Abstract
Design features that ensure reproducible and invariant embryonic processes are major characteristics of current gene regulatory network models. New cis-regulatory studies on a gene regulatory network subcircuit activated early in the development of the sea urchin embryo reveal a sequence of encoded “fail-safe” regulatory devices. These ensure the maintenance of fate separation between skeletogenic and nonskeletogenic mesoderm lineages. An unexpected consequence of the network design revealed in the course of these experiments is that it enables the embryo to “recover” from regulatory interference that has catastrophic effects if this feature is disarmed. A reengineered regulatory system inserted into the embryo was used to prove how this system operates in vivo. Genomically encoded backup control circuitry thus provides the mechanism underlying a specific example of the regulative development for which the sea urchin embryo has long been famous.
Keywords: gene regulatory, embryonic development, regulatory subcircuit topology, pmar/hesC
Micromeres of the sea urchin embryo are formed at the vegetal pole of the egg by means of the unequal fourth cleavage. Their four larger fifth cleavage daughter cells are the founders of a cell lineage, the sole later product of which is skeletogenic mesenchyme (SM). The gene regulatory network (GRN) determining the processes of specification of this lineage provides a causal explanation of its prominent developmental functions, up to the ingression of the skeletogenic cells in the late blastula (24 h). Thus, the GRN explains how the micromeres initially acquire their regulatory identity; how they emit the signals that they do, a function essential for development of the surrounding endomesodermal lineages; and what are the regulatory interactions by which they activate skeletogenic gene batteries (1). In terms of causality, the pmar1 gene lies at the top of the SM specification GRN. This gene is activated immediately upon the birth of the fourth cleavage micromeres in response to two maternal transcriptional inputs at this stage confined to these cells (2, 3). Perturbation experiments showed that these are Otx, a transcription factor nuclearized in the fourth cleavage micromeres, but later in other cells as well (4), and maternal β-catenin, which is nuclearized downstream of a maternal anisotropy in activated Dishevelled protein, tethered at the vegetal pole of the egg (5, 6). As described earlier (1, 7, 8), pmar1 transcription activates a double-negative logic gate, which accounts for the institution of the whole downstream skeletogenic regulatory state, formulated by transcription of a specific set of regulatory and signaling genes. Thus, the pmar1 gene encodes a repressor (2), the role of which is to prohibit transcription of the hesC gene (8), which also encodes a repressor. The HesC repressor specifically clamps down on transcription of the skeletogenic lineage regulatory state genes. Because the hesC gene is activated zygotically all over the embryo, it keeps these genes silent except where pmar1 is expressed. If hesC mRNA translation is globally blocked or if pmar1 mRNA is presented globally, then the whole embryo turns into cells expressing the skeletogenic program (refs. 8 and 9 and reviewed in ref. 10). The double-negative gate operates to ensure that the skeletogenic regulatory state genes may respond to their widely expressed activators only in the skeletogenic lineage, when and where the gate is unlocked by the encoded pmar1 repressor.
However, the pmar1-hesC gate is not the only regulatory function activated in the newly born SM founder cells. The β-catenin:Tcf input also is used within the fourth to fifth cleavage cycle to set up a transcriptional feedback between the wnt8 gene and the blimp1 regulatory gene in the SM founder cells (11–13). Blimp1 activates the wnt8 gene, but the blimp1 gene also requires the activated Tcf input produced in response to reception of the Wnt8 signal. Because Wnt8 is a ligand for the Tcf signal transduction system, the result is to drive further activated β-catenin:Tcf into the nuclei of the skeletogenic micromere lineage. The pmar1 and blimp1 genes are two of the initial regulatory genes to be turned on in these cells. As shown by cis-regulatory analysis (12), the blimp1 gene also responds to the same Otx plus β-catenin:Tcf inputs as does pmar1. The expression of these two regulatory genes and of the wnt8 ligand gene, uniquely defines the fifth cleavage founder cells of the SM lineage (9, 11–13).
In considering the SM specification GRN in the larger context of the surrounding nonskeltogenic mesoderm (NSM), new questions arise, because many of the same regulatory genes are ultimately expressed in both this and the skeletogenic territory. Indeed, the same inputs that activate pmar1, the regulatory gene at the top of the skeletogenic specification network, also later appear in the NSM. Because ectopic expression of pmar1 suffices to convert any blastomeres expressing it into the skeletogenic regulatory state (2), why the NSM does not eventually activate pmar1 and become skeletogenic or, more generally, what mechanisms ensure the separation of fates in these adjacent mesodermal lineages remain to be explained. We know that the separation mechanisms can be reversed, because after gastrulation NSM cells can acquire skeletogenic function if the normal complement of SM is depleted experimentally (reviewed in ref. 10). However, in the normal pregastrular embryo, the SM and NSM lineages remain rigidly discrete. Here, we show that an unexpected feature of the pmar1 cis-regulatory system provides at least part of the functional mechanism of fate exclusion, encoding part of a simple but remarkably designed regulatory switch function. Furthermore, the genomically encoded circuitry into which pmar1 is locked endows the embryo with a capacity to execute regulative skeletogenic specification, even if pmar1 function is experimentally blocked.
Results
Genomic Basis for Restriction of pmar1 Expression to the Skeletogenic Lineage.
To determine the cis-regulatory basis for spatial expression of pmar1, including both its initial micromere expression and the subsequent exclusion of its expression from the NSM, we had first to resolve the regulatory sequence organization of this locus. When pmar1 activity was initially discovered by Oliveri et al. (2), they noted that on the basis of genome blots there appeared to be several similar such genes, and this was later confirmed in the Strongylocentrotus purpuratus genomic sequence. The current annotated genome assembly [version 2.1 (14)] indicates two linked genes, pmar1a and pmar1b, which are very closely related, plus at least two others. The pmar1a and pmar1b genes share a ≈2.5-kb upstream duplicated sequence together with the duplicated gene coding regions plus a complex set of additional smaller duplications and inversions (an analysis is shown in Fig. S1). However, not one of the key pmar1a transcription factor target sites revealed in the following functional assays is present in the pmar1b sequence, due both to the presence of an additional sequence in pmar1a (the result of an indel) and to sequence divergence in the duplicated upstream sequence. Furthermore, quantitative PCR primer pairs designed specifically to target the 3′ UTR of pmar1b failed to detect expression at any embryonic stage. Thus, we focused on the pmar1a gene.
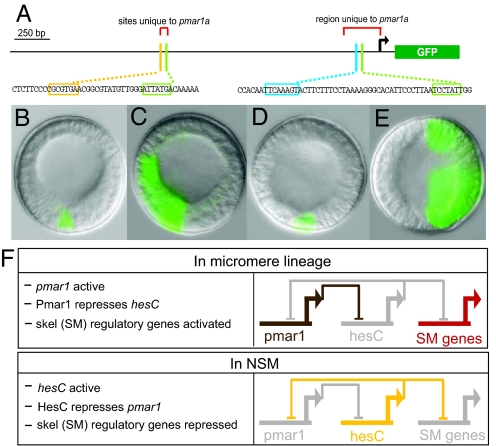
Transcripts recognized by a pmar1a probe (which also would cross-react with transcripts of the other possible pmar1 genes) appear abruptly and exclusively in the micromeres during the fourth cleavage cycle (a 20-min interval high-density time course is shown in Fig. S2; see also ref. 2). To explore the cis-regulatory control of pmar1a, a GFP expression construct was built that contained the pmar1a-specific fragment just upstream of the start site plus the duplicated upstream region (Fig. S1). This construct included two putative Tcf target sites (TTCAAAG) plus two Otx sites (TAATCC). These are the inputs predicted from prior analysis (3) to activate pmar1. When eggs were injected, this construct expressed faithfully only in cells of the SM lineage, exactly resembling the endogenous pmar1 expression (e.g., Fig. 1B). Mutational disruption of the distal Otx target site (Fig. 1A) decreased the number of GFP mRNA molecules and number of incorporated construct DNA molecules to 23 ± 18% (SEM) of those of the wild-type control (three experiments on each mutation); mutation of the proximal Otx site reduced it to 14 ± 11% of those of the control. Both Otx sites apparently are required. Mutation of the single Tcf included in Fig. 1A decreased expression to 38 ± 32%. (Disruption of the second candidate Tcf site produced no significant effect and is not included in Fig. 1A.) The predicted positive Otx and β-catenin:Tcf inputs thus are confirmed, and we can see by experimental test that these sites constitute a sufficient genomic code for transduction of the initial maternal spatial inputs present in the newly born micromeres.
Fig. 1.
Exclusive regulatory domains established by reciprocal repression between pmar1 and hesC. (A) Diagram of a 2.59-kb fragment upstream of the pmar1a start of translation fused with the coding sequence for the GFP reporter. Small portions of the sequence are reproduced below to indicate highlighted transcription factor target sites discussed in the text. The bent arrow indicates the start of transcription. (cf. Fig. S1). (B–E) Typical results in embryos bearing pmar1a expression constructs at 16 h postfertilization: (B) wild-type pmar1 reporter activity (this construct expressed only in micromere descendants in 124 of 135 GFP+ embryos); (C) pmar1 construct but with HesC target site shown in A disrupted (result is gross ectopic activity in 77 of 92 embryos harboring construct). (D) Normal skeletogenic mesenchyme (SM) expression of wild-type pmar1 construct in presence of coinjected control morpholino antisense oligonucleotide (MASO). (E) Wild-type pmar1 construct in presence of coinjected HesC MASO resulted in ectopic expression in nearly all embryos (56 of 61). (F) Summary of regulatory interactions from refs. 1 and 8 and these experiments.
By the fifth to sixth cleavage, β-catenin can be visualized in all endomesodermal nuclei (i.e., future NSM plus future endoderm and in the SM lineage nuclei) (5). Nuclear Otx also becomes widely available (4), and yet our pmar1 construct, like the endogenous pmar1 gene, continues to be expressed only in SM. Fig. 1A shows a putative site (CGCGTG) for the dominant HesC repressor located 1.4-kb upstream of the start site, and this single 6-bp sequence turns out to be responsible for the continued accuracy of expression of the pmar1 construct. When it was mutated, ectopic expression of the reporter spread to all domains of the embryo in 77 of 92 GFP+ embryos in three separate experiments (e.g., Fig. 1C), although this pattern rarely was seen for the wild-type reporter (11 of 135 embryos). To confirm this, we coinjected the wild-type pmar1 construct with either a randomized control morpholino antisense oligonucleotide (MASO) or a MASO targeting hesC mRNA, with typical results as illustrated in Fig. 1 D and E, respectively.
Thus, the pmar1 gene product represses the hesC gene (8), but the hesC gene product also represses pmar1 (Fig. 1). However, this reciprocal repression mechanism is not deployed developmentally as a “bidirectional switch”: It is entirely unidirectional, depending on the invariant, spatially controlled temporal sequence of repressor availability. There is a very small amount of maternal hesC mRNA, ≈100 molecules per egg (8). If, like every other maternal mRNA so far investigated in the sea urchin egg, it is uniformly distributed, only approximately eight molecules would be included in the four micromeres (i.e., an insignificant two molecules per cell). The transcriptional activation of pmar1 is in contrast measurably elevated, the slope of the quantitation plot indicating that transcription of pmar1 operates at close to the biochemical maximum for this system of approximately six to nine transcription initiation events per minute. Thus, as the SM lineage is founded, the Pmar1 repressor is first on the scene, and it prevents the activation of the hesC gene in these cells. Everywhere else the hesC gene is transcriptionally activated (8), so by the time the Otx and Tcf drivers become available outside the SM, HesC repressor is present; despite the availability of the activators, the repressor is dominant, and it prevents ectopic pmar1 expression in these cells. This relation is summarized in Fig. 1F, which describes the establishment of two exclusive repressive regulatory domains, one Pmar1+ the other HesC+.
Morphological and Molecular Effects of pmar1 MASO and Regulative Recovery of the SM Lineage.
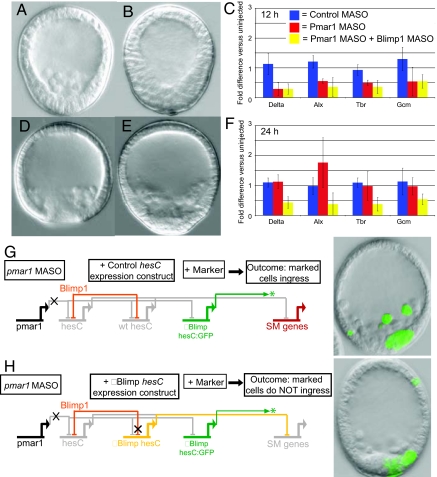
A MASO was prepared complementary to the pmar1a mRNA sequence. The morphological effects of this MASO were striking, if not unexpected. In 24-h mesenchyme blastula control embryos, the SM cells have ingressed into the blastocoel and are a prominent feature of normal sea urchin embryo morphology (Fig. 2A, control MASO injection). However, in 75% of pmar1 MASO embryos, ingression is abolished, resulting in the phenotype shown in Fig. 2B. The mechanistic reason for this is that expression of the regulatory genes downstream of the pmar1 double-negative gate are required for the cell biological and differentiation functions of the SM (1, 15). We can see directly in Fig. 1C that at 12 h expression of direct target genes of the double- negative gate (delta, alx1, and tbr) and of the indirect target gcm, which requires Delta signaling (16), is severely depressed by pmar1 MASO, exactly as predicted from GRN topology (1, 8). However, most surprisingly, the embryos recover fully in the succeeding hours, and by 31 h, as gastrulation is beginning, the morphologies of control (Fig. 2D) and pmar1 MASO embryos (Fig. 2E) are identical. Many additional examples of these phenotypes are reproduced in Fig. S3 A–C, including perfectly formed pluteus stage embryos with mature skeletons and pigment cells (Fig. S3F). Again, the reason lies in the regulatory state: Fig. 2F shows that the expression of the target genes at 24 h is back at normal or even overshot levels. This observation confronts us with the mechanistic problem of understanding the interactions allowing return of the SM regulatory state despite the absence of pmar1 function. We have here a classic example of regulative developmental adjustment and the opportunity to determine how it works.
Fig. 2.
pmar1 morpholino antisense oligonucleotide (MASO) effects and mechanism of regulative recovery. (A) Control MASO embryo at 24 h postfertilization, showing normal ingression of skeletogenic mesenchyme (SM). (B) pmar1 MASO-injected embryo at 24 h: Ingression is totally blocked. (C) Severe decrease in levels of expression of genes downstream of the double-negative gate at 12 h after injection of pmar1 MASO or pmar1 MASO + blimp1 MASO. (D) Control MASO-injected embryo at 31 h. (E) Recovery from pmar1 MASO by 31 h: normal SM ingression, similar to control in D. (F) Regulative recovery is seen at the molecular level by 24 h but not when the blimp1 MASO is also present. (G, H) Regulative recovery depends on specific repression of hesC by Blimp1. (G) Control experiment where in addition to pmar1 MASO a wild-type hesC BAC (“wt hesC”) is coinjected as a hesC expression vector; an additional hesC GFP reporter lacking the previously characterized intronic Blimp1 target site (“ΔBlimp hesC:GFP”) is coinjected as a marker of incorporation of the expression constructs. Reporter activity is seen in ingressed SM cells as regulative recovery takes place. (H) A hesC BAC in which the single intronic Blimp1 target site is mutated (“ΔBlimp hesC”) is introduced together with the same GFP marker construct and pmar1 MASO. Clones harboring the transgenes now fail to ingress, although SM cells lacking the transgenes recover and ingress.
Fail-Safe GRN Subcircuit That Endows the Embryo with Regulative Capability.
Early blimp1 gene expression is controlled by a cis-regulatory module that includes two autorepressive target sites responsible for shutting down blimp1 transcription wherever it had been active after a certain accumulation of its gene product (11, 17). Recently, we found that this repression also extends to the hesC gene. After ≈18 h, blimp1 expression in the NSM is extinguished by autorepression, and at the same time, hesC transcription in the NSM also is extinguished, for which a hesC cis-regulatory target site for Blimp1 is required (18). Because earlier blimp1 also is expressed and then represses itself in the SM, the possibility thus arose that this independent mechanism of abolishing the HesC repressor might be involved in the regulative recovery of embryos after pmar1 MASO treatment. Indeed, if embryos are treated with blimp1 and with pmar1 MASOs, then they never recover, even after 3 days (the disastrous developmental arrest phenotype is illustrated in Fig. S3D), despite the fact that treatment with the blimp1 MASO alone displays no effect on SM ingression (17). In embryos treated with both blimp1 and pmar1 MASOs, target gene expression never recovers, as would be expected (Fig. 2F). The effects of the double MASO treatment are consistent with the idea that blimp1 expression is necessary for regulative recovery, but because Blimp1 is central to the deployment of two central signal transduction systems (17, 18), we could not rule out pleiotropic effects.
We thus turned to a cis-reengineering approach using recombineered BAC knock-in constructs to allow us to test in isolation a single Blimp1 target site found previously to be responsible for shutting off hesC (18). In the earlier studies, a hesC:GFP BAC construct had been shown to function as does the endogenous hesC gene (18). Therefore, the parental BAC and the knock-in construct contain the endogenous hesC cis-regulatory system. However, the GFP construct does not produce any HesC, only GFP. In the first of the following series of experiments, this reporter construct was introduced together with pmar1 MASO (in three separate trials). The embryos displayed expression of GFP in the presumptive SM, but these cells were all resident in the vegetal wall of the temporarily arrested embryos, because there is no Pmar1 repression of HesC, and the cells at the early stage were unable to invaginate. Later, during the period of regulative recovery, GFP transcription turns off in ingressed cells. Thus, the wild-type hesC:GFP BAC reporter responds in accordance to the Blimp1 repression that we propose underlies regulative recovery. Next, the hesC:GFP BAC was altered by mutation of the Blimp1 binding site (18), so that it could no longer respond to Blimp1 repression. The mutated GFP construct then was coinjected with pmar1 MASO. After 30 h, the embryos underwent regulative recovery of ingression as previously. In those embryos where the incorporated GFP clone included SM cells, some of the newly ingressed cells continue to display green fluorescence, because the altered hesC:GFP construct is not subject to the blimp1 repression that would normally clear its transcription from these cells. Injection in addition of the normal parental hesC BAC gives the same result, as it should, because this exogenous source of hesC transcript is under the same regulatory control as the endogenous gene. That is, the exogenous and the endogenous hesC genes will be turned off by Blimp1 repression. This control experiment is illustrated in Fig. 2G: 46 of 51 GFP+ embryos displayed ingression of fluorescent cells (i.e., recovered SM cells expressing the exogenous vector). In considering this protocol, recall that in sea urchin embryos exogenous DNA is concatenated and stably incorporated after injection into a given early cleavage nucleus (19), so that whatever constructs are introduced will be taken up together into the same cells in a mosaic pattern with respect to cell lineage.
The stage is now set for a direct test of the idea that Blimp1 repression of the hesC gene is the mechanism of regulative recovery from pmar1 MASO. If, instead of the normal parental hesC BAC, a mutated hesC BAC lacking the Blimp1 repression sites is injected together with pmar1 MASO plus the similarly mutated hesC:GFP BAC knock-in, then the result should be different from that in Fig. 2G. The crucial difference is that there will now be a continuing source of hesC in the SM cells. Thus, recovery should fail in all of those cells bearing the exogenous constructs and fluorescing green. No green cells should be seen in the ingressed population, which should consist exclusively of those SM cells lacking the exogenous constructs. This experiment is illustrated in Fig. 2H, and further examples are shown in Fig. S4. In 26 of 29 GFP+ embryos, none of the recovered, ingressed cells expressed GFP, all of the fluorescent cells remaining in the wall of the embryo, as in the illustrated cases. Only in three embryos (all appearing abnormal) were there a few fluorescent ingressed cells. In summary, the experiment demonstrates that recovery and ingression depend on the single intact Blimp1 repression site in the cis-regulatory module controlling hesC gene expression.
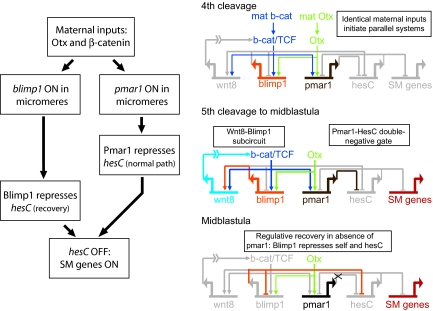
This is a clear example of fail-safe circuitry, as summarized in Fig. 3. Here, we see that under normal conditions pmar1 repression of hesC allows SM expression of the immediately downstream genes, which build the SM regulatory state, but there is a backup. The efficient delayed recovery observed when pmar1 expression is poisoned is due to an additional device: hesC repression by Blimp1. An interesting feature of this system is that the primary and the backup effectors, pmar1 and blimp1, are wired perfectly in parallel and are activated by the same inputs at almost the same time (Fig. 3). The canonical repressor encoded by pmar1 (2) is active first, because it takes some hours for the blimp1 product to accumulate and achieve repressor activity. This may be due to the specific mechanism of Blimp1-mediated repression, which depends on heterochromatin induction (20) or perhaps for a corepressor to appear; in any case, the blimp1 gene does not shut itself off in the SM by interaction with its own autorepression sites until ≈6 h after it is activated. This would indicate the timing for the transcriptional turndown of hesC expression as well.
Fig. 3.
Fail-safe function and underlying gene regulatory network circuitry.
Discussion
The depth of experimental knowledge about this regulatory system makes it possible to disentangle the fundamental sources of causality in the process that it mediates. By the test of necessity and sufficiency, we know that early on pmar1 expression is the cause of expression (derepression) of the SM regulatory state (1, 2, 8, 9). The cause of SM-specific expression of pmar1, as the present work shows, is unequivocally the encoded target sites in its cis-regulatory sequence. These enable the cis-regulatory system to read the specific inputs in the SM founder cells. These in turn are produced by maternal anisotropies generated during oogenesis. Development of almost all animal eggs begins with maternal anisotropies of regulatory significance (21, 22), and the off-the-DNA mechanisms by which these topological anisotropies are formulated are the causes of the initial state of the developmental mechanism. However, after that, everything depends causally on the genomic cis-regulatory program. Thus, to continue, one cause of the separation of SM and NSM is the establishment of Pmar1+ vs. HesC+ repression domains due to the additional, unanticipated feature of the pmar1 cis-regulatory system discovered here, namely, the functional HesC site that prevents pmar1 expression outside the SM lineage. Finally, the cause of the robust but late regulative recovery phenomenon that we observed in pmar1 MASO poisoned eggs again lies in a cis-regulatory sequence, this time the Blimp1 response elements of the HesC cis-regulatory system, as demonstrated in the reengineering experiment of Fig. 2 G and H. Although the biochemical parameters of protein and mRNA synthesis and accumulation, target site occupancy, and transcriptional response will determine the actual kinetics of all of these events (23, 24), their causal basis, the informational inputs that specify a reliable outcome, lies in the cis-regulatory apparatus of the respective genes.
General “Fail-Safe Quality” of the GRN.
The regulative fail-safe recovery apparatus is only the last, and perhaps the most spectacular, of the network circuitry devices that we now know about that function to ensure absolutely the correct developmental outcome. These devices are worth enumerating: (i) The pmar1-hesC double-negative gate is itself a global control mechanism, because it both activates the SM genes and specifically precludes their expression everywhere else (7, 8). (ii) Dynamic feedback lockdown of the SM specification state ensures its stability: Just downstream of the double-negative gate target genes, a three-gene feedback system is set up after which the skeletogenic regulatory state is locked in a positive regulatory embrace and cannot further change so long as these genes continue to be expressed (1). (iii) An exclusion function downstream of alx1 expression (alx1 is a primary double-negative gate target) causes a direct or indirect repression in the SM domain of the major early NSM regulator, gcm (1). This additional device independently excludes the alternative NSM fate. (iv) A spatially dynamic signaling subcircuit ensures exclusive adjacent domains of Wnt8 and Delta signaling (18). (v) Reciprocal repression, as described above, establishes mutually exclusive Pmar1+ (i.e., SM) and HesC+ (e.g., NSM) domains. (vi) The backup blimp1 fail-safe regulative recovery mechanism discovered in this work ensures skeletogenic specification even if something goes wrong with the first step in the process. Note that the presence of this device is assured by the parallel wiring of blimp1 and pmar1: When one goes on, so does the other.
There are additional reciprocal layers to the fate stabilization system as well. In the normal late blastula, the NSM is prohibited from assuming SM fate even though blimp1-wnt8 subcircuit expression eventually spreads to the NSM and turns off hesC expression there (18). There are just two regulatory genes of the SM, which are never expressed in NSM, and these are tbr and alx1; tbr expression is forbidden in NSM by a special repression device (25). Both of these genes are required for skeletogenesis to occur (1, 15). Furthermore, in the postgastrular embryo, an intercellular communication system operates to ensure alx1 repression if the normal complement of SM cells is present (10).
Our knowledge of the SM specification GRN and surrounding network relationships is more complete at the moment than that for other components of the embryonic regulatory system. However, there is unlikely anything special about the overall character of the SM GRN, so a safe generalization is that we probably will see more and more fail-safe wiring as we delve deeper into the networks controlling specification and development of other territories of the embryo. Indeed, several such devices already have been revealed. Among them, for example, are the multiple feedbacks installed in a kernel of the endomesoderm specification GRN among the otx, gatae, blimp1, and brachyury genes (24, 26). On top of this is layered two additional potent devices to ensure correct outcome: Many of the primary endoderm specification genes use a Tcf feed, and it has been proved at the cis-regulatory level for key genes such as foxa and evenskipped (12) that the repressive function of Tcf in cells not receiving Wnt signaling independently ensures accurate expression. Furthermore, foxa expression (endoderm) excludes gcm expression (NSM), probably by direct cis-regulatory input (27). Developmental networks involving interlocking layers of repression are found elsewhere, for example, among gap genes in Drosophila (28, 29). To see if in other settings these subcircuits similarly function as part of the fate stabilization system ensuring an invariant developmental outcome will be interesting.
These observations imply a major feature of developmental GRN organization. Perhaps it is a definition of embryonic specification in crown group animals that the underlying GRNs are interwoven with diverse, genomically encoded mechanisms to ensure as absolutely as possible that spatial specification in development will not fail and will not vary. Fail-safe wiring characterizes these networks, and never have the system design paradigms of “Ockham's razor” or “maximization of parsimony” been less applicable as predictive guides to structure, replaced by “reliability” and “reproducibility.”
Evolution.
How can fail-safe wiring evolve? That fail-safe devices like that in Fig. 3 exist shows that they are not to be regarded simply as redundant devices. One key to the evolutionary origins of this type of circuitry is its modularity. As discussed elsewhere, the diverse modules of a GRNs have separate evolutionary histories; they change at different rates and are apparently loaded into the GRN at different stages in the evolution of the crown group lineages (30). The implication in the present case is that the blimp1 subcircuit could be older, because it has multiple other functions in other domains [e.g., in the NSM, where its control of hesC is causal to the initiation of delta expression there according to cis-regulatory analysis of both genes (18)]. This prediction can be tested easily by comparative observations. The pmar1 initiation system has been proposed to be particular to the modern echinoids (31). Thus, the pmar1-hesC double-negative gate may be layered atop the blimp-hesC double-negative gate in evolutionary sequence, in temporal operation, and in GRN topology.
Materials and Methods
Microinjection and Quantitative PCR Measurements.
The PCR products were purified with the Qiagen Qiaquick PCR purification kit and microinjected into fertilized S. purpuratus eggs as described in refs. 8 and 12). Linearized BAC constructs were desalted by drop dialysis into TE buffer (10 mM Tris, 1 mM EDTA) on a 0.025-μm filter (VSWP-02500, Millipore). Approximately 1,500 molecules of the desired reporter construct were injected along with a sixfold molar excess of HindIII-digested carrier sea urchin DNA per egg in 4 pL of 0.12 M KCl. A similar injection solution was made for BAC reporters but with 400 copies of the BAC per 4 pL and no carrier DNA. Embryos were collected at different stages for observation by fluorescence microscopy for qualitative assessment of spatial activity. For high-density cDNA time course experiments, gametes were harvested from three females and three males, pooled, and cultured at 14 °C. Three separate samples were removed at 20-min intervals for independent processing and quantitative PCR (QPCR) analysis. Data points represent the average of the three samples. All experimental and control constructs were tested in multiple batches of eggs. Microinjection and measurement of GFP mRNA by QPCR was performed as described in ref. 32. DCt was computed by taking the change in cycle number of an internal standard (Spz12) mRNA in control condition minus the change in cycle number of Spz12 mRNA and target gene in experimental condition.
Constructs and pmar1a MASO.
The sequence of the MASO targeting pmar1a (and potentially other pmar1 genes) was 5′-GTGATCATGGTGTAATCTGCCATTC-3′. This MASO sequence also would target any other annotated pmar1 sequence with one or two mismatches. The BAC clone 132 M17, containing pmar1a (GLEAN3_14721; Gene ID 373266) within 3 kb of the T7 end and excluding pmar1b, was isolated for pmar1a cis-regulatory analysis. Standard PCR and fusion PCR techniques using the High Fidelity PCR Kit (Roche) were used to build constructs. The pmar1 reporter constructs were cloned using the CopyControl Cloning System (EPICENTRE) in the case of large inserts (>5 kb) or the pGEM-T Easy Vector System from Promega and were confirmed by sequencing. Binding site sequences were mutated by PCR, and the resulting constructs were checked by sequencing. The PCR primers were designed with tailed nonpriming sequences, including the mutant form of the candidate transcription factor binding sites. Mutations were designed by swapping A to C or T to G, and vice versa.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grants HD37105 and GM75089. J.S. was supported by a fellowship from the California Institute of Regenerative Medicine.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910007106/DCSupplemental.
References
- 1.Oliveri PO, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci USA. 2008;105:5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oliveri P, Carrick DM, Davidson EH. A regulatory gene network that directs micromere specification in the sea urchin embryo. Dev Biol. 2002;246:209–228. doi: 10.1006/dbio.2002.0627. [DOI] [PubMed] [Google Scholar]
- 3.Davidson EH, et al. A provisional regulatory gene network for specification of endomesoderm in the sea urchin embryo. Dev Biol. 2002;246:162–190. doi: 10.1006/dbio.2002.0635. [DOI] [PubMed] [Google Scholar]
- 4.Chuang CK, Wikramanayake AH, Mao CA, Li X, Klein WH. Transient appearance of Strongylocentrotus purpuratus Otx in micromere nuclei: Cytoplasmic retention of SpOtx possibly mediated through an α-actinin interaction. Dev Genet. 1996;19:231–237. doi: 10.1002/(SICI)1520-6408(1996)19:3<231::AID-DVG6>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 5.Logan CY, Miller JR, Ferkowicz MJ, McClay DR. Nuclear β-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development. 1999;126:345–357. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- 6.Weitzel HE, et al. Differential stability of β-catenin along the animal-vegetal axis of the sea urchin embryo mediated by dishevelled. Development. 2004;131:2947–2956. doi: 10.1242/dev.01152. [DOI] [PubMed] [Google Scholar]
- 7.Levine M, Davidson EH. Gene regulatory networks for development. Proc Natl Acad Sci USA. 2005;102:4936–4942. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Revilla-i-Domingo R, Oliveri P, Davidson EH. A missing link in the sea urchin embryo gene regulatory network: hesC and the double-negative specification of micromeres. Proc Natl Acad Sci USA. 2007;104:12383–12388. doi: 10.1073/pnas.0705324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliveri P, Davidson EH, McClay DR. Activation of pmar1 controls specification of micromeres in the sea urchin embryo. Dev Biol. 2003;258:32–43. doi: 10.1016/s0012-1606(03)00108-8. [DOI] [PubMed] [Google Scholar]
- 10.Ettensohn CA. Lessons from a gene regulatory network: Echinoderm skeletogenesis provides insights into evolution, plasticity and morphogenesis. Development. 2009;136:11–21. doi: 10.1242/dev.023564. [DOI] [PubMed] [Google Scholar]
- 11.Smith J, Theodoris C, Davidson EH. A gene regulatory network subcircuit drives a dynamic pattern of gene expression. Science. 2007;318:794–797. doi: 10.1126/science.1146524. [DOI] [PubMed] [Google Scholar]
- 12.Smith J, Kraemer E, Liu H, Theodoris C, Davidson EH. A spatially dynamic cohort of regulatory genes in the endomesodermal gene network of the sea urchin embyro. Dev Biol. 2008;313:863–875. doi: 10.1016/j.ydbio.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minokawa T, Wikramanayake AH, Davidson EH. cis-Regulatory inputs of the wnt8 gene in the sea urchin endomesoderm network. Dev Biol. 2005;288:545–558. doi: 10.1016/j.ydbio.2005.09.047. [DOI] [PubMed] [Google Scholar]
- 14.Sea Urchin Genome Sequencing Consortium et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ettensohn CA, Illies MR, Oliveri P, De Jong DL. Alx1, a member of the Cart1/Alx3/Alx4 subfamily of Paired-class homeodomain proteins, is an essential component of the gene network controlling skeletogenic fate specification in the sea urchin embryo. Development. 2003;130:2917–2928. doi: 10.1242/dev.00511. [DOI] [PubMed] [Google Scholar]
- 16.Ransick A, Davidson EH. cis-Regulatory processing of Notch signaling input to the sea urchin glial cells missing gene during mesoderm specification. Dev Biol. 2006;297:587–602. doi: 10.1016/j.ydbio.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 17.Livi CB, Davidson EH. Expression and function of blimp1/krox, an alternatively transcribed regulatory gene of the sea urchin endomesoderm network. Dev Biol. 2006;293:513–525. doi: 10.1016/j.ydbio.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 18.Smith J, Davidson EH. Gene regulatory network subcircuit controlling a dynamic spatial pattern of signaling in the sea urchin embryo. Proc Natl Acad Sci USA. 2008;105:20089–20094. doi: 10.1073/pnas.0806442105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMahon AP, et al. Introduction of cloned DNA into sea urchin egg cytoplasm: Replication and persistence during embryogenesis. Dev Biol. 1985;108:420–430. doi: 10.1016/0012-1606(85)90045-4. [DOI] [PubMed] [Google Scholar]
- 20.Gyory I, Wu J, Fejér G, Seto E, Wright KL. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol. 2004;5:299–308. doi: 10.1038/ni1046. [DOI] [PubMed] [Google Scholar]
- 21.Davidson EH. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. San Diego, CA: Academic; 2006. [Google Scholar]
- 22.Davidson EH. How embryos work: A comparative view of diverse modes of cell fate specification. Development. 1990;108:365–389. doi: 10.1242/dev.108.3.365. [DOI] [PubMed] [Google Scholar]
- 23.Bolouri H, Davidson EH. Transcriptional regulatory cascades in development: Initial rates, not steady state, determine network kinetics. Proc Natl Acad Sci USA. 2003;100:9371–9376. doi: 10.1073/pnas.1533293100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben-Tabou de-Leon S, Davidson EH. Gene regulation: Gene control network in development. Annu Rev Biophys Biomol Struct. 2007;36:191–212. doi: 10.1146/annurev.biophys.35.040405.102002. [DOI] [PubMed] [Google Scholar]
- 24a.Davidson EH, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 25.Wahl ME, Hahn J, Gora K, Davidson EH, Oliveri P. The cis-regulatory system of the tbrain gene: Alternative use of multiple modules to promote skeletogenic expression in the sea urchin embryo. Dev Biol. doi: 10.1016/j.ydbio.2009.08.005. doi: 10.1016/j.ydbio.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuh CH, Dorman ER, Howard ML, Davidson EH. An otx cis-regulatory module: A key node in the sea urchin endomesoderm gene regulatory network. Dev Biol. 2004;269:536–551. doi: 10.1016/j.ydbio.2004.02.025. [DOI] [PubMed] [Google Scholar]
- 27.Oliveri P, Walton KD, Davidson EH, McClay DR. Repression of mesodermal fate by foxa, a key endoderm regulator of the sea urchin embryo. Development. 2006;133:4173–4181. doi: 10.1242/dev.02577. [DOI] [PubMed] [Google Scholar]
- 28.Clyde DE, et al. A self-organizing system of repressor gradients establishes segmental complexity in Drosophila. Nature. 2003;426:849–853. doi: 10.1038/nature02189. [DOI] [PubMed] [Google Scholar]
- 29.Andrioli LP, Oberstein AL, Corado MS, Yu D, Small S. Groucho-dependent repression by sloppy-paired 1 differentially positions anterior pair-rule stripes in the Drosophila embryo. Dev Biol. 2004;276:541–551. doi: 10.1016/j.ydbio.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 30.Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 31.Gao F, Davidson EH. Transfer of a large gene regulatory apparatus to a new developmental address in echinoid evolution. Proc Natl Acad Sci USA. 2008;105:6091–6096. doi: 10.1073/pnas.0801201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Revilla-i-Domingo R, Minokawa T, Davidson EH. R11: A cis-regulatory node of the sea urchin embryo gene network that controls early expression of SpDelta in micromeres. Dev Biol. 2004;274:438–451. doi: 10.1016/j.ydbio.2004.07.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.