations (Fig. 5). PDE2A cycles between the “open” and “closed” conformations described above, with cGMP binding to GAF-B favoring the “open” state by a mechanism proposed as follows. cGMP binding to the GAF-B domain leads to a relative reorientation of helix α5 and the linker helix LH2, and thereby the appended catalytic domains. Specifically, the linker helix (LH2) that connects GAF-B to the catalytic domain is an extension of the C-terminal helix α5 of GAF-B. The entire length of this helix participates in dimer contacts. The N-terminal halves make mostly hydrophobic contacts. The C-terminal halves (E559-M572) are in contact with helices H10 and H11 of the catalytic domain of the dimer partner. Such interactions could be the basis of a cooperative mechanism whereby conformational changes in the regulatory domain of one molecule are transmitted to the catalytic domain of the second molecule. Thus, cGMP binding to the GAF-B domain leads to a relative reorientation of helix α5 and the linker helix LH2. This, in turn, disrupts the dimer interface between the catalytic domains, and the H-loop is freed to swing away from the binding pocket, allowing substrate access to this “open” configuration. Functionally, this is observed as enzyme activation.
Fig. 5.
Proposed mechanism of activation. Cyclic nucleotide binding to the GAF-B domain is accompanied by an ordering of the α4 helix, which “closes down” on the nucleotide, and by a relative rotation of the two linker helices, which causes the two catalytic domains to swing out relative to each other. The H-loop, which was held in a position to occlude the substrate binding site in the catalytic domain, now swings out, making the substrate binding site accessible.
The proposed model is consistent with an early observation (32) that cGMP binding to native bovine PDE2A leads to an increased susceptibility to proteolytic attack by chymotrypsin in the region of Tyrosine 553. This corresponds to Tyrosine 573 in the human sequence, which is at the C-terminal end of LH2, buried at the dimer interface with the catalytic domain of the neighboring molecule. When these contacts are disrupted by cGMP binding in GAF-B, this region would be disordered, and more susceptible to proteolytic attack.
A General Mechanism for Activation of PDEs.
We have proposed a model for the regulation of PDE2 activity whereby access to the substrate-binding pocket is blocked by the dimer partner, and allosteric binding of cGMP to one of the GAF domains causes a relative motion of the catalytic domains, relieving this blockage. This specific mechanism suggests elements of a more general scheme for PDE regulation. The key “effector” in this mechanism is the H-loop, which in PDE2A occludes the substrate binding pocket when the enzyme is in the closed configuration. The H-loop is a feature common to all of the PDEs, and within this region, two residues, glycine 702 and alanine 716 (hPDE2A numbering; Fig. S5) are completely conserved. These residues occur roughly at the beginnings and ends of the various H-loops. Due to their small size, glycines and alanines have greater conformational freedom than any other amino acids. Thus, these residues are strategically placed to confer H-loop flexibility as a common functional element. Such conformational flexibility is inferred from the structures presented here and has been reported previously based on the structure of the catalytic domain of PDE5 (34). Furthermore, in PDE5, mutation of the conserved glycine to alanine (G659A) was reported (34) to cause a 17-fold drop of kcat and 24-fold weaker affinity for cGMP, supporting a key role for the H-loop in regulation of enzyme activity and the flexibility of the H-loop at the conserved hinge residues as a specific element in this regulation.
The present study draws attention to the importance of dimerization in the proposed regulatory role for the H-loop. In the conformation of PDE2A (215–900), the dimer interface formed between the catalytic domains necessitates that the H-loops are locked into the closed configuration. Indeed, the H-loops appear to contribute to this dimer interface. Our data are consistent with rearrangement of the dimer interface being a necessary step to allow movement of the H-loop for access to the catalytic pocket. Thus, dimerization is proposed as a mechanism to constrain the flexibility of the H-loop. Given that dimerization is common across the PDE gene family, it is not unreasonable to speculate that this feature may be part of a common regulatory mechanism. Based on our structural data, we propose a mechanism by which this dimer interface is regulated in PDE2A upon binding of cGMP to GAF-B. We note, based on the fact that PDE2A is enzymatically active in absence of cGMP binding to GAF-B (i.e., when cAMP is used as substrate in vitro), that the PDE2A catalytic site must cycle between open and closed configurations in the absence of cGMP binding to GAF-B. This implies that activation of PDE2A by cGMP binding to GAF-B most likely reflects a stabilization of the open configuration over the closed configuration. Insight into the nature of the molecular movements involved await further study, including crystallization of a full-length construct of PDE2A in the presence of a GAF-B domain ligand. The mechanisms for regulation of the activity of different PDE isozymes are quite diverse. Thus, it remains to be determined the extent to which the scheme presented here based on PDE2A generalizes across the PDE gene family.
Materials and Methods
Expression, Purification, and Crystallization.
C-terminally histidine-tagged PDE2A (215–900) was expressed in Sf21 insect cells and purified by Ni2+ affinity chromatography, followed by thrombin cleavage to remove the tags, followed by a second Ni2+ affinity step and an ion exchange step. Crystals were grown by hanging drop vapor diffusion at room temperature.
The catalytic domain, amino acids 579–919, of human PDE2A1 was expressed in Sf21 insect cells with an N-terminal 6×His tag and thrombin protease site. Purification followed the same protocol as described for PDE2A (215–900), except that all buffers had 10% glycerol added.
Apo crystals were grown by hanging drop vapor diffusion at both room temperature and 4 °C over a precipitant solution comprised of 20% PEG 3350 and 0.2 M tri-sodium citrate.
For co-crystals with IBMX, the protein was concentrated with 1 mM IBMX in the crystallization buffer. The precipitant solution was 25% PEG 3350, 0.1 M Tris, pH 8.5, and 0.2 M MgCl2. Clusters of needle-like crystals first appeared, which were crushed and used as seeds to get diffraction-quality crystals.
X-Ray Structure Determination and Refinement.
X-ray diffraction data for PDE2A (215–900) crystals were collected at the microfocus beamline ID14 at European Synchrotron Radiation Facility (ESRF) Grenoble, with the wavelength tuned to the Zn absorption edge (1.28 Å). The structure was solved by molecular replacement, using structures of the individual subdomains as search models. Detailed protocols for expression, purification, crystallization, and structure determination can be found in the SI Experimental Procedures.
Supplementary Material
Acknowlegements.
The authors thank Pat Loulakis for help with the catalytic domain purification, Mark Tardie for light scattering data, Chris Phillips and Dave Brown for data collection at the European Synchrotron Radiation Facility (ESRF), Dennis Danley, Dave Nettleton, and Chris Schmidt for discussions. Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with the Center for Advanced Radiation Sources at the University of Chicago.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3IBJ, 3ITM, and 3ITU).
This article contains supporting information online at www.pnas.org/cgi/content/full/0907635106/DCSupplemental.
References
- 1.Conti M, Beavo J. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: Essential components in cyclic nucleotide signaling. Annu Rev Biochem. 2007;76:481–511. doi: 10.1146/annurev.biochem.76.060305.150444. [DOI] [PubMed] [Google Scholar]
- 2.Huai Q. Three-dimensional structures of PDE4D in complex with roliprams and implication on inhibitor selectivity. Structure. 2003;11:865–873. doi: 10.1016/s0969-2126(03)00123-0. [DOI] [PubMed] [Google Scholar]
- 3.Wang H, et al. Kinetic and structural studies of phosphodiesterase-8A and implication on the inhibitor selectivity. Biochemistry. 2008;47:12760–12768. doi: 10.1021/bi801487x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huai Q, Colicelli J, Ke H. The crystal structure of AMP-bound PDE4 suggests a mechanism for phosphodiesterase catalysis. Biochemistry. 2003;42:13220–13226. doi: 10.1021/bi034653e. [DOI] [PubMed] [Google Scholar]
- 5.Huai Q, Liu Y, Francis SH, Corbin JD, Ke H. Crystal structures of phosphodiesterases 4 and 5 in complex with inhibitor 3-isobutyl-1-methylxanthine suggest a conformation determinant of inhibitor selectivity. J Biol Chem. 2004;279:13095–13101. doi: 10.1074/jbc.M311556200. [DOI] [PubMed] [Google Scholar]
- 6.Huai Q, et al. Crystal structure of phosphodiesterase 9 shows orientation variation of inhibitor 3-isobutyl-1-methylxanthine binding. Proc Natl Acad Sci USA. 2004;101:9624–9629. doi: 10.1073/pnas.0401120101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iffland A, et al. Structural determinants for inhibitor specificity and selectivity in PDE2A using the wheat germ in vitro translation system. Biochemistry. 2005;44:8312–8325. doi: 10.1021/bi047313h. [DOI] [PubMed] [Google Scholar]
- 8.Lee ME, Markowitz J, Lee J-O, Lee H. Crystal structure of phosphodiesterase 4D and inhibitor complex1. FEBS Lett. 2002;530:53–58. doi: 10.1016/s0014-5793(02)03396-3. [DOI] [PubMed] [Google Scholar]
- 9.Scapin G, et al. Crystal structure of human phosphodiesterase 3B: Atomic basis for substrate and inhibitor specificity. Biochemistry. 2004;43:6091–6100. doi: 10.1021/bi049868i. [DOI] [PubMed] [Google Scholar]
- 10.Sung B-J, et al. Structure of the catalytic domain of human phosphodiesterase 5 with bound drug molecules. Nature. 2003;425:98–102. doi: 10.1038/nature01914. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Liu Y, Chen Y, Robinson H, Ke H. Multiple elements jointly determine inhibitor selectivity of cyclic nucleotide phosphodiesterases 4 and 7. J Biol Chem. 2005;280:30949–30955. doi: 10.1074/jbc.M504398200. [DOI] [PubMed] [Google Scholar]
- 12.Wang H, et al. Structural insight into substrate specificity of phosphodiesterase 10. Proc Natl Acad Sci USA. 2007;104:5782–5787. doi: 10.1073/pnas.0700279104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu RX, et al. Atomic structure of PDE4: Insights into phosphodiesterase mechanism and specificity. Science. 2000;288:1822–1825. doi: 10.1126/science.288.5472.1822. [DOI] [PubMed] [Google Scholar]
- 14.Xu RX, et al. Crystal structures of the catalytic domain of phosphodiesterase 4B complexed with AMP, 8-Br-AMP, and rolipram. J Mol Biol. 2004;337:355–365. doi: 10.1016/j.jmb.2004.01.040. [DOI] [PubMed] [Google Scholar]
- 15.Zhang KYJ, et al. A glutamine switch mechanism for nucleotide selectivity by phosphodiesterases. Mol Cell. 2004;15:279–286. doi: 10.1016/j.molcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Houslay MD, Adams DR. PDE4 cAMP phosphodiesterases: Modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem J. 2003;370:1–18. doi: 10.1042/BJ20021698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sette C, Conti M. Phosphorylation and activation of a cAMP-specific phosphodiesterase by the cAMP-dependent protein kinase. Involvement of serine 54 in the enzyme activation. J Biol Chem. 1996;271:16526–16534. doi: 10.1074/jbc.271.28.16526. [DOI] [PubMed] [Google Scholar]
- 18.Corbin JD, Francis SH. Cyclic GMP phosphodiesterase-5: Target of sildenafil. J Biol Chem. 1999;274:13729–13732. doi: 10.1074/jbc.274.20.13729. [DOI] [PubMed] [Google Scholar]
- 19.Muradov KG, Boyd KK, Martinez SE, Beavo JA, Artemyev NO. The GAFa domains of rod cGMP-phosphodiesterase 6 determine the selectivity of the enzyme dimerization. J Biol Chem. 2003;278:10594–10601. doi: 10.1074/jbc.M208456200. [DOI] [PubMed] [Google Scholar]
- 20.Song J, et al. Intrinsically disordered Î3-subunit of cGMP phosphodiesterase encodes functionally relevant transient secondary and tertiary structure. Proc Natl Acad Sci USA. 2008;105:1505–1510. doi: 10.1073/pnas.0709558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gross-Langenhoff M, Hofbauer K, Weber J, Schultz A, Schultz JE. cAMP is a ligand for the tandem GAF domain of human phosphodiesterase 10 and cGMP for the tandem GAF domain of phosphodiesterase 11. J Biol Chem. 2006;281:2841–2846. doi: 10.1074/jbc.M511468200. [DOI] [PubMed] [Google Scholar]
- 22.Martins TJ, Mumby MC, Beavo JA. Purification and characterization of a cyclic GMP-stimulated cyclic nucleotide phosphodiesterase from bovine tissues. J Biol Chem. 1982;257:1973–1979. [PubMed] [Google Scholar]
- 23.Wu AY, Tang X-B, Martinez SE, Ikeda K, Beavo JA. Molecular determinants for cyclic nucleotide binding to the regulatory domains of phosphodiesterase 2A. J Biol Chem. 2004;279:37928–37938. doi: 10.1074/jbc.M404287200. [DOI] [PubMed] [Google Scholar]
- 24.Tcheudji JFK, et al. Molecular organization of bovine rod cGMP-phosphodiesterase 6. J Mol Biol. 2001;310:781–791. doi: 10.1006/jmbi.2001.4813. [DOI] [PubMed] [Google Scholar]
- 25.Ho Y-SJ, Burden LM, Hurley JH. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 2000;19:5288–5299. doi: 10.1093/emboj/19.20.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez SE, et al. The two GAF domains in phosphodiesterase 2A have distinct roles in dimerization and in cGMP binding. Proc Natl Acad Sci USA. 2002;99:13260–13265. doi: 10.1073/pnas.192374899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez SE, et al. Crystal structure of the tandem GAF domains from a cyanobacterial adenylyl cyclase: Modes of ligand binding and dimerization. Proc Natl Acad Sci USA. 2005;102:3082–3087. doi: 10.1073/pnas.0409913102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heikaus CC, et al. Solution structure of the cGMP binding GAF domain from phosphodiesterase 5: Insights into nucleotide specificity, dimerization, and cGMP-dependent conformational change. J Biol Chem. 2008;283:22749–22759. doi: 10.1074/jbc.M801577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez SE, Heikaus CC, Klevit RE, Beavo JA. The structure of the GAF A domain from phosphodiesterase 6C reveals determinants of cGMP binding, a conserved binding surface, and a large cGMP-dependent conformational change. J Biol Chem. 2008;283:25913–25919. doi: 10.1074/jbc.M802891200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Handa N, et al. Crystal structure of the GAF-B domain from human phosphodiesterase 10A complexed with its ligand, cAMP. J Biol Chem. 2008;283:19657–19664. doi: 10.1074/jbc.M800595200. [DOI] [PubMed] [Google Scholar]
- 31.Liu S, et al. Structural basis for the catalytic mechanism of human phosphodiesterase 9. Proc Natl Acad Sci USA. 2008;105:13309–13314. doi: 10.1073/pnas.0708850105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stroop SD, Beavo JA. Structure and function studies of the cGMP-stimulated phosphodiesterase. J Biol Chem. 1991;266:23802–23809. [PubMed] [Google Scholar]
- 33.Erneux C, Miot F, Boeynaems J-M, Dumont JE. Paradoxical stimulation by 1-methyl-3-isobutylxanthine of rat liver cyclic AMP phosphodiesterase activity. FEBS Lett. 1982;142:251–254. doi: 10.1016/0014-5793(82)80146-4. [DOI] [PubMed] [Google Scholar]
- 34.Wang H, et al. Multiple conformations of phosphodiesterase-5: Implications for enzyme function and drug development. J Biol Chem. 2006;281:21469–21479. doi: 10.1074/jbc.M512527200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.