Abstract
We examined the MLL genomic translocation breakpoint in acute myeloid leukemia of infant twins. Southern blot analysis in both cases showed two identical MLL gene rearrangements indicating chromosomal translocation. The rearrangements were detectable in the second twin before signs of clinical disease and the intensity relative to the normal fragment indicated that the translocation was not constitutional. Fluorescence in situ hybridization with an MLL-specific probe and karyotype analyses suggested t(11;22)(q23;q11.2) disrupting MLL. Known 5′ sequence from MLL but unknown 3′ sequence from chromosome band 22q11.2 formed the breakpoint junction on the der(11) chromosome. We used panhandle variant PCR to clone the translocation breakpoint. By ligating a single-stranded oligonucleotide that was homologous to known 5′ MLL genomic sequence to the 5′ ends of BamHI-digested DNA through a bridging oligonucleotide, we formed the stem–loop template for panhandle variant PCR which yielded products of 3.9 kb. The MLL genomic breakpoint was in intron 7. The sequence of the partner DNA from band 22q11.2 was identical to the hCDCrel (human cell division cycle related) gene that maps to the region commonly deleted in DiGeorge and velocardiofacial syndromes. Both MLL and hCDCrel contained homologous CT, TTTGTG, and GAA sequences within a few base pairs of their respective breakpoints, which may have been important in uniting these two genes by translocation. Reverse transcriptase-PCR amplified an in-frame fusion of MLL exon 7 to hCDCrel exon 3, indicating that an MLL-hCDCrel chimeric mRNA had been transcribed. Panhandle variant PCR is a powerful strategy for cloning translocation breakpoints where the partner gene is undetermined. This application of the method identified a region of chromosome band 22q11.2 involved in both leukemia and a constitutional disorder.
The incidence of leukemia in infants less than 1 year old in the United States is 20 per million per year for acute lymphoblastic leukemia (ALL) and 10.6 per million per year for acute myeloid leukemia (AML) (1). The rate of twin births in the United States is 24.6 per thousand live-born infants (2). If one monozygous twin develops leukemia, estimated probabilities of leukemia in the other twin range from 5 to 25% (3–5). Although leukemia in twins is rare, the molecular analysis of leukemia in monozygous twins is central to understanding the etiology and pathogenesis of leukemia in infants.
Rearrangements of the MLL gene at chromosome band 11q23 are the most common molecular abnormalities in leukemia in infants (6). Ford et al. (7) used Southern blot analysis to detect identical unique MLL gene rearrangements in ALLs in three pairs of infant twins. The karyotypes were not determined in one case and were 46,XY and 46,XX, t(11;19)(q23;p13) in the other two (7). Shortly afterwards, Mahmoud et al. (8) and Gill Super et al. (9) found identical, unique MLL gene rearrangements in ALLs of infant twins with the t(11;19)(q23;p13). The identical, clonal, nonconstitutional rearrangements suggested that the translocations occurred in utero and that there had been intraplacental metastasis from one twin to the other (7–9). Monozygous twins with congenital ALL and t(4;11)(q21;q23) have also been described (10).
Although MLL gene translocations involve about 30 different partner genes (9), the cytogenetic locations of the partner genes of MLL in leukemias of twins have been more restricted. No MLL rearrangement breakpoints have been cloned in leukemias of twins and detection of concordant rearrangements by Southern blot analysis has been limited to ALL (7–9). We adapted the panhandle PCR variant strategy (11) to clone the translocation breakpoint on the der(11) chromosome in a pair of infant twins with AML. Molecular cloning of MLL genomic breakpoints can be formidable because many breakpoint junctions are composed of known 5′ sequence from MLL, but 3′ sequence from uncharacterized intronic regions of known partner genes or from partner genes that have not yet been cloned. We formed the desired template with an intrastrand loop for panhandle variant PCR by ligating an oligonucleotide homologous to known 5′ MLL genomic sequence to the unknown partner DNA. With this methodology, we identified the human cell division cycle-related gene (hCDCrel) in the genomic region of deletion in DiGeorge and velocardiofacial syndromes at chromosome band 22q11.2, as a partner gene of MLL in the leukemias of the infant twins.
MATERIALS AND METHODS
The Institutional Review Board at the Children’s Hospital of Philadelphia approved the use of patient specimens as described below.
Detection of MLL Gene Rearrangements by Southern Blot Analysis.
Peripheral blood mononuclear cells or leukemic marrow cells were obtained at times of diagnosis from monozygous infant twins designated patients 68 and 72. Peripheral blood mononuclear cells from patient 72 also were obtained 7 weeks before the onset leukemia. High molecular weight genomic DNAs were isolated by ultracentrifugation on 4 M guanidine isothiocyanate/5.7 M CsCl gradients (12). BamHI-digested DNAs were hybridized with the B859 fragment of MLL cDNA, which contains exons 5–11 encompassing the breakpoint cluster region (bcr) and detects a germ-line BamHI fragment of 8.3 kb (13). The size of the rearrangement on the Southern blot representing the der(11) chromosome was the approximate size of the target sequence for panhandle variant PCR.
Panhandle Variant PCR Amplification of MLL Genomic Translocation Breakpoints.
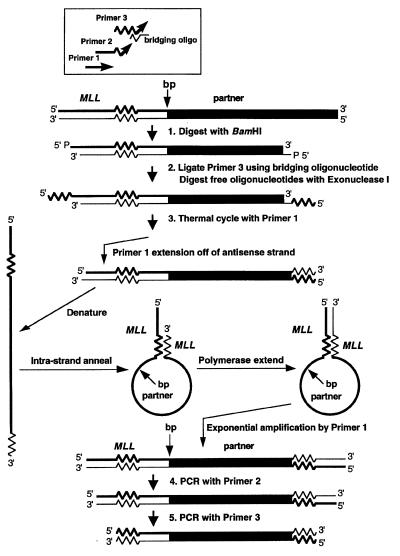
As shown in Fig. 1, we adapted panhandle variant PCR to amplify the breakpoint from a der(11) chromosome with known 5′ MLL sequence juxtaposed to 3′ unknown partner DNA.
Figure 1.
Schematic of template generation and amplification of genomic translocation breakpoint on a der(11) chromosome by panhandle variant PCR.
Step 1.
Genomic DNA was digested to completion with BamHI to create a restriction fragment with a 5′ overhang. Each 48-μl reaction contained 2.4 μg of genomic DNA, 30 units of BamHI, 1× buffer, and 1× BSA (New England Biolabs).
Step 2.
A single-stranded oligonucleotide that was homologous to known MLL sequence at positions 51–81 in exon 5 in the 5′ bcr (13) (primer 3) was ligated to the 5′ ends of the BamHI-digested DNA. The sequence was 5′-TCGAGGAAAAGAGTGAAGAAGGGAATGTCTC-3′. Primer 3 also later served as the nested primer in the last PCR (see below). A bridging oligonucleotide, the purpose of which was to position the 3′ end of primer 3 at the 5′ ends of the BamHI-digested DNA, directed the ligation. The 5′ end of the bridging oligonucleotide was complementary to the 4-base 5′ overhang of BamHI-digested DNA; the 3′ end was complementary to the 3′ end of the ligated oligonucleotide. The sequence was 5′-GATCGAGACATTCCCTTCT-3′.
The 24-μl ligation reaction mixture contained 0.05 μg of the BamHI-digested DNA, a 50-fold molar excess of primer 3 (96.2 ng), a 50-fold molar excess of the bridging oligonucleotide (59.1 ng), 1 Weiss unit of T4 DNA ligase and 1× ligase buffer (Boehringer Mannheim). Ligations were performed overnight at 17°C. BamHI-digested DNAs were added directly to ligation reaction mixtures without purification after the digestions. Excess BamHI-digested DNAs were reserved for use as unligated controls in panhandle variant PCRs. After the ligation, the bridging oligonucleotide and free primer 3 that remained unligated were eliminated by exonuclease I digestion. One microliter (20 units) of exonuclease I (Epicentre Technologies, Madison, WI) was added to each reaction mixture followed by incubation at 37°C for 30 min and heat inactivation at 75°C for 15 min (Fig. 1).
Step 3.
The antisense DNA strand (Fig. 1, lower strand, step 3) then contained the translocation breakpoint and unknown partner DNA bracketed with an inverted repeat of known MLL sequence. The purpose of step 3 was to complete formation of the template during thermal cycling and then to amplify the template with a single PCR primer (primer 1). Primer 1 annealed to the antisense DNA strand immediately 3′ to the complement of the ligated oligonucleotide. Primer 1 was homologous to positions 34–55 in MLL exon 5. The sequence was 5′-TCCTCCACGAAAGCCCGTCGAG-3′. Template-directed primer extension generated a new 5′–3′ strand with known MLL sequence at the 5′ end, the translocation breakpoint and unknown partner DNA, and a complement of 5′ MLL genomic sequence (i.e., the complement of the ligated oligonucleotide) at the 3′ end. This strand would become the template. Heat denaturation during thermal cycling made the DNA single-stranded. Intrastrand annealing of the 5′ MLL genomic sequence to its 3′ complement formed an intrastrand loop or pan-like structure, and template-directed polymerase extension of the recessed 3′ end completed formation of the handle. The intrastrand loop contained the translocation breakpoint and unknown partner DNA; the handle contained known 5′ sequence from MLL and a complement to that sequence. Thermal cycling with primer 1, which then annealed at both ends of the template, would amplify the target sequence.
Each 50-μl panhandle variant PCR mixture contained 20 ng of the digested, ligated, exonuclease I-treated DNA or unligated control DNA, 0.5 μl (1.75 units) of Taq/Pwo DNA polymerase mixture, all four dNTPs (each at 350 μM), 1× Expand buffer 1 (Expand long template PCR system, Boehringer Mannheim), and 12.5 pmol of primer 1. Primer 1 was added to reaction mixtures while they were preheated at 80°C to prevent nonspecific annealing and polymerization. After initial denaturation at 94°C for 1 min, 10 cycles at 94°C for 10 sec and 68°C for 7 min, and 20 cycles at 94°C for 10 sec and 68°C for 7 min (increment 20 sec per cycle) were used, followed by a final elongation at 68°C for 7 min. Panhandle variant PCR amplifications of DNA-negative control reactions also were included.
Steps 4 and 5.
Two sequential nested single-primer PCR amplifications using primer 2 and then primer 3 were then performed. The box at the top of Fig. 1 shows the relative positions of the primers with respect to each other and their orientations with respect to the genomic template. Primer 2 was homologous to positions 38–61 in MLL exon 5 (5′-CCACGAAAGCCCGTCGAGGAAAAG-3′). The location and sequence of primer 3, which also was the ligated oligonucleotide, were described above (see step 2).
The products of panhandle variant PCR were directly sequenced by automated methods or analyzed by sequencing after subcloning into pUC19 (GIBCO/BRL) by recombination PCR.
Subcloning by Recombination PCR.
Recombination PCR is based on the observation that DNA ends containing short regions of homology can undergo intra- and intermolecular recombination in vivo in Escherichia coli, and uses E. coli itself to mediate DNA recombination (14). First, 0.5 μg of pUC19 was linearized by digestion with 10 units of HindIII (GIBCO/BRL). Two nanograms of the digested plasmid template was amplified in a 50-μl PCR mixture containing 1.25 units of AmpliTaq DNA polymerase (Perkin–Elmer), 12.5 pmol of each primer, all four dNTPs (each at 200 μM), and 1× PCR buffer (Perkin–Elmer) to generate a linearized plasmid with ends complementary to the ends of the product of panhandle variant PCR that was to be inserted (14). The sequences of the forward and reverse primers used to amplify the HindIII-digested pUC19 were 5′-TCCCTTCTTCACTCTTTTCCTCGATGGCGTAATCATGGTCATAGC-3′ and 5′-TCCCTTCTTCACTCTTTTCCTCGACATGCCTGCAGGTCG- ACTCTAGAG-3′, respectively. After initial denaturation at 94°C for 1 min, 25 cycles at 94°C for 30 sec, 50°C for 30 sec, and 72°C for 2 min 42 sec were performed, followed by a final elongation at 72°C for 7 min.
The products from PCR amplification of the plasmid and from panhandle variant PCR were purified with a Geneclean III kit (Bio 101) according to the manufacturer’s instructions and resuspended in 10 μl of elution buffer provided in the kit. Purified PCR products from amplification of the plasmid (2.5 μl) and purified panhandle variant PCR products (2.5 μl) were combined and added to 50 μl of MAX efficiency DH5α competent cells (Life Technologies) to bring about in vivo recombination. The transformation procedure was exactly as described in the manufacturer’s instructions (Life Technologies), except that we plated the entire 1-ml reaction. Individual transformants were grown overnight in 4 ml of LB broth containing ampicillin (100 μg/ml).
We used PCR to identify recombinant plasmids containing products of panhandle variant PCR. Two microliters of the saturated 4-ml cultures were amplified in PCRs containing 0.5 μl (1.75 units) of Taq/Pwo DNA polymerase mixture, all four dNTPs (each at 350 μM), 1× Expand buffer 1, and 12.5 pmol of primer 3, which had been used in panhandle variant PCR. The PCR conditions were the same as those for panhandle variant PCR. Agarose gel electrophoresis of the products identified the transformants containing the recombinant plasmid DNA of interest, which then were grown in 25-ml cultures for plasmid preparation and automated sequencing.
Validation of MLL Genomic Translocation Breakpoint by Conventional PCR and Direct Genomic Sequencing.
To detect the translocation breakpoint by an independent method, we amplified fresh aliquots of genomic DNA from the leukemic cells of patients 68 and 72 with primers encompassing the translocation breakpoint, which were designed from sequences of the products of panhandle variant PCR. Forward and reverse primers derived from MLL and from the partner DNA were 5′-GCAGATGGAGTCCACAGGAT-3′ and 5′-CAAAGCCTTTCTTCACCGAC-3′, respectively, and would yield a 344-bp product. Fifty nanograms of genomic DNA were amplified in 100-μl reaction mixtures containing 2.5 units of AmpliTaq DNA polymerase, all four dNTPs (each at 250 μM), PCR buffer at 1× final concentration, and 5 pmol of each primer. After initial denaturation at 94°C for 9 min, 35 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min were used, followed by a final elongation at 72°C for 7 min. Products of reactions performed in quadruplicate for each patient were isolated from a 1.2% agarose gel with a Geneclean III kit (Bio 101). Approximately 40 ng of purified PCR products (10 ng/100 bp) were used for each sequencing reaction. Sequencing was performed in both directions by automated methods.
Reverse Transcriptase-PCR (RT–PCR) Analysis.
RT–PCR analysis was performed to evaluate whether the translocation generated an MLL-hCDCrel chimeric mRNA. We used the Superscript preamplification system and random hexamers for synthesis of cDNA from 1 μg of total RNA from the leukemic cells of patient 68 according to the manufacturer’s directions (GIBCO/BRL). The 100-μl RT–PCR mixtures contained 2 μl of random hexamer-primed cDNA, 2.5 units of AmpliTaq DNA polymerase, all four dNTPs (each at 200 μM), PCR buffer at 1× final concentration (Perkin–Elmer), and 100 pmol of each primer. The sequences of the forward primer from MLL exon 6 and the reverse primer from hCDCrel exon 3 were 5′-CGCCCAAGTATCCCTGTAAA-3′ and 5′-CAAAGCCTTTCTTCACCGAC-3′, respectively. After initial denaturation at 95°C for 2 min, 35 cycles at 95°C for 1 min, 55°C for 2 min, and 72°C for 1 min were used, followed by a final elongation at 72°C for 10 min. As the positive control, random hexamer-primed cDNA was amplified with β-actin primers, which would yield a 250-bp product (15). RNA-negative (dH2O) controls with and without reverse transcriptase were included for each primer set.
Ten microliters of the RT–PCR mixtures were electrophoresed in a 1.5% agarose gel to visualize the products. Products of four reactions were electrophoresed in 2% agarose and gel-purified with a Geneclean III kit (Bio 101) for automated sequencing. Twenty-five nanograms of purified RT–PCR products were used for direct automated sequencing with the same forward and reverse primers used for RT–PCR.
RESULTS
Case Histories.
Patient 68 presented at 11.5 months of age with fever, bruising, thrombocytopenia, a white blood cell count of 228 × 109 per liter and leukemia in the central nervous system. The bone marrow was replaced by blasts of French–American–British (FAB) M2 morphology that expressed CD33 and CD45. The G banded karyotype was 46,XX, t(11;22)(q23;q11.2)[15], and fluorescence in situ hybridization (FISH) analysis with an MLL-specific probe (Oncor) showed hybridization with the normal chromosome 11 and split signals on the der(11) chromosome and chromosome 22. The patient was a monozygous twin. Seven weeks later, the twin of patient 68, designated patient 72, was diagnosed with AML. Patient 72 presented with bruising and a white blood cell count of 20.6 × 109 per liter. There were 67% abnormal blasts of FAB M1 morphology on marrow differential. The blasts expressed HLA-DR, CD13, and CD33. The G banded karyotype of the diagnostic marrow was 46,XX[5]/46,XX, t(11;22)(q23;q11) [15]. On FISH analysis, the MLL-specific probe hybridized with the normal and der(11) chromosomes and chromosome 22, suggesting that the t(11;22)(q23;q11.2) disrupted MLL.
Southern Blot Analysis Identifies Identical MLL Gene Rearrangements in Infant Twins.
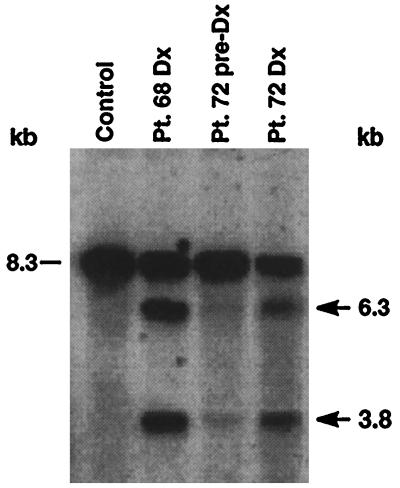
We examined peripheral blood mononuclear cells from patient 68 and leukemic marrow cells from patient 72 for MLL gene rearrangement at times of diagnosis. In both cases, the B859 probe showed the 8.3-kb germ-line band and identical rearranged BamHI restriction fragments 3.8 kb and 6.3 kb, indicating chromosomal translocation (Fig. 2). Prediagnosis peripheral blood mononuclear cells were obtained from patient 72 at time of diagnosis of leukemia in her twin. On 2-week exposure, Southern blot analysis of the prediagnosis specimen detected both the germ-line band and faint 3.8-kb and 6.3-kb rearrangements (Fig. 2), showing presence of cells with the translocation before clinical leukemia appeared. The intensity of the rearrangements relative to the germ-line band increased from prediagnosis to the time of diagnosis (Fig. 2).
Figure 2.
Identification of rearrangements within the MLL bcr by Southern blot in leukemias of infant twins 68 and 72 and in peripheral blood mononuclear cells (PBMCs) of twin 72 7 weeks before the onset of clinical leukemia. Intensity of identical rearrangements in preleukemic PBMCs of twin 72 relative to the germ-line band suggests that the translocation was not constitutional. BamHI-digested DNAs were hybridized with B859 human MLL cDNA probe spanning exons 5–11 (13). The 8.3-kb fragment is from the unrearranged allele (dash marks). Control DNA from PBMCs of a normal individual shows the germ-line pattern. Arrows show rearrangements.
Panhandle PCR Variant Amplifies MLL Genomic Translocation Breakpoint.
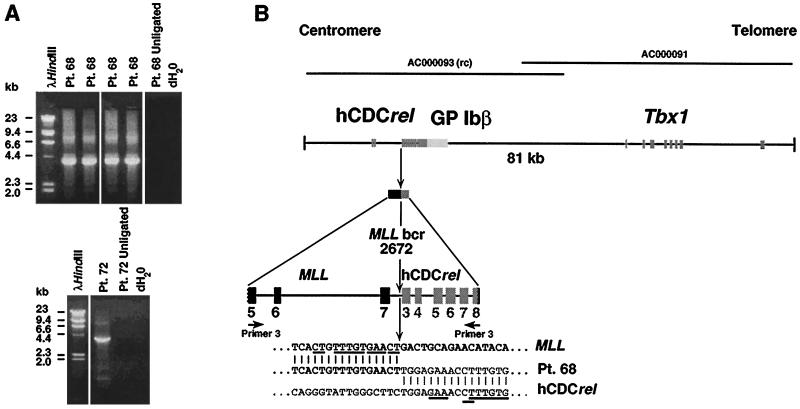
We first used panhandle variant PCR to clone the MLL genomic breakpoint on the der(11) chromosome in the leukemia of patient 68. Six reactions yielded products ∼3.9 kb (Fig. 3), indicating that the 3.8-kb rearrangement on Southern blot analysis was from the der(11) chromosome. There was sufficient material for direct genomic sequencing of the breakpoint junction without subcloning of the products. To confirm the translocation breakpoint and obtain additional information on the partner DNA, we performed recombination PCR using the products of one panhandle variant PCR reaction. Six of eight recombination PCR-generated subclones contained the desired 3.9-kb insert, and we sequenced two subclones in entirety.
Figure 3.
(A) Panhandle variant PCR products from der (11) chromosome of t(11;22)(q23;q11.2) in AMLs of infant twins 68 and 72. (B) Summary of t(11;22) breakpoint junction and partner DNA identified by directly sequencing panhandle variant PCR products from AML of patient 68 and sequencing products of panhandle variant PCR from leukemias of patients 68 and 72 subcloned by recombination PCR. Comparison with normal sequence identified the breakpoint at nucleotide 2,672 in MLL intron 7 (bold arrow). Primer 3 serves as the primer in the final PCR (see Fig. 1). The 5′ 2,622 bp include primer 3 and MLL bcr sequence to breakpoint at position 2,672 in intron 7. The 1,240 bp of 3′ sequence are partner DNA, and the most 3′ 31 bp of sequence are the complement of the ligated oligonucleotide (i.e., the complement of primer 3). Underlines show short segments of homology between MLL and hCDCrel at respective breakpoint junctions. Positions of relevant cosmid clones in chromosome band 22q11.2 deletion syndromes are shown at top (GenBank accession nos. AC000093 and AC000091). rc indicates that GenBank entry is reverse complement of hCDCrel.
The t(11;22)(q23;q11.2) Fuses MLL with hCDCrel, a Cell Division Cycle Gene in the Genomic Region of Deletion in DiGeorge and Velocardiofacial Syndromes.
Direct automated sequencing identified the genomic breakpoint at nucleotide 2,672 in MLL intron 7 and partial sequence of the partner DNA (Fig. 3). Sequencing of the subcloned products confirmed the translocation breakpoint and yielded additional sequence of the partner gene. A blast search against the nucleotide database indicated that the sequence of the partner DNA was identical to an intronic region of the hCDCrel (human cell division cycle related) gene (GenBank accession no. AC000093), which is a member of a gene family involved in cell division cycle that includes the Drosophila peanut-like protein 1 gene (16). The hCDCrel gene maps to the central portion of a 1.3-megabase sequence contig on chromosome band 22q11.2 that is commonly deleted in DiGeorge and velocardiofacial syndromes (B. R. Roe, G. Zhang, B. S. Emanuel, and M.L.B., unpublished results).
Comparison of the cDNA to the genomic sequence indicates that hCDCrel contains 11 exons that span approximately 9 kb. The genomic breakpoint in hCDCrel in the leukemia of patient 68 was in intron 2 at nucleotide 26,510 relative to cosmid carlaa, although the orientation of the GenBank entry (accession no. AC000093) is in opposite orientation to the ORF of the cDNA. Consistent with the size of the rearrangement on Southern blot and with the size of the panhandle variant PCR product, the next BamHI site in the hCDCrel gene 3′ of the breakpoint is located in exon 8 at position 25,275 of cosmid carlaa (GenBank accession no. AC000093). Thus, 2,622 bp of sequence in the panhandle variant PCR product were from MLL and 1,240 bp were from hCDCrel. Fig. 3 summarizes the panhandle variant PCR products from the der(11) chromosome in relation to the chromosome band 22q11.2 genomic region. The breakpoint region in hCDCrel was rich in simple repeats and low complexity repeats. Both MLL and hCDCrel contained homologous CT, TTTGTG, and GAA sequences (Fig. 3, underlined) within a few base pairs of their respective breakpoints.
Independent Confirmation of MLL Genomic Breakpoint in Leukemia of Patient 68.
Fresh aliquots of genomic DNA from the leukemic cells of patient 68 were amplified with primers encompassing the translocation breakpoint, which were designed from sequences of the products of panhandle variant PCR. Four reactions gave the predicted 344-bp product (data not shown). Direct sequencing was performed on the products of two reactions and verified the breakpoint.
RT–PCR Analysis Shows MLL-hCDCrel Chimeric mRNA.
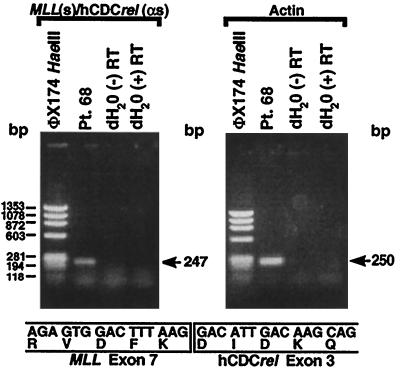
We synthesized randomly primed cDNA from the leukemic cells of patient 68 and performed RT-PCR to evaluate whether the translocation produced a fusion mRNA. The RT–PCR with sense and antisense primers from MLL exon 6 and hCDCrel exon 3, respectively, gave the predicted 247-bp product (Fig. 4). Direct sequencing showed an in-frame fusion of MLL exon 7 to hCDCrel exon 3 at position 142 of the 2,032-bp full-length hCDCrel cDNA (GenBank accession no. U74628) (16) (Fig. 4).
Figure 4.
RT–PCR analysis of total RNA from leukemic cells of patient 68 indicating MLL-hCDCrel chimeric mRNA. RT–PCRs with sense and antisense primers from MLL exon 6 and hCDCrel exon 3, respectively, and randomly primed cDNA template gave a single 247-bp product (Upper). Direct sequencing of the products of four reactions revealed an in-frame fusion of MLL exon 7 to hCDCrel exon 3 at position 142 of the 2,032-bp full-length hCDCrel cDNA (GenBank accession no. U74628) (16) (Lower). Reactions using β-actin primers and RNA negative control (distilled H2O) are also shown (Upper).
Panhandle Variant PCR Amplifies Identical MLL Genomic Breakpoint in AML of Patient 72.
We used panhandle variant PCR to isolate the translocation breakpoint junction in the AML of patient 72, the twin of patient 68. The products of one reaction (Fig. 3A) were subcloned by recombination PCR. The desired 3.9-kb insert was present in six of seven subclones and two positive subclones were sequenced in entirety. The sequence showed the same MLL intron 7 breakpoint at nucleotide 2,672 and the same hCDCrel partner DNA as in the leukemia of patient 68 (Fig. 3B). For independent confirmation, we amplified fresh aliquots of genomic DNA from the leukemic cells with primers encompassing the breakpoint junction. Four PCRs gave the predicted 344-bp product. We directly sequenced the products of two of the reactions, which verified the translocation breakpoint.
DISCUSSION
By using panhandle variant PCR, we determined that the t(11;22)(q23;q11.2) in concordant AMLs of monozygous infant twins was the result of fusion of MLL with hCDCrel and identified a partner gene of MLL at chromosome band 22q11.2. The results were validated independently by direct genomic sequencing of products of conventional PCR and by RT–PCR analysis. The genomic sequence of the partner DNA at the breakpoint junction of the der(11) chromosome was identical to intron 2 of the hCDCrel gene at chromosome band 22q11.2 (GenBank accession no. AC000093). hCDCrel is a member of a gene family involved in cell division cycle that includes the Drosophila peanut-like protein 1 gene (16). The hCDCrel gene contains 11 exons that span approximately 9 kb (Fig. 3) and yields two transcripts of ∼2.5 and ∼3.5 kb (16). The smaller transcript terminates at an imperfect polyadenylylation site, and the longer transcript is produced by alternative use of the polyadenylylation site of glycoprotein (GP) Ibβ, the adjacent 3′ gene (16). The putative protein product of hCDCrel is a GTP-binding protein (16).
The hCDCrel gene is in the central portion of a 1.3-megabase sequence contig that is part of the region on chromosome band 22q11.2 commonly deleted in both DiGeorge and velocardiofacial syndromes (B. R. Roe, G. Zhang, B. S. Emanuel, and M.L.B., unpublished results). DiGeorge syndrome is a constitutional disorder characterized by cardiac anomalies, thymic and parathyroid hypoplasia and dysmorphic craniofacial features, whereas the major features of velocardiofacial syndrome are palatal and cardiac defects, facial dysmorphia, and learning disabilities (17). hCDCrel is the second partner gene of MLL located in a region of the genome involved in both leukemia and a constitutional disorder. CBP is the partner gene of MLL in myelodysplastic syndrome with the t(11;16)(q23;p13.3) (18, 19) and the partner gene of MOZ in AML with the t(8;16)(p11;p13) (20). CBP encodes a histone acetyltransferase that functions as a transcriptional coactivator (20). The Rubinstein–Taybi syndrome, a constitutional disorder that includes mental retardation, dysmorphic facial features, and broad thumbs and toes (21), is characterized by chromosomal translocations, microdeletions and point mutations of CBP (22).
We detected short homologous sequences 2–6 bp long at the breakpoint junctions in MLL and hCDCrel (Fig. 3). Similarly, we found short segments of homology between MLL and AF-4 or AF-9 at t(4;11) and t(9;11) breakpoint junctions (ref. 23; C.A.F., M. Hosler, D. Slater, M.D.M., T.M.W., P. Nowell, J. Hoxie, B. Lange, and E.R., unpublished data). On the basis of these findings, we proposed base pairing of homologous DNA ends of MLL and partner gene as one step in the translocation process (C.A.F., M. Hosler, D. Slater, M.D.M., T.M.W., P. Nowell, J. Hoxie, B. Lange, and E.F.R., unpublished data). The cloning of a constitutional balanced t(2;22)(q14;q11.21) associated with DiGeorge syndrome identified several small segments of nucleotides (∼6 bp) repeated on chromosomes 2 and 22 (24), suggesting that the same phenomenon may occur in constitutional and somatic translocations.
The t(11;22)(q23;q11.2) that fused MLL with hCDCrel in the leukemias of infant twins is distinct from the constitutional t(11;22)(q23;q11), which is the most frequent, recurrent, non-Robertsonian translocation (25). In the constitutional t(11;22), the phenotype is normal, the translocation is present in all cells, and the chromosome 11 breakpoints are proximal to leukemia-associated breakpoints (26). Although mosaicism has not been excluded, two lines of evidence argue that the t(11;22)(q23;q11.2) that we observed was not constitutional. In patient 72, the intensity of MLL gene rearrangements relative to the germ-line band on Southern blot analysis progressively increased from prediagnosis to the time of diagnosis and the karyotype of the diagnostic marrow in 5 of 20 cells was normal.
Concordance of the unique, clonal, nonconstitutional MLL gene rearrangements suggests that the t(11;22) occurred in utero and that there was metastasis from one twin to the other via the placenta (7). The ages of the twins at diagnosis of leukemia were similar, 11.5 months and 13 months. Within pairs of twins, the ages at onset of leukemia have generally been concordant (3, 7–10), suggesting similar times of latency before disease appears. The delineation of MLL gene rearrangements in twins as in utero events complements research efforts on prenatal exposures to environmental toxins as etiologic factors in leukemia in infants. One line of investigation involves maternal dietary DNA topoisomerase II inhibitors, because leukemias in infants resemble treatment-related leukemias linked to chemotherapy that targets DNA topoisomerase II (27). Moreover, the latency to onset of disease suggests secondary alterations in addition to the translocations (7), but the influence of various translocation partners on sufficiency of MLL gene translocations for full leukemogenesis has not been addressed.
By using panhandle variant PCR, we amplified a 3.9-kb product, identified the t(11;22) translocation breakpoint and distinguished hCDCrel as a partner gene of MLL in AML of infant twins. The method was devised to simplify the PCR-based cloning of genomic DNAs with unknown 3′ flanking sequences (11), precisely the situation with many MLL genomic breakpoints. Beyond the finding of a partner gene of MLL, this work introduces a particular PCR technology that expedites translocation breakpoint cloning. We recently used the original panhandle PCR as another strategy (15, 28, 29). Increased use of both methods should test whether one or the other is more advantageous in specific situations. For retreival and detection of the MLL genomic breakpoint, we used recombination PCR, which uses E. coli itself to mediate DNA recombination (14) and obviates the ligation step in subcloning.
Including hCDCrel, 13 partner genes of MLL have been cloned to date (18, 19, 30–40). In addition, MLL may fuse with self in partial tandem duplications (28, 41–44). The joining of the MLL bcr and several different partner genes renders PCR-based cloning of the breakpoints difficult. Panhandle variant PCR offers another strategy to surmount this challenge. Identification of a genomic region at chromosome band 22q11.2 involved in AML and in the constitutional DiGeorge and velocardiofacial syndromes also is of interest.
Acknowledgments
C.A.F. supported by American Cancer Society Grant DHP143, National Institutes of Health Grant 1R29CA66140–03, Leukemia Society of America Scholar Award (1996–2001), Children’s Cancer Group, National Leukemia Research Association, and Children’s Hospital of Philadelphia High Risk High Impact Grant. M.L.B. was supported by National Institutes of Health Grants HL51533 and DC02027.
Note Added in Proof:
Since the original submission, we performed PCR analysis on buccal mucosa genomic DNA from patient 72 by using clonotypic primers that would amplify the t(11;22) translocation breakpoint. No product was obtained, indicating that there was no evidence of mosaicism and confirming that the translocation was not constitutional.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: AML, acute myeloid leukemia; hCDCrel, human cell division cycle-related gene; RT–PCR, reverse transcriptase-PCR; ALL, acute lymphoblastic leukemia.
References
- 1.Gurney J G, Severson R K, Davis S, Robison L L. Cancer. 1995;75:2186–2195. doi: 10.1002/1097-0142(19950415)75:8<2186::aid-cncr2820750825>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 2.CDC. Morb Mortal Wkly Rep. 1997;46:121–125. [PubMed] [Google Scholar]
- 3.Clarkson B D, Boyse E A. Lancet. 1971;1:699–701. doi: 10.1016/s0140-6736(71)92705-x. [DOI] [PubMed] [Google Scholar]
- 4.Zueler W W, Cox D E. Semin Hematol. 1969;6:228–249. [PubMed] [Google Scholar]
- 5.Buckley J, Buckley C, Breslow N, Draper G, Roberson P, Mack T. Med Pediatr Oncol. 1996;26:223–229. doi: 10.1002/(SICI)1096-911X(199604)26:4<223::AID-MPO1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 6.Pui C-H, Kane J R, Crist W M. Leukemia. 1995;9:762–769. [PubMed] [Google Scholar]
- 7.Ford A M, Ridge S A, Cabrera M E, Mahmoud H, Steel C M, Chan L C, Greaves M. Nature (London) 1993;363:358–360. doi: 10.1038/363358a0. [DOI] [PubMed] [Google Scholar]
- 8.Mahmoud H H, Ridge S A, Behm F G, Pui C-H, Ford A M, Raimondi S C, Greaves M F. Med Pediatr Oncol. 1995;24:77–81. doi: 10.1002/mpo.2950240203. [DOI] [PubMed] [Google Scholar]
- 9.Gill Super H J, Rothberg P G, Kobayashi H, Freeman A I, Diaz M O, Rowley J D. Blood. 1994;83:641–644. [PubMed] [Google Scholar]
- 10.Bayar E, Kurczynski T W, Robinson M G, Tyrkus M, Al Saadi A. Cancer Genet Cytogenet. 1996;89:177–180. doi: 10.1016/0165-4608(94)00293-2. [DOI] [PubMed] [Google Scholar]
- 11.Jones D, Winistorfer S. Biotechniques. 1997;23:132–138. doi: 10.2144/97231rr01. [DOI] [PubMed] [Google Scholar]
- 12.Felix C A, Poplack D G, Reaman G H, Steinberg S M, Cole D E, Taylor B J, Begley C G, Kirsch I R. J Clin Oncol. 1990;8:431–442. doi: 10.1200/JCO.1990.8.3.431. [DOI] [PubMed] [Google Scholar]
- 13.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce C M, Canaani E. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 14.Jones D, Howard B. Biotechniques. 1991;10:62–66. [PubMed] [Google Scholar]
- 15.Felix C, Kim C, Megonigal M, Slater D, Jones D, Spinner N, Stump T, Hosler M, Nowell P, Lange B, Rappaport E. Blood. 1997;90:4679–4686. [PubMed] [Google Scholar]
- 16.Zieger B, Hashimoto Y, Ware J. J Clin Invest. 1997;99:520–525. doi: 10.1172/JCI119188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Budarf M, Emanuel B. Hum Mol Genet. 1997;6:1657–1665. doi: 10.1093/hmg/6.10.1657. [DOI] [PubMed] [Google Scholar]
- 18.Sobulo O M, Borrow J, Tomek R, Reshmi S, Harden A, Schlegelberger B, Housman D, Doggett N A, Rowley J D, Zeleznik-Le N J. Proc Natl Acad Sci USA. 1997;94:8732–8737. doi: 10.1073/pnas.94.16.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taki T, Sako M, Tsuchida M, Hayashi Y. Blood. 1997;89:3945–3950. [PubMed] [Google Scholar]
- 20.Borrow J, Stanton V P, Jr, Andresen M, Becher R, Behm F G, Chaganti R S K, Civin C I, Disteche C, Dube I, Frischauf A M, et al. Nat Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 21.Rubinstein J H, Taybi H. Am J Dis Child. 1963;105:588–608. doi: 10.1001/archpedi.1963.02080040590010. [DOI] [PubMed] [Google Scholar]
- 22.Petrij F, Giles R, Dauwerse H, Saris J, Hennekam R, Masuno M, Tommerup N, van Ommen G-J, Goodman R, Peters D, Breuning M. Nature (London) 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 23.Felix C A, Lange B J, Hosler M R, Fertala J, Bjornsti M-A. Cancer Res. 1995;55:4287–4292. [PubMed] [Google Scholar]
- 24.Budarf M, Collins J, Gong W, Roe B, Wang Z, Bailey L, Sellinger B, Michaud D, Driscoll D, Emanuel B. Nat Genet. 1995;10:269–278. doi: 10.1038/ng0795-269. [DOI] [PubMed] [Google Scholar]
- 25.Budarf M, Sellinger B, Griffin C, Emanuel B. Am J Hum Genet. 1989;45:128–139. [PMC free article] [PubMed] [Google Scholar]
- 26.Eleventh International Workshop on Human Gene Mapping (1991) 58, 469.
- 27.Ross J A, Potter J D, Reaman G H, Pendergrass T W, Robison L L. Cancer Causes and Control. 1996;7:581–590. doi: 10.1007/BF00051700. [DOI] [PubMed] [Google Scholar]
- 28.Megonigal M D, Rappaport E F, Jones D H, Kim C S, Nowell P C, Lange B J, Felix C A. Proc Natl Acad Sci USA. 1997;94:11583–11588. doi: 10.1073/pnas.94.21.11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Felix, C. & Jones, D. (1998) Leukemia, in press. [DOI] [PubMed]
- 30.Bernard O, Mauchauffe M, Mecucci C, Van Den Berghe H, Berger R. Oncogene. 1994;9:1039–1045. [PubMed] [Google Scholar]
- 31.Nakamura T, Alder H, Gu Y, Prasad R, Canaani O, Kamada N, Gale R P, Lange B, Crist W M, Nowell P C, et al. Proc Natl Acad Sci USA. 1993;90:4631–4635. doi: 10.1073/pnas.90.10.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rubnitz J E, Morrissey J, Savage P A, Cleary M L. Blood. 1994;84:1747–1752. [PubMed] [Google Scholar]
- 33.Prasad R, Gu Y, Alder H, Nakamura T, Canaani O, Saito H, Huebner K, Gale R P, Nowell P C, Kuriyama K, et al. Cancer Res. 1993;53:5624–5628. [PubMed] [Google Scholar]
- 34.Thirman M J, Levitan D A, Kobayashi H, Simon M C, Rowley J D. Proc Natl Acad Sci USA. 1994;91:12110–12114. doi: 10.1073/pnas.91.25.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tse W, Zhu W, Chen H S, Cohen A. Blood. 1995;85:650–656. [PubMed] [Google Scholar]
- 36.Chaplin T, Bernard O, Beverloo H B, Saha V, Hagemeijer A, Berger R, Young B D. Blood. 1995;86:2073–2076. [PubMed] [Google Scholar]
- 37.Chaplin T, Ayton P, Bernard O A, Saha V, Valle V D, Hillion J, Gregorini A, Lillington D, Berger R, Young B D. Blood. 1995;85:1435–1441. [PubMed] [Google Scholar]
- 38.Parry P, Wei Y, Evans G. Genes Chromosomes Cancer. 1994;11:79–84. doi: 10.1002/gcc.2870110203. [DOI] [PubMed] [Google Scholar]
- 39.Hillion J, Le Coniat M, Jonveaux P, Berger R, Bernard O. Blood. 1997;9:3714–3719. [PubMed] [Google Scholar]
- 40.So C, Caldas C, Liu M-M, Chen S-J, Huang Q-H, Gu L-J, Sham M, Wiedemann L, Chan L. Proc Natl Acad Sci USA. 1997;99:2563–2568. doi: 10.1073/pnas.94.6.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schichman S A, Caligiuri M A, Gu Y, Strout M P, Canaani E, Bloomfield C D, Croce C M. Proc Natl Acad Sci. 1994;91:6236–6239. doi: 10.1073/pnas.91.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caligiuri M A, Strout M P, Schichman S A, Mrozek K, Arthur D C, Herzig G P, Baer M R, Schiffer C A, Heinonen K, Knuutila S, et al. Cancer Res. 1996;56:1418–1425. [PubMed] [Google Scholar]
- 43.Yamamoto K, Hamaguchi H, Nagata K, Kobayashi M, Taniwaki M. Am J Hematol. 1997;55:41–45. doi: 10.1002/(sici)1096-8652(199705)55:1<41::aid-ajh8>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 44.Yu M, Honoki K, Andersen J, Paietta E, Nam D, Yunis J. Leukemia. 1996;10:774–780. [PubMed] [Google Scholar]