Abstract
The proinflammatory cytokine TNF-α exerts its pleiotropic functions through activation of multiple downstream effectors, including JNK1. Yet, the underlying regulatory mechanism is incompletely understood. Here, we report that the transcription factor Myc-interacting zinc-finger protein 1 (Miz1) selectively suppresses TNF-α-induced JNK1 activation and cell death independently of its transcription activity. Proteomics analysis and yeast two-hybrid screening reveal that Miz1 is a JNK-associated protein. The TNF-α-induced activation of JNK1 is augmented in Miz1-deficient mouse embryonic fibroblasts (Miz1−/− MEFs), but the augmentation is abrogated by reintroduction of Miz1 or its transcription-deficient mutant. The regulation by Miz1 is highly specific, because it regulates TNF-α-induced TRAF2 K63-linked polyubiquitination. Neither JNK1 activation by IL-1β or UV nor TNF-α-induced activation of p38, ERK, or IκB kinase complex is affected by the loss of Miz1. The TNF-α-induced cell death also is accelerated in Miz1−/− MEFs. Upon TNF-α stimulation, Miz1 is degraded rapidly by the proteasome, relieving its suppression on JNK1 activation. Thus, our results show that in addition to being a transcription factor Miz1 acts as a signal- and pathway-specific modulator or regulator that specifically regulates TNF-α-induced JNK1 activation and cell death.
JNK (c-Jun N-terminal kinase; also known as stress-activated protein kinase) is a member of the MAPK superfamily (1, 2). The JNK MAPK subfamily has two ubiquitously expressed isoforms, JNK1 and JNK2, and a tissue-specific isoform, JNK3, with 10 splicing variants (3–5). JNK is activated by sequential protein phosphorylation through the MAPK module (i.e., MAPKKK → MAPKK → MAPK) in response to a variety of extracellular stimuli, including TNF-α, IL-1β, UV, and oncogenes (4, 6, 7). Between JNK1 and JNK2, JNK1 is the major c-Jun kinase that is significantly activated by most known JNK inducers and is responsible for most known JNK functions (8–16).
JNK1 plays a critical role in TNF-α-regulated cellular activities, such as cell death, inflammation, and tumorigenesis. Prolonged activation of JNK1 is essential for TNF-α-induced cell death when NF-κB activation is impaired (5, 7, 12, 17–22), likely through activation of the E3 ligase Itch, which ubiquitinates the caspase-8 inhibitor c-FLIPL and triggers its proteasomal degradation (8). JNK1 also regulates TNF-α-induced inflammation through c-Jun-induced expression of several proinflammatory genes (23). Recent studies show that JNK1 activation contributes to cell-death-induced “compensatory proliferation,” which may be related to inflammation-driven types of cancer (10, 11, 14). Not surprisingly, JNK1 has become an attractive therapeutic target (24). However, regulation of JNK1 is highly complex, because it is activated by numerous extracellular stimuli, such as TNF-α, IL-1β, UV, and oncogenes, which also activate many other signaling pathways, such as MAPKs p38, ERK, and IκB kinase (IKK) (3, 5, 7). Whether JNK1 activation can be regulated in a signal- and pathway-specific manner is not clear.
Myc-interacting zinc-finger protein 1 (Miz1), also known as Z13, is member of the poxvirus and zinc-finger (POZ) domain/zinc-finger transcription factor family (25–27). Miz1 has an N-terminal POZ domain, which is required for its transcriptional activation and repression activity (28) and 13 zinc fingers (25–27). Miz1 localizes in both the cytoplasm and the nucleus (29). Unlike its well-studied transcription activation and repression functions in the nucleus (28, 30), the cytoplasmic function of Miz1 is largely unknown, although microtubule depolymerization agent T113242 has been reported to induce the translocation of Miz1 from the cytoplasm to the nucleus for activating gene expression (29). Genetic disruption of Miz1 alleles in mice results in early embryonic lethality caused by massive apoptosis with no known mechanisms (31). We report here the identification of Miz1 as a signal- and pathway-specific modulator or regulator (SMOR) (7) that selectively suppresses TNF-α-induced JNK1 activation and cell death through inhibition of TRAF2 K63-linked polyubiquitination.
Results and Discussion
Identification of Miz1 as a JNK1-Associated Protein.
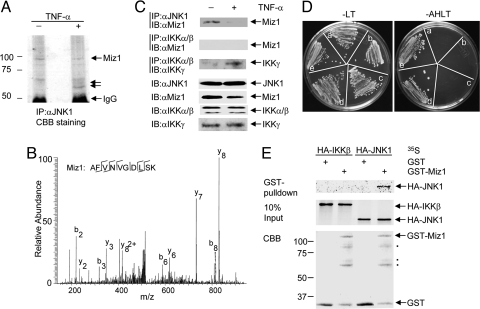
Activation of JNK1 by extracellular stimuli is mediated likely by distinct JNK1 signalsomes, whose activities could be specifically regulated by SMORs (7). To identify the SMORs that selectively regulate TNF-α-induced JNK1 activation, endogenous JNK1 was immunoprecipitated with anti-JNK1 antibody from control or TNF-α-stimulated HeLa cells. Several putative JNK1-associated proteins (p85, p68, and p64), which either dissociated from or associated with JNK1 upon TNF-α stimulation as detected by Coomassie brilliant blue staining (Fig. 1A), were analyzed by nano-HPLC electrospray ion trap mass spectrometry (MS/MS). Protein sequence database searching of the MS/MS spectra revealed that the band corresponding to p85 contained multiple proteins, among which is the zinc-finger transcription factor Miz1 (Fig. 1 A and B). Immunoblotting analysis of the JNK1 immune complex using anti-Miz1 antibody revealed that Miz1 was coimmunoprecipitated with JNK1 from nonstimulated HEK293 cells and the amount of coimmunoprecipitated Miz1 was reduced significantly 15 min after TNF-α stimulation (Fig. 1C). In contrast, Miz1 was not coimmunoprecipitated with IKKα/β in nonstimulated or stimulated cells, although the regulatory subunit of the IKK complex IKKγ/NF-κB essential modulator (NEMO) was coimmunoprecipitated readily with IKKα/β upon stimulation (Fig. 1C). Thus, Miz1 may interact selectively with JNK in cells. Miz1 was able to interact with JNK1 directly, because a truncated Miz1 fragment (amino acids 556–713, corresponding to the 10th though 12th zinc fingers of Miz1) interacted with JNK1 in a yeast two-hybrid screening (Fig. 1C). Miz1 also interacted with JNK2, which shares 84% of its amino acid sequence with JNK1 (32), in a yeast cotransformation assay (Fig. S1). The interaction between Miz1 and JNK 1 was highly specific, as demonstrated by an in vitro GST pulldown assay, in which purified GST-Miz1 proteins interacted with in vitro-translated 35S-labeled JNK1 but not [35S]-IKKβ (Fig. 1D). These results identify Miz1 as a JNK-associated protein. More importantly, Miz1 may be involved in regulation of TNF-α-induced JNK1 activation, because its association with JNK1 is negatively regulated by TNF-α.
Fig. 1.
Identification of Miz1 as a JNK1-associated protein. (A) JNK1-associated proteins were isolated from control or TNF-α-stimulated (5 ng/mL, 15 min) HeLa cell extracts (10 mg of each) by immunoprecipitation with anti-JNK1 antibody and visualized by Coomassie brilliant blue (CBB) staining. Putative JNK1-associated proteins, p85, p68, and p64, are indicated by the arrows. (B) Mass spectrometry spectrum of a Miz1 tryptic peptide identified. The b and y ions designate N- and C-terminal fragment ions produced by collision-induced backbone fragmentation in the mass spectrometer. Another Miz1 tryptic peptide identified was GGQAQSAASGAEQTEK. (C) Analysis of the association between Miz1 and JNK1 or IKK in control or TNF-α-stimulated HEK293 cells by coimmunoprecipitation (co-IP) in combination with immunoblotting (IB). (D) Detection of the interaction between Miz1 and JNK1 in yeast two-hybrid screening. a, pGBKT7 + pACT2; b, pGBKT7/JNK1 + pACT2; c, pGBKT7 + pACT2/Miz1; d, pGBKT7/JNK1 + pACT2/Miz1; e, pGBKT7/JNK1 + pACT2/JIP1. A, adenine; L, leucine; H, histidine; T, tryptophan. (E) Miz1 interacted with JNK1 but not IKKβ in an in vitro binding assay. The amount of GST and GST-Miz1 was 2 μg of each.
Miz1 Suppresses TNF-α-Induced JNK1 Activation.
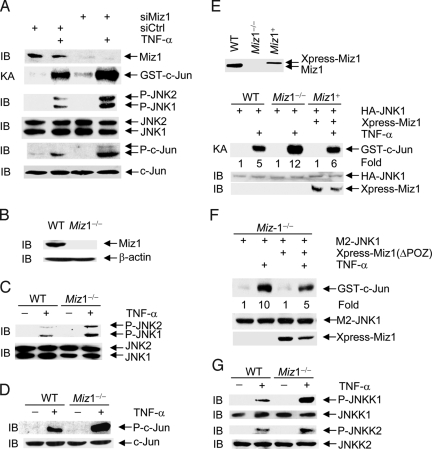
To determine whether Miz1 is involved in regulation of TNF-α-induced JNK1 activation, WT mouse embryonic fibroblasts (MEFs) were transfected with the control scramble siRNA or siRNA of Miz1 (siMiz1), followed by stimulation without or with TNF-α. Silencing of Miz1 by siRNA significantly augmented TNF-α-induced JNK1 activation and phosphorylation of c-Jun at Ser63 and Ser73 (Fig. 2A). Similar results were obtained with a second siMiz1 that targets Miz1 at a different region (Fig. S2). Consistently, TNF-α-induced JNK1 activation and c-Jun phosphorylation were augmented significantly in Miz1−/− MEFs when compared with those in WT MEFs (Fig. 2 B–D). When Xpress-tagged Miz1 was reintroduced into Miz1−/− MEFs at levels similar to that of endogenous Miz1, it abrogated the augmentation by the loss of Miz1 (Fig. 2E). Similar results were obtained with reintroduction of Xpress-ΔPOZ-Miz1 (Fig. 2F). Because the POZ domain deletion mutant can no longer activate or repress transcription (28), this result indicates that the transcription activity of Miz1 is likely dispensable for Miz1 to inhibit TNF-α-induced JNK1 activation. In addition, TNF-α-induced activation of MKK4/JNKK1 and MKK7/JNKK2, which are upstream kinases of JNK1 (33, 34), was augmented in Miz1−/− MEFs (Fig. 2G). This suggests that Miz1 may target upstream regulators of JNK1 to inhibit TNF-α-induced JNK1 activation. Taken together, Miz1 is a suppressor of TNF-α-induced JNK1 activation.
Fig. 2.
Miz1 inhibits TNF-α-induced JNK activation. (A) WT fibroblasts were transfected with the control scramble siRNA or siRNA of Miz1 (siMiz1) (100 nM for each). Expression of Miz1, phosphorylation, and expression of JNK and c-Jun, and JNK activity in response to TNF-α (5 ng/mL, 15 min) were analyzed by immunoblotting and immune complex kinase assays, respectively. (B) Immunoblotting analysis of Miz1 expression in WT and Miz1−/− MEFs with anti-Miz1 antibody. (C and D) WT and Miz1−/− MEFs were treated without or with TNF-α (5 ng/mL, 15 min). Expression and phosphorylation of JNK (C) and c-Jun (D) were analyzed by immunoblotting with corresponding anti-phospho- or pan-antibodies. (E) Ectopic expression of Xpress-Miz1 in Miz1−/− MEFs abrogated the augmented TNF-α-induced JNK activation by the loss of Miz1. The transfection efficiency was ≈30%, and the amount of Xpress-Miz1 was one-third that of endogenous Miz1 so that the expression level of Xpress-Miz1 was similar to that of endogenous Miz1. (F) Ectopic expression of the Xpress-Miz1(ΔPOZ) mutant in Miz1−/− MEFs abrogated the augmented TNF-α-induced JNK1 activation by the loss of Miz1.
Miz1 Does Not Inhibit IL-1β- or UV-Induced JNK1 Activation or TNF-α-Induced Activation of p38, ERK, or IKK.
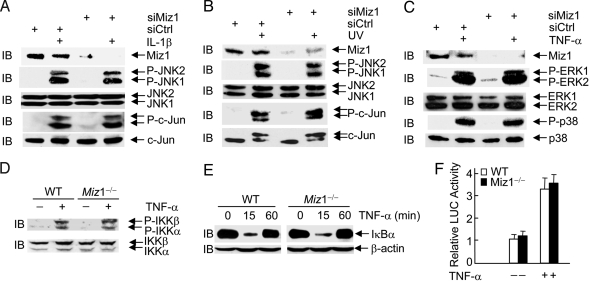
A major challenge in JNK biology is to understand whether JNK activation by a specific extracellular signal can be regulated without affecting activation of other signaling effectors by the signal or activation of JNK by other signals. We were curious whether Miz1 can regulate TNF-α-induced JNK1 activation in a signal- and pathway-specific manner. In contrast to its inhibitory effect on TNF-α-induced JNK1 activation (Fig. 2), the silencing of Miz1 had no detectable effects on phosphorylation of JNK or c-Jun by IL-1β or UV (Fig. 3 A and B).
Fig. 3.
Miz1 does not inhibit JNK1 activation by IL-1β and UV or activation of ERK, p38, and IKK by TNF-α. (A and B) WT fibroblasts were transfected with the control scramble siRNA or siRNA of Miz1 (siMiz1) (100 nM of each). Expression of Miz1, phosphorylation and expression of JNK and c-Jun, and JNK activity in response to IL-1β (2 ng/mL, 15 min) (A) or UV (20 J/m2, 30 min) (B) were analyzed by immunoblotting and immune complex kinase assays, respectively. (C) The effect of Miz1 silencing on TNF-α-induced activation of ERK and p38 was determined as described in A and B. (D–F) Effects of the loss of Miz1 on TNF-α-induced IKK phosphorylation (D) and activation (IκBα degradation) were analyzed by immunoblotting using corresponding antibodies. The effect of the loss of Miz1 on TNF-α-induced NF-κB activation was measured by a NF-κB reporter gene assay, as described (13) (F).
In addition to activation of JNK1, TNF-α activates several other downstream effectors, such as p38, ERK, and IKK (7). Interestingly, the silencing of Miz1 had no detectable effects on TNF-α-induced activation of p38 and ERK (Fig. 3C). There were also no detectable changes in TNF-α-induced IKK activation (Fig. 3C), IκBα degradation (Fig. 3D), and NF-κB reporter gene activity (Fig. 3E) in Miz1 null MEFs. Taken together, these results demonstrate that Miz1 is a SMOR in TNF-α–JNK1 signaling, because it can regulate TNF-α-induced activation of JNK1 in a signal-specific (Fig. 2) and pathway-specific (Fig. 3) manner.
Miz1 Is a Negative Regulator of TRAF2 K63-Linked Polyubiquitination.
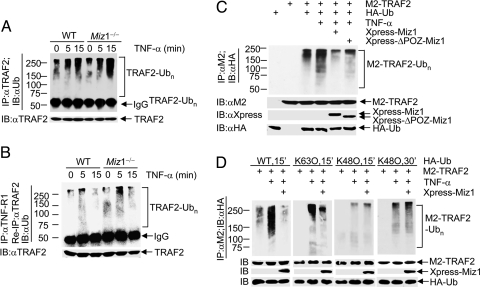
Upon TNF-α stimulation, TRAF2 is recruited rapidly to TNF-Receptor 1 (TNF-R1) and becomes polyubiquitinated. Although TRAF2 protein is involved in TNF-α-induced activation of MAPKs (JNK, p38, and ERK) and IKK (35–37), TRAF2 K63-linked polyubiquitination is required for activation of only JNK but not p38 or IKK (38, 39). We were curious about whether Miz1 suppresses JNK activation by regulating TRAF2 K63-linked polyubiquitination. We found that the loss of Miz1 significantly augmented TNF-α-induced polyubiquitination of total TRAF2, as early as 5 min and lasted for 15 min after TNF-α stimulation (Fig. 4A). More importantly, polyubiquitination of TNF-R1-associated TRAF2 also was augmented by the loss of Miz1 (Fig. 4B). The polyubiquitination of TNF-R1-associated TRAF2 peaked at 5 min of TNF-α stimulation and decreased 15 min later, which is likely caused by the dissociation of complex 1 from the receptor. Although the loss of Miz1 already resulted in augmentation of polyubiquitination of TNF-R1-associated TRAF2 in resting cells (Fig. 4B, lane 4), it did not activate JNK1 (Fig. 2 A and C), suggesting that TRAF2 polyubiquitination may be necessary but not sufficient for activation of JNK. Conversely, ectopic expression of Xpress-Miz1 or the POZ domain deletion mutant of Xpress-Miz1, which is deficient in Miz1-mediated transcription activation or repression (28), suppressed TNF-α-induced polyubiquitination of M2-TRAF2 (Fig. 4C). These data suggest that Miz1 negatively regulates TNF-α-induced polyubiquitination of TNF-R1-associated TRAF2 in a transcription-independent manner.
Fig. 4.
Miz1 inhibits TNF-α-induced TRAF2 K63-ubiquitination. (A and B) Ubiquitination of immunoprecipitated TRAF2 was analyzed by immunoblotting using anti-ubiquitin antibody. (A) Ubiquitination of total TRAF2. (B) TNF-R1-assoicated TRAF2 was reimmunoprecipitated with anti-TRAF2 antibody, and its ubiquitination was analyzed by immunoblotting using anti-Ub antibody (see Materials and Methods for details). (C and D) WT fibroblasts were transfected with expression vectors encoding M2-TRAF2 or HA-Ub (5 μg of each), along with Xpress-Miz1 or Xpress-ΔPOZ-Miz1 (C), hemagglutinin-ubiquitin (HA-Ub) (WT, K63O, or K48O) (D), or empty vector (3 μg of each). The effect of Xpress-Miz1 on TNF-α-induced ubiquitination of immunoprecipitated M2-TRAF2 was analyzed by immunoblotting with anti-HA antibody (see Materials and Methods for details).
We then examined which ubiquitin linkage of the polyubiquitin chains on TRAF2 (i.e., K63-linked or K48-linked) was suppressed by Miz1. Treatment with TNF-α for 15 min induced robust polyubiquitination of M2-TRAF2 in cells transfected with hemagglutinin-ubiquitin (HA-Ub) or HA-Ub(K63O) but not with HA-Ub(K48O) (Fig. 4D, lanes 2, 5, and 8). Ectopic coexpression of Xpress-Miz1 significantly suppressed TNF-α-induced polyubiquitination of M2-TRAF2 with HA-Ub or HA-Ub(K63O) but not with HA-Ub(K48O) (Fig. 4D, lanes 3, 6, and 9). In contrast, treatment with TNF-α for 30 min induced a significant increase in K48-linked polyubiquitination of M2-TRAF2, but it was still not affected by Xpress-Miz1 (Fig. 4D). Thus, Miz1 selectively suppresses TNF-α-induced K63-linked polyubiquitination of TRAF2. Future studies are needed to determine how Miz1 suppresses TRAF2 K63-linked polyubiquitination.
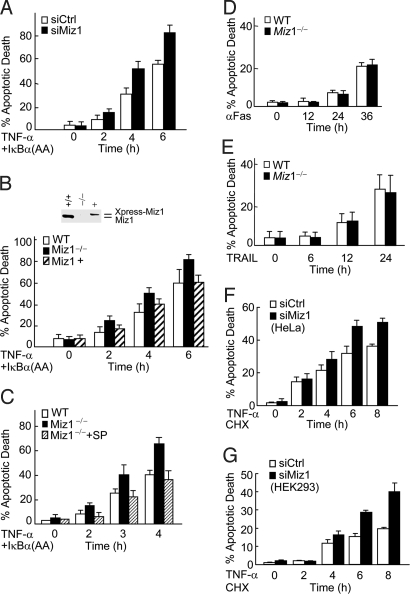
Loss of Miz1 Accelerates TNF-α-Induced Apoptosis.
JNK1 is essential for TNF-α-induced cell death when NF-κB activation is impaired (5, 7, 8, 11, 12, 17, 21, 22). To test whether Miz1 also can regulate TNF-α-induced cell death, WT fibroblasts were infected with adenoviral vector encoding IκBα(AA) [Ad-IκBα(AA)], a superrepressor of NF-κB that specifically blocks TNF-α-induced NF-κB activation and thereby sensitizes cells to TNF-α-induced death (22), followed by transfection with the control siRNA or siMiz1. The silence of Miz1 profoundly accelerated TNF-α-induced cell death, as analyzed by cell death assays (Fig. 5A). Similar results were obtained when Miz1 null MEFs were examined (Fig. 5B). Reintroduction of Xpress-Miz1 into Miz1 null MEFs at a level similar to that of endogenous Miz1 reversed the hypersensitivity (Fig. 5B). The loss of Miz1 also accelerated TNF-α-induced cell death when de novo protein synthesis was blocked by the protein synthesis inhibitor cycloheximide (Fig. S3). The hypersensitivity of Miz1 null MEFs to TNF-α-induced cell death depended on JNK1 activity, because it was blocked by a subdose of the JNK inhibitor SP600125 (Fig. 5C). This notion was supported further by the observations that the loss of Miz1 did not affect cell death induced by other death stimuli, such as anti-Fas antibody or TNF-related apoptosis-inducing ligand (TRAIL), both of which induce cell death independent of JNK1 (40, 41) (Fig. 5 D and E). The inhibition by Miz1 of TNF-α-induced cell death was not limited to fibroblasts, because the silencing of Miz1 also augmented TNF-α-induced cell death in other types of cells, such as HeLa and HEK293 (Fig. 5 F and G). Taken together, these results demonstrate that Miz1 selectively regulates TNF-α-induced, JNK1-dependent cell death.
Fig. 5.
Loss of Miz1 selectively accelerates TNF-α-induced cell death. (A) WT fibroblasts were transfected with siRNA of Miz1 (siMiz1) or control scramble siRNA (siCtrl) (100 nM of each) and then infected with adenoviral vector encoding the superrepressor of NF-κB, IκBα(SS32/36AA) [Ad/IκBα(AA)] [200 MOI (multiplicity of infection)], followed by TNF-α treatment (5 ng/mL) for various periods of time. Cell death was monitored by Hoechst staining. (B and C) WT and Miz1−/− MEFs were transfected without (C) or with (B) expression vector encoding Xpress-Miz1 or empty vector (3.2 μg of each) along with EGFP (0.8 μg), as indicated. Cells then were infected with Ad/IκBα(AA) (200 MOI), followed by without or with the treatment of TNF-α (5 ng/mL) for various periods of time in the absence (B) or presence (C) of subdose of the JNK inhibitor SP600125 (2.5 μM). Apoptotic death of EGFP-positive cells (B) or total cell populations (C) were determined as in A. The transfection efficiency was ≈30%, and the amount of Xpress-Miz1 was one-third of that of endogenous Miz1, so the expression level of Xpress-Miz1 was similar to that of endogenous Miz1. (D and E) WT and Miz1−/− MEFs were treated without or with anti-Fas antibody (0.5 μg/mL) or TRAIL (10 ng/mL) for various times, as indicated. Apoptotic death was determined as in A. (F and G) HeLa or HEK293 cells were transfected with siMiz1 or siCtrl (100 nM each), then treated without or with TNF-α (5 ng/mL) in the presence of the protein synthesis inhibitor cycloheximide (CHX) (1 μg/mL) for various periods of time, as indicated. Apoptotic death was determined as in A. The results were presented as mean standard error and represented three separate experiments.
TNF-α Induces Proteasomal Degradation of Miz1.
TNF-α typically induces JNK1 activation in its target cells within minutes (12, 33). If Miz1 is a physiologically relevant inhibitor of TNF-α-induced JNK1 activation, then the suppression by Miz1 has to be rapidly relieved upon TNF-α stimulation. In support of this hypothesis, the association of Miz1 with JNK1 was reduced rapidly and dramatically upon TNF-α stimulation (Fig. 1C). To understand the mechanism by which TNF-α rapidly regulates Miz1 association with JNK1, we analyzed the stability of Miz1 proteins. Immunoblotting analysis revealed that Miz1 protein levels in whole-cell extracts were reduced rapidly upon TNF-α stimulation, which occurred as early as 5 min and reached a maximum 10 min after TNF-α stimulation (Fig. 6A). TNF-α-induced reduction of cytoplasmic Miz1 proteins could be caused by the translocation of Miz1 to other cellular compartments or its proteasomal degradation. To distinguish between these possibilities, cells were pretreated with the proteasome inhibitor MG-132. Under this condition, the amount of Miz1 proteins in whole-cell extracts was not affected by TNF-α treatment (Fig. 6B). Thus, TNF-α-induced rapid proteasomal degradation of Miz1 should relieve its inhibition on TRAF2 K63-linked polyubiquitination and JNK1 activation.
Fig. 6.
TNF-α induces Miz1 proteasomal degradation, which also controls the rate of JNK1 activation. (A and B) WT fibroblasts pretreated without (A) or with (B) the proteasomal inhibitor MG-132 (50 μM, 2 h) and then treated with TNF-α (5 ng/mL) for various periods of time, as indicated. The expression levels of Miz1 were analyzed by immunoblotting using anti-Miz1 antibody. (C) WT and Miz1−/− MEFs were treated with TNF-α (5 ng/mL) for various periods of time, as indicated. Phosphorylation and expression of JNK were analyzed by immunoblotting using corresponding antibodies. (D) Schematic illustration showing that Miz1 inhibits JNK1 activation by TNF-α in a signal- and pathway-specific manner and the inhibition by Miz1 is relieved by TNF-α-induced proteasomal degradation.
The observation above prompted us to test whether Miz1 also controls the rate of TNF-α-induced JNK1 activation. We found that JNK1 activation in Miz1 null MEFs occurred as early as 3 min after TNF-α stimulation, whereas in WT fibroblasts it took 5 min after TNF-α stimulation (Fig. 6C). This result demonstrates that the rate of Miz1 degradation controls the kinetics of TNF-α-induced JNK1 activation, consistent with the observation that TNF-α-induced cell death also was accelerated in Miz1 null MEFs (Figs. 5 and 6C). Taken together, Miz1 negatively regulates both strength and rate of TNF-α-induced JNK1 activation and cell death, and the inhibition is relieved by TNF-α-induced proteasomal degradation of Miz1 (Fig. 6D).
Conclusions
The cell signaling circuitry is composed of numerous signaling pathways that mediate the responses of a cell to various extracellular stimuli. Within the signaling circuitry, a signaling pathway is activated usually by a variety of stimuli that simultaneously activate many other signaling pathways as well. For instance, JNK1 is activated by various stimuli, including TNF-α, IL-1β, and UV, all of which also activate other downstream effectors, such as p38 and NF-κB (Fig. 6D). Whether JNK1 activation by a given signal, such as TNF-α, can be regulated without affecting JNK1 activation by other stimuli, such as IL-1β and UV, without affecting TNF-α-induced activation of p38, ERK, and NF-κB is unknown. Understanding how the activity of a signaling pathway in the complex signaling circuitry is regulated in a signal- and pathway-specific manner may pave the way for selectively targeting a specific signaling pathway in future cell signaling-based therapy, in which the therapeutic target will be subjected most likely to regulation by multiple stimuli that also regulate many other signaling events in vivo.
We hypothesize that the specificity of signaling pathways can be regulated by a SMOR (7). We found that the zinc-finger transcription factor Miz1 selectively regulates TNF-α-induced JNK1 activation. The inhibition by Miz1 is signal- and pathway-specific (Figs. 2 and 3), demonstrating that Miz1 is a SMOR for TNF-α–JNK1 signaling. Although Miz1 is a component of the JNK1 signalsome through its interaction with JNK1 (Fig. 1), it does not inhibit directly JNK1 itself (Figs. 2G and 3 A and B). Rather, Miz1 targets TRAF2 K63-linked polyubiquitination (Fig. 4), which is required for activation of JNK1 but not other MAPKs or IKK by TNF-α (38). This explains how Miz1 could inhibit selectively JNK1 activation by TNF-α but not IL-1β or UV. Furthermore, the inhibition by Miz1 is independent of its transcription activity (Fig. 2F), uncovering its nontranscription function. The augmentation and acceleration by the loss of Miz1 of TNF-α-induced, JNK1-dependent cell death support the notion that Miz1 is a physiologically relevant SMOR for TNF-α signaling. Finally, the rapid proteasomal degradation of Miz1 upon TNF-α stimulation (Fig. 6 A and B) suggests that Miz1 also may play a critical role in regulation of the kinetics of TNF-α-induced JNK1 activation and cell death (Figs. 5 and 6C). Taken together, our results show that in addition to being a transcription factor Miz1 can function as a physiologically relevant SMOR that provides both strength and temporal control of JNK1 activation to ensure proper cellular responses to TNF-α. Determining whether Miz1 is deregulated in certain types of cancer and other human diseases will be of interest.
Materials and Methods
Reagents, Plasmids, and siRNA.
Antibodies against JNK (333 for immunoprecipitation and 666 for immunoblotting) were from Pharmingen. Antibodies against JNK, phospho-JNK, JNKK1, JNKK2, phospho-JNKK1, phospho-JNKK2, p38, phospho-p38, ERK, phospho-ERK, phospho-IKKα/β, and phospho-c-Jun(Ser63) were from Cell Signaling. Antibodies against Miz1 were generated by injecting a KLH-conjugated peptide (EPPEENEESAGTDSG) into rabbits (Synbiosci) and isolated (Fig. S4). Antibodies against IKKα/β, TRAF2, Xpress, and HA were from Santa Cruz Biotechnology. All siRNAs were purchased from Dharmacon. Hoechst 33258 and antibodies against the M2 tag and β-actin were from Sigma. Anti-Fas antibody, IL-1β, and murine TNF-α were from R & D Systems. TRAIL was from Biomol, [γ-32P]ATP (3,000 mCi/nmol) and [35S]methionine were from DuPont/NEN. HA-JNK1, M2-JNK1, and GST-c-Jun have been described (13). M2-TRAF2 was kindly provided by Zhenggang Liu (National Cancer Institute, National Institutes of Health, Bethesda), and HA-Ub (WT, K63O, and K48O) was kindly provided by James Z. Chen (University of Texas Southwestern Medical Center, Dallas). Xpress-Miz1 and GST-Miz1 were generated by PCR and subcloned into a pcDNA3.1 vector (Invitrogen). All constructs were verified by DNA sequencing. The sequences of the two siMiz1s are UGCUGAACCUGCAUAGAAdTdT (Figs. 2–4) and CGAGACGGAAGUACUCAAAdTdT (Fig. S2).
Cell Culture, Transfection, Protein Kinase Assays, and Apoptosis Assays.
Cell culture conditions and transfections have been described (12). Immortalized WT and Miz1−/− MEFs have been described (31). siRNAs were transfected by using Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen). Immune complex kinase assays and apoptosis assays were performed as described (13, 33).
Mass Spectrometry.
Protein in-gel digestion and nano-HPLC MS/MS were carried out as described (42, 43). The acquired MS/MS spectra were searched against the National Center for Biotechnology Information nonredundant protein sequence database. Two Miz1 tryptic peptides were identified, AFVNVGDLSK and GGQAQSAASGAEQTEK.
Yeast Two-Hybrid Screening.
Yeast two-hybrid assay was performed withmatchmaker GAL4 Two-Hybrid System 3 from Clontech, according to the manufacturer's instructions. pGBKT7-JNK1 was used as the bait, and a mouse brain cDNA library constructed in pATC2 was used as the prey.
In Vitro Binding Assays.
HA-JNK1 or HA-IKKβ plasmid (8 μg of each) was translated in vitro in the presence of [35S]methionine in a 100-μL reaction mixture using the TNT system (Promega). One-tenth of the synthesized [35S]HA-JNK1 or [35S]HA-IKKβ was incubated with purified GST-Miz1 or GST (2 μg of each) and glutathione beads (30 μL, 50% vol/vol) in a binding buffer [20 mM Tris (pH 8.0), 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 1 mM DTT, 1 mM PMSF, 1 μg/mL leupeptin, 1 μg/mL aprotinin, and 1 μg/mL pepstatin] for 4 h at 4 °C. GST- or GST-Miz1-associated [35S]HA-JNK1 or [35S]HA-IKKβ was pulled down by glutathione beads and analyzed by SDS/PAGE, followed by autoradiography. Ten percent of the input [35S]HA-JNK1 or [35S]HA-IKKβ also was analyzed.
Supplementary Material
Acknowledgments.
We thank Zhenggang Liu and James Chen for valuable reagents that made this work possible and Mrs. Hanh Chi Do and Mr. Yue Chen for excellent technical support. This work was partially supported by National Institutes of Health Grants CA100460, GM071409, and ES015868 (to A.L.) and GM081603 (to J.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906328106/DCSupplemental.
References
- 1.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 2.Kyriakis JM, et al. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 3.Chang L, Karin M. Mammalian MAP kinase signaling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 4.Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 5.Lin A. Activation of the JNK signaling pathway: Breaking the brake on apoptosis. BioEssays. 2003;25:1–8. doi: 10.1002/bies.10204. [DOI] [PubMed] [Google Scholar]
- 6.Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. J Biol Chem. 1995;270:16483–16486. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, Lin A. Wiring the cell signaling circuitry by the NF-κB and JNK1 cross-talk and its applications in human diseases. Oncogene. 2007;26:3267–3278. doi: 10.1038/sj.onc.1210417. [DOI] [PubMed] [Google Scholar]
- 8.Chang L, et al. The E3 ubiquitin ligase itch couples JNK activation to TNFα-induced cell death by inducing c-FLIPL turnover. Cell. 2006;124:601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 9.Deng H, et al. Phosphorylation of Bad at Thr-201 by JNK1 promotes glycolysis through activation of phosphofructokinase-1. J Biol Chem. 2008;283:20754–20760. doi: 10.1074/jbc.M800024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greten FR, et al. IKKβ links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 11.Maeda S, et al. IKKβ couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121:977–990. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Liu J, Minemoto Y, Lin A. c-Jun N-terminal protein kinase 1 (JNK1), but not JNK2, is essential for tumor necrosis factor α-induced c-Jun kinase activation and apoptosis. Mol Cell Biol. 2004;24:10844–10856. doi: 10.1128/MCB.24.24.10844-10856.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, et al. NF-κB is required for UV-induced JNK activation via induction of PKCδ. Mol Cell. 2006;21:467–480. doi: 10.1016/j.molcel.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 14.Sakurai T, Maeda S, Chang L, Karin M. Loss of hepatic NF-κB activity enhances chemical hepatocarcinogenesis through sustained c-Jun N-terminal kinase 1 activation. Proc Natl Acad Sci USA. 2006;103:10544–10551. doi: 10.1073/pnas.0603499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabpathy K, et al. Distinct roles for JNK1 and JNK2 in regulating JNK activity and c-Jun-dependent cell proliferation. Mol Cell. 2004;15:713–725. doi: 10.1016/j.molcel.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 16.Yu C, et al. JNK suppresses apoptosis via phosphorylation of the proapoptotic Bcl-2 family protein BAD. Mol Cell. 2004;13:329–340. doi: 10.1016/s1097-2765(04)00028-0. [DOI] [PubMed] [Google Scholar]
- 17.De Smaele E, et al. Induction of gadd45β by NF-κB down-regulates proapoptotic JNK signaling. Nature. 2001;414:308–313. doi: 10.1038/35104560. [DOI] [PubMed] [Google Scholar]
- 18.Kamata H, et al. Reactive oxygen species promote TNFα-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell. 2005;120:649–661. doi: 10.1016/j.cell.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 19.Karin M, Lin A. NF-κB at the crossroads of life and death. Nat Immunol. 2002;3:221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- 20.Maeda S, et al. IKKβ is required for prevention of apoptosis mediated by cell-bound but not by circulating TNFα. Immunity. 2003;19:725–737. doi: 10.1016/s1074-7613(03)00301-7. [DOI] [PubMed] [Google Scholar]
- 21.Tang G, et al. Inhibition of JNK activation through NF-κB target genes. Nature. 2001;414:313–317. doi: 10.1038/35104568. [DOI] [PubMed] [Google Scholar]
- 22.Tang F, et al. Absence of NF-κB-mediated inhibition of c-Jun N-terminal kinase activation contributes to tumor necrosis factor α-induced apoptosis. Mol Cell Biol. 2002;22:8571–8579. doi: 10.1128/MCB.22.24.8571-8579.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ventura J-J, Kennedy NJ, Lamb JA, Flavell RA, Davis RJ. c-Jun NH2-terminal kinase is essential for the regulation of AP-1 by tumor necrosis factor. Mol Cell Biol. 2003;23:2871–2882. doi: 10.1128/MCB.23.8.2871-2882.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karin M, Gallagher E. From JNK to pay dirt: Jun kinases, their biochemistry, physiology and clinical importance. IUBMB Life. 2005;57:283–295. doi: 10.1080/15216540500097111. [DOI] [PubMed] [Google Scholar]
- 25.Bardwell VJ, Treisman R. The POZ domain: A conserved protein–protein interaction motif. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 26.Schulz TC, Hopwood B, Rathjen PD, Wells JR. An unusual arrangement of 13 zinc fingers in the vertebrate gene Z13. Biochem J. 1995;311:219–224. doi: 10.1042/bj3110219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peukert K, et al. An alternative pathway for gene regulation by Myc. EMBO J. 1997;16:5672–5686. doi: 10.1093/emboj/16.18.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herold S, et al. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol Cell. 2002;10:509–521. doi: 10.1016/s1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- 29.Ziegelbauer J, et al. Transcription factor MIZ-1 is regulated via microtubule association. Mol Cell. 2001;8:339–349. doi: 10.1016/s1097-2765(01)00313-6. [DOI] [PubMed] [Google Scholar]
- 30.Wanzel M, Herold S, Eilers M. Transcriptional repression by Myc. Trends Cell Biol. 2003;13:146–150. doi: 10.1016/s0962-8924(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 31.Adhikary S, et al. Miz1 is required for early embryonic development during gastrulation. Mol Cell Biol. 2003;23:7648–7657. doi: 10.1128/MCB.23.21.7648-7657.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kallunki T, et al. JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes Dev. 1994;8:2996–3007. doi: 10.1101/gad.8.24.2996. [DOI] [PubMed] [Google Scholar]
- 33.Lin A, et al. Identification of a dual specificity kinase that activates the Jun kinases and p38-Mpk2. Science. 1995;268:286–290. doi: 10.1126/science.7716521. [DOI] [PubMed] [Google Scholar]
- 34.Lu X, Nemoto S, Lin A. Identification of c-Jun NH2-terminal protein kinase (JNK)-activating kinase 2 as an activator of JNK but not p38. J Biol Chem. 1997;272:24751–24754. doi: 10.1074/jbc.272.40.24751. [DOI] [PubMed] [Google Scholar]
- 35.Liu ZG, Hsu H, Goeddel DV, Karin M. Dissection of TNF receptor 1 effector functions: JNK activation is not linked to apoptosis while NF-κB activation prevents cell death. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 36.Tada K, et al. Critical roles of TRAF2 and TRAF5 in tumor necrosis factor-induced NF-κB activation and protection from cell death. J Biol Chem. 2001;276:36530–36534. doi: 10.1074/jbc.M104837200. [DOI] [PubMed] [Google Scholar]
- 37.Yeh W-C, et al. Early lethality, functional NF-κB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity. 1997;7:715–725. doi: 10.1016/s1074-7613(00)80391-x. [DOI] [PubMed] [Google Scholar]
- 38.Shi C-S, Kehrl JH. Tumor necrosis factor (TNF)-induced germinal center kinase-related (GCKR) and stress-activated protein kinase (SAPK) activation depends upon the E2/E3 complex Ubc13-Uev1A/TNF receptor-associated factor 2 (TRAF2) J Biol Chem. 2003;278:15429–15434. doi: 10.1074/jbc.M211796200. [DOI] [PubMed] [Google Scholar]
- 39.Habelhah H, et al. Ubiquitination and translocation of TRAF2 is required for activation of JNK but not of p38 or NF-κB. EMBO J. 2004;23:322–332. doi: 10.1038/sj.emboj.7600044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tournier C, et al. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science. 2000;288:870–874. doi: 10.1126/science.288.5467.870. [DOI] [PubMed] [Google Scholar]
- 41.Lin Y, et al. The death domain kinase RIP is essential for TRAIL (Apo2L)-induced activation of IκB kinase and c-Jun N-terminal kinase. Mol Cell Biol. 2000;20:6638–6645. doi: 10.1128/mcb.20.18.6638-6645.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raman M, Earnest S, Zhang K, Zhao Y, Cobb MH. TAO kinases mediate activation of p38 in response to DNA damage. EMBO J. 2007;26:2005–2014. doi: 10.1038/sj.emboj.7601668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y, Kwon SW, Kim SC, Zhao Y. Integrated approach for manual evaluation of peptides identified by searching protein sequence databases with tandem mass spectra. J Proteome Res. 2005;4:998–1005. doi: 10.1021/pr049754t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.