Abstract
Myosin-Va transports intracellular cargos along actin filaments in cells1.
This processive two-headed motor takes multiple 36-nm steps in which the two heads swing forward alternately towards the barbed end of actin driven by ATP hydrolysis2. The ability of myosin-Va to move processively is a function of its long lever arm, the high duty ratio of its kinetic cycle and the gating of the kinetics between the two heads such that ADP release from the lead head is greatly retarded3-10. Mechanical studies at the multiple and single molecule level suggest that there is tight coupling (i.e. one ATP is hydrolyzed per power stroke), but this has not been directly demonstrated4,5,11. We therefore investigated the coordination between the ATPase mechanism of the two heads of myosin-Va and directly visualized the binding and dissociation of single fluorescently-labelled nucleotide molecules while simultaneously observing the stepping motion of the fluorescently labelled myosin-V as it moves along an actin filament. Here we show that preferential ADP dissociation from the trail head of myosin Va is followed by ATP binding and a synchronous 36-nm step. Even at low ATP concentrations, the myosin-V molecule retains at least one nucleotide (ADP in the lead head position) while moving. Thus we directly demonstrate tight coupling between myosin Va movement and the binding and dissociation of nucleotide by simultaneously imaging with nanometer precision.
The ability to visualize the binding of fluorescent nucleotides to myosin in the light microscope has been limited by technical problems such as the nonspecific binding of the fluorescent nucleotde to the coverslip, low quantum yield and rapid photobleaching. This has limited the the maximum nucleotide concentration that could be used with analogs such as Cy3ATP to less than 100 nM12-15. To overcome these problems, we utilized a fluorescent ATP analog, (3′-(7-diethyaminocoumarin-3-carbonylamino)-3′-deoxyadenosine-5′-triphosphate (deac-aminoATP) whose fluorescence emission increases ∼25-fold (Supplementary Fig.1 online) when bound to a heavy meromyosin-like fragment of myosin Va (MyoV-HMM) in solution16,17. The kinetic mechanism of MyoV-HMM using deac-aminoATP as a substrate has been thoroughly studied including the extent of gating that occurs between the two heads during movment10,17. In brief, deac-aminoATP binds 3 fold faster to MyoV-HMM than ATP does and deac-aminoADP dissociates 10-20-fold slower than ADP17. When MyoV-HMM is bound to actin by both heads, the release of deac-aminoADP from the lead head is decreased about 30-fold compared to the unstrained rate10. The processive run length of MyoV-HMM on actin using deac-aminoATP as a substrate is shorter (1050±80 nm) than when using ATP alone (1950 ±160 nm)(Supplementary Fig. 2a). The maximal velocity of movement on actin at saturating deac-aminoATP is 120 nms-1, approximately 8-10 fold less than observed with ATP10 (Supplementary Fig.2b).
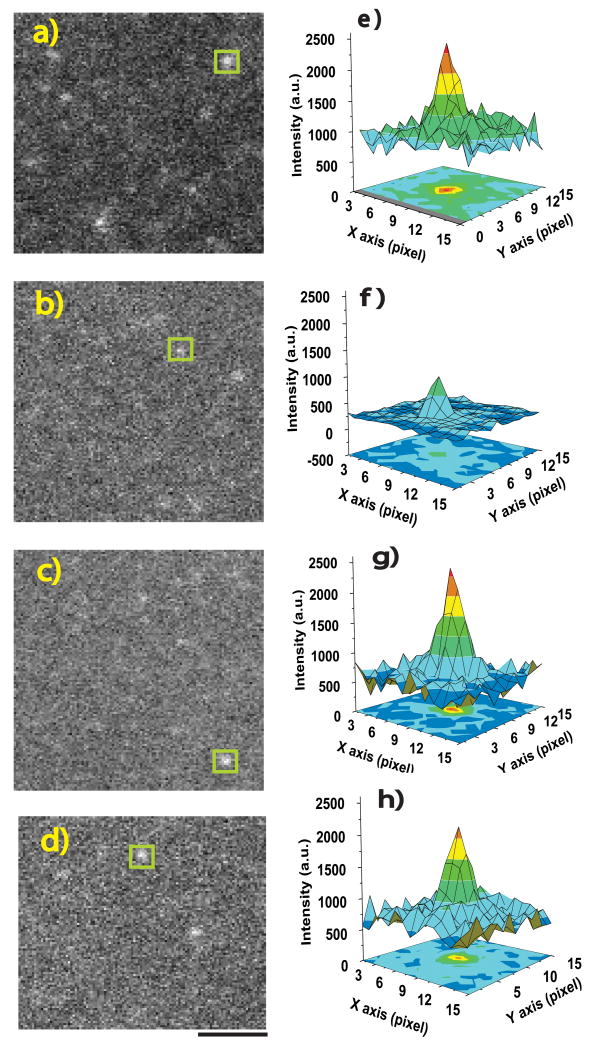
Deac-aminoADP that was nonspecifically bound to a coverslip surface in the absence of MyoV-HMM was visualized using an electron multiplying charged coupled device (EMCCD) camera at a camera gain level of 1000 (scale for gain is 0-1000)(Fig.1a and e). The gain on the camera chip was then reduced to 400, at which the intensity of the non-specifically bound deac-aminoADP spots was considerably reduced (Fig.1b and f). However, at the same gain (400) and collection of 330 ms, deac-aminoADP that was bound to MyoV-HMM on the cover slip (Fig.1c and g) has a sufficiently high intensity (>10,000 photons) to fit the point spread function of a single spot and so determine its precise nanometer localization18 (Supplementary Fig.3). At the single molecule level, we find a 4-fold enhancement of the fluorescent intensity of deac-aminoADP upon binding to MyoV-HMM.
Figure 1. Imaging deac-aminoADP and Alexa-Fluor-568-MyoV-HMM.
a-d TIRF-microscopic images (110 x 100 pixels) are shown. Deac-aminoADP was imaged at 442 nm (a-c); Alexa-Fluor-568-MyoV-HMM was imaged at 568 nm (d). Two-dimensional intensity profiles from each white square in a-d are shown in e-h.a,e, Deac-aminoADP bound directly on the coverslip, with maximum camera gain (1000). b,f, Deac-aminoADP bound directly on the coverslip surface at a gain of 400 on camera. c,g, Deac-aminoADP bound to MyoV-HMM at a gain of 400. d,h, Alexa-Fluor-568- MyoV-HMM bound to surface at a gain of 400. All data were taken with an iXon+ camera (DV897, Andor technology) with at 10 MHz readout at a constant laser power. The background level was fixed at about 750 (arbitrary units, a.u..) intensity. Scale bar, 2 μm
We exchanged Alexa-Fluor-568-labelled calmodulins for the endogenous calmodulin bound to the neck region of MyoV-HMM. On average each calmodulin contained 1.8 Alexa-Fluor-568 moities and three Alexa-Fluor-568-labelled calmodulins were exchanged per MyoV-HMM making it much brighter than either myosin fused to GFP molecules or containing a single Cy3 or rhodamine-labelled calmodulin that were previously used for single molecule studies3,18. Similar estimates for labeling ratios were obtained using spectrophotometric methods in solution or by examining the photobleaching kinetics of the molecules in the microscope (Supplementary Fig.4). This allows the Alexa-Fluor-568-MyoV-HMM to be as bright as the deac-aminonucleotides and permits the same camera and camera settings to be used to image both (Fig.1d and h; Supplementary Fig.3).
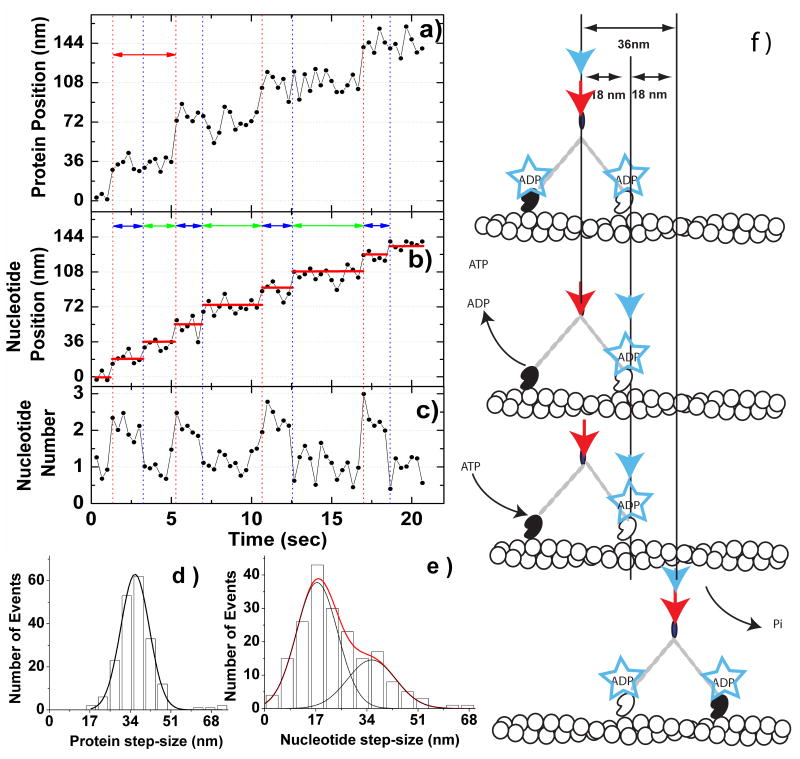
We simultaneously visualized Alexa-Fluor-568-MyoV-HMM and deac-aminonucleotide during processive movement on actin filaments in vitro (Fig.2a and b: Supplementary Fig.5: and Supplementary Movie). The Alexa-Fluor-568-MyoV-HMM and the deac-aminonucleotide fluorescence moved in the same direction at the same rate and on the same actin filaments (Fig.2a and b; see Supplemental Figs 5 and 6). The fluorescence signal from Alexa-Fluor-568-MyoV-HMM moved in 36 nm steps as would be expected from a molecule, in which both heads were labelled (Fig.2a), albeit there is the possibility of minor differences between the alternating steps sizes due to unevenness in the labeling of the two heads (see Supplementary Fig.7 for an example of “limping” movement). The deac-aminonucleotide moved with 18 nm steps. One step occurred simultaneously with the MyoV-HMM step, whereas the other step occurred during a dwell in the MyoV-HMM movement (Fig.2b). These observations from a single trace are reinforced by examining histograms of the MyoV-HMM step-size (which show a peak of 36±7 nm; Fig.2d), and of the deac-aminonucleotide step-size (which shows two peaks of 18±7 nm and 36±9 nm;Fig.2e). The larger, 36 nm values for deac-aminonucleotide movement are expected to result of cases whene two 18-nm movements occurred without a discernable dwell between them. This is calculated to occur in 22-37% of the time (1 − e-kt) on the basis of the deac-aminonucleotide association and dissociation rate constants measured in Fig.3 and the 330 ms data acquisition time. The intensity of the deac-aminonucleotide signal integrated from a 12 x 12 pixel (i.e. 840x840 nm2) area surrounding the molecule at each frame showed a bimodal distribution where one peak contained a factor of two more photons per frame than the other (Supplementary Fig.8). The photon count in the smaller peak represents one deac-aminonucleotide per MyoV-HMM, whereas that in the other represents two per MyoV-HMM. Note that this nucleotide has similar fluorescence intensity when bound as MyoV-HMM-ADP, MyoV-HMM-ADP-Pi or MyoV-HMM–ATP, and thus, we cannot discriminate between different nucleotide states of a single head by intensity17. Using this criterion, the normalized intensity of the deac-aminonucleotide signal was also plotted as a function of time (Fig.2c) and was shown to change from a value of one to two during each MyoV-HMM step and then decrease from a value of two to one during the MyoV-HMM dwell period.
Figure 2. Correlation between the movement of MyoV-HMM and the binding/dissociation of deac-aminonucleotide.
Images of Alexa-Fluor-568-MyoV-HMM and deac-aminoATP fluorescence were acquired simultaneously with a Dual-View system. The excitation/emission wavelengths of Alexa Fluor 568 are distinct from those of deac-aminoATP, allowing simultaneous visualization of the MyoV-HMM and the nucleotide. The photons from the spots were acquired in 330 ms integrations and the point spread function from each spot was fit with a two-dimensional Gaussian to determine the location of the fluorophor(es) at each time point18. a, The protein fluorescence data shows ∼36 nm steps, which are marked by red vertical dotted lines at 200 nM deac-aminoATP. Red double-ended arrows delineate the dwell time of a step. b, The deac-aminonucleotide stepping events are marked by alternating red and blue vertical dotted lines. Red vertical dotted lines are steps of both MyoV-HMM and deac-aminonucleotide, whereas blue vertical dotted lines show only stepping of deac-aminonucleotide. Individual spots move in a stepwise manner in the same direction as the MyoV-HMM. Dwell times are marked by blue and green double arrows. Red horizontal lines mark the average position of the spot during a pause. c, Normalized intensity of the deac-aminonucleotide fluorescence. d, Step-size histogram for the movement of Alexa-Fluor-568-MyoV-HMM (n = 145 steps, 38 MyoV-HMM molecules). The curve represents the fit to a Gaussian, (mean ± S.D.: 36.3 ± 7.2 nm.) e, Step-size histogram for the movement of the deac-aminonucleotide (n = 267 steps, 38 MyoV-HMM molecules). The red curve represents the fit to the sum of two Gaussians (shown individually in black lines) (mean ± S.D: 17.5 ± 7.1 nm, 36.0 ± 8.6 nm). F, Model for correlation of movement of Alexa-Fluor-568-MyoV-HMM and deac-aminonucleotide binding and dissociation. The red and blue arrows show the position of the centroid of the Alexa-Fluor-568-MyoV-HMM and of the deac-aminonucleotide fluorescence, respectively. See text for description of the model.
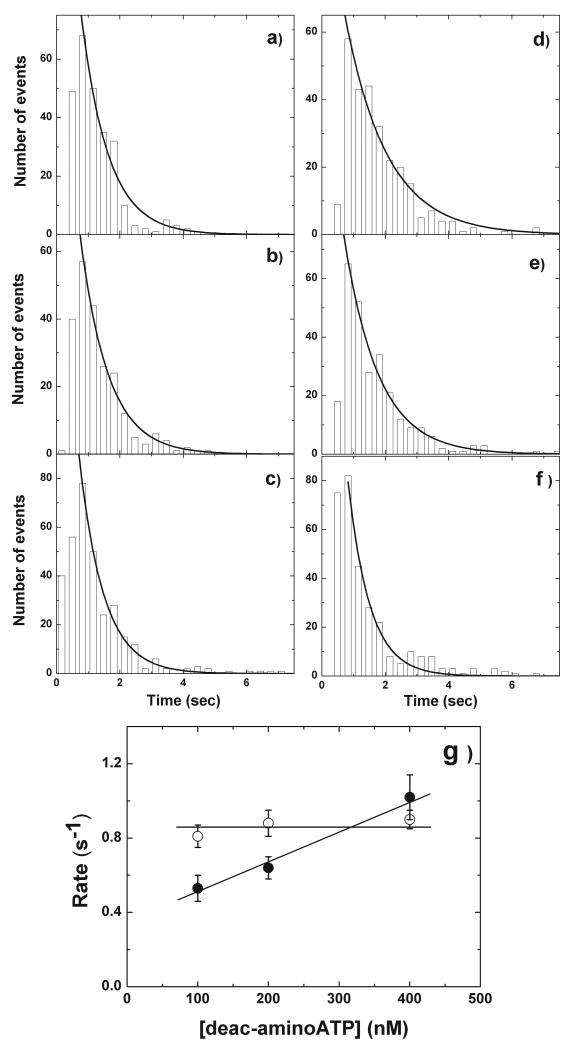
Figure 3. Histogram of lifetimes of deac-aminonucleotide association and dissociation.
a-c, Histograms of the lifetimes prior to deac-aminonucleotide dissociation at 100 nM (n = 261 steps, 44 MyoV-HMM molecules), 200 nM (n = 262 steps, 38 myosin-V molecule), and 400 nM (n = 310 steps, 35 MyoV-HMM molecules) deac-aminoATP. The solid lines represent the exponential fit of the dwell time distribution. The fitted lifetimes at 100 nM (a), 200 nM (b), and 400 nM (c) deac-aminoATP are 0.85 ± 0.06 s (r2 = 0.98), 0.88 ± 0.07 s (r2 = 0.98), and 0.77 ± 0.04 s (r2 = 0.98; all mean ± S.D.), respectively, corresponding to rate constants of 0.82 ± 0.06 s-1, 0..79 ± 0.07 s-1 and 0.90 ± 0.05 s-1. d-f, Histograms of the lifetimes of deac-amino ATP binding. The fitted lifetimes at 100 nM (n = 296); d), 200 nM (n = 267); e) and 400 nM (n = 309); f) deac-aminoATP are 1.32 ± 0.17 s (r2 = 0.98), 1.08 ± 0.11 s (r2 = 0.98), and, 0.68 ± 0.08 s (r2 = 0.98) (mean ± S.D.), respectively. Note that the number of spots is the same as a-c. This corresponds to rate constants of 0.53 ± 0.07 s-1, 0.64 ± 0.06 s-1, and 1.02 ± 0.12 s-1, respectively. Statistical analysis (t-test) between each experimental points revealed that the data of ADP dissociation rate from three conditions are not significantly different (P(T≤t) = 0.68, 0.09, 0.16 of 100 vs. 200, 100 vs 400, and 200 vs. 400 nM deac-aminoATP, respectively). In contrast, P values of ATP binding rates are significantly different as shown (P(T≤t) = 0.01, 0.028, and 0.02 for 100 vs. 200, 100 vs 400, and 200 vs. 400 nM deac-aminoATP, respectively). g, Concentration dependence of the rates of deac-aminoADP dissociation (open circles) and deac-aminoATP binding (filled circles). The deac-aminoATP binding data were fit by linear regression which gave a slope corresponding to a second order rate constant of 1.67 μM-1s-1. Note the nonzero intercept which is also present in the solution kinetic measurements of deac-aminoATP binding to acto-HMM in Supplementary Fig. 9. A non-zero intercept is predicted by modeling the kinetic mechanism and is seen in some published studies (Rief et al5) but is readily because of the large extrapolation from the high nucleotide values typically used. The horizontal line through the deac-aminoADP dissociation data represents the average of the mean value for the three nucleotide concentrations (0.86 s-1).
The model to account for the 36 nm Alexa-Fluor-568-MyoV-HMM steps and the 18 nm deac-aminonucleotide steps is shown in Fig.2f. Initially MyoV-HMM has deac-aminoADP bound to both heads and the position of the Alexa-Fluor-568-MyoV-HMM spot and the deac-aminonucleotide spot are coincident (step 1). Deac-aminoADP is released from the trail head, which results in the position of the deac-aminonucleotide signal advancing by 18 nm (step 2). After deac-aminoATP binds to the nucleotide-free trailing head, this head rapidly dissociates and swings forward to rebind and become the new lead head (steps 3 and 4). Single molecule and bulk solution kinetic studies suggest that the time between detachment of the trailing head, followed by its forward swing and reattachment is a few milliseconds and is thus much faster than the sampling rate (330 ms) used in our experiments7,10,11,19. Therefore ATP binding to the trail head, dissociation of that head, stepping and rebinding, are all associated with a 36 nm movement of the MyoV-HMM molecule and a simultaneous 18 nm movement of the deac-aminonucleotide signal. The binding of deac-aminoATP to the trail head might be expected to produce a transient backward movement of the nucleotide fluorescence centroid, but this is not seen because the trail head quickly detaches and is rapidly moved forward by the powerstroke occurring on the lead head.
To confirm the model, lifetimes during the two and one deac-aminonucleotide signal levels were analyzed at three different deac-aminoATP concentrations (Fig.3). We interpret the two to one nucleotide signal decrease to be associated with deac-aminoADP release from the trail head, whereas the one to two nucleotide signal increase is associated with deac-aminoATP binding to that head. Thus, fitting the lifetimes of the high nucleotide signal at 100, 200, and 400 nM deac-aminoATP showed no statistical difference in the rate of deac-aminoADP dissociation (0.82 s-1, 0.79 s-1, and 0.90 s-1, respectively; Fig.3a-c, left handed panels). This is similar to the deac-aminoADP dissociation rate constants measured in solution under identical conditions using stopped-flow spectrofluorimetry (1.2 s-1;Supplementary Fig.9c). This would indicate that in our experiments there is no acceleration of the deac-aminoADP release from the trail head and is consistent with stopped-flow kinetic results of previously reported10. An acceleration of the rate of ADP release from a positively strained trail head of up to 50-fold was previously predicted if the lead head were to complete its powerstroke when both heads were attached9. However, an earlier study found that the lead heads were only at the start of their powerstroke20, which is consistent with the lack of acceleration of ADP release from the rear head observed in our study. On the other hand, the observed deac-aminoATP binding rates determined by fitting the lifetimes of the low signal level intermediate increased as the deac-aminoATP concentration used was increased from 100 to 200 to 400 nM (0.53 s-1, 0.64 s-1 and 1.02 s-1, respectively; Fig.3d-f). This corresponds to a second order association rate constant of 1.67 μM-1s-1 which is very similar to a value of 2.48 μM-1s-1 measured in solution under identical conditions (Supplementary Fig.9b).
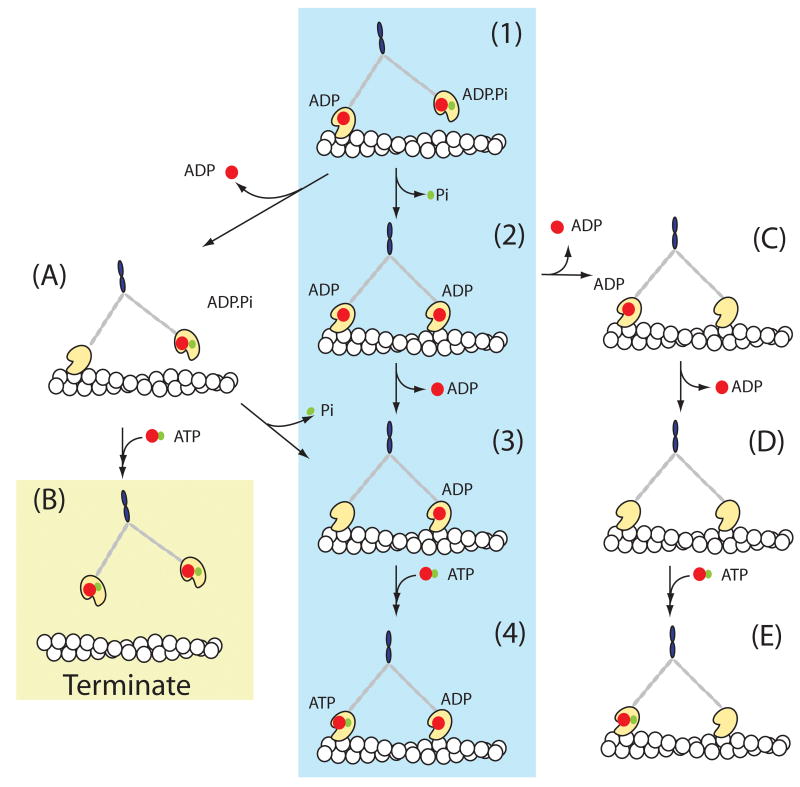
These results support a model in which the trailing head of the MyoV-HMM molecule releases ADP much more rapidly than ADP is released from the leading head2,9,10. In fact, solution kinetics studies at 20° C demonstrated that the deac-aminoADP dissociation rate (0.48 s-1) from the (presumably) trailing head was 32 times faster than that of the leading head (0.015 s-1) and a similar mechanism occurs with ADP10,21. Inhibition of ADP dissociation from the lead head is essential for long processive movements. Our results indicate that the main pathway of the MyoV-HMM ATPase is via the central shaded line of intermediates in Fig.4. The recently detached (formerly rear) head containing ATP or ADP·Pi rapidly swings forward to the leading position, where it binds actin (state 1). On binding to a new forward actin, this head quickly releases Pi (state (1) to (2) in Fig.4)7. ADP then dissociates from the trailing head (state(2) to (3) which allows a new ATP to bind. This results in a rapid detachment of that head, allowing the lead head to undergo its power stroke and repositions the detached head to become the new lead head (state (3) to (4). This model accounts for the 36-nm forward steps taken by the Alexa-Fluor-568-MyoV-HMM that occur coincidently with the 18-nm movement and a doubling of the intensity of the nucleotide fluorescence. An 18-nm backward step of nucleotide fluorescence would occur if deac-aminoADP dissociated first from a lead head (state (2) to (C). We did not observe such steps, which attest to the high level of strain-dependent gating between the kinetics of the two heads of MyoV-HMM. Termination of runs occurs principally by the route (1) to (A) to (B). This is consistent with a majority of termination cases in which the myosin has only one deac-aminonucleotide bound.
Figure 4. A scheme of tight coupling pathway of myosin-V.
The mechanism shown by intermediates (1) through (4) is the main pathway of stepping during a MyoV-HMM processive run. Termination of runs would occur via pathway (1) to (A) to (B) or more rarely by (C) to (D) to (E) to (A) to (B). Red spots represent ADP and green spots represent Pi.
Here we have directly observed the substrate binding and product dissociation steps of single motors moving along their tracks. The data show the relationship between these steps and the mechanism of processive movement of myosin Va on actin. These observations directly show that, as previously proposed, myosin Va is a tightly coupled motor2,6,7,10,19,4,5,8,9,11. Each step in a processive run involves the binding of an ATP molecule to the trail head of the myosin Va, which is rapidly followed by a 36-nm step along the actin and subsequently by the dissociation of ADP from the trail head. Deac-aminoATP should be a useful analog for other single-molecule studies such as combined optical trapping and total internal refection fluorescence (TIRF) microscopy.
Methods Summary
Protein purification and labeling
Mouse MyoV-HMM, MyoV-S1 and calmodulin were purified as previously described22. Calmodulin was labelled with Alexa Fluor 568 and exchanged for endogenous calmodulins into MyoV-HMM in a similar method to that previously described3,22.
Data acquisition and analysis
The single molecule in vitro motility assay was carried out essentially as previously described22 Dual imaging of deac-aminonucleotide and Alexa-Fluor-568-labelled MyoV-HMM were conducted using an Olympus IX81 microscope equipped for two fibre optic input cables using the DualView system23. Images were taken at a frame rate of 330 ms and the position of each fluorescent spot was determined using the FIONA method18
Transient kinetic data
Measurement of the deac-aminoATP binding and deac-aminADP dissociation were performed on a KinTek stopped-flow spectrofluorimeter as previously described10,17
Methods
Preparation of Proteins
Mouse MyoV-HMM or myoV-S1 were purified from Sf9 cells following infection with baculoviruses driving the expression of the HMM (or S1) and calmodulin24. Calmodulin was purified from bovine testes and labelled with Alexa-Fluor-568 succimidyl ester (Invitrogen)22. The molar ratio of Alexa-Fluor-568 per calmodulin was determined to be 1.8 by measuring the absorbance in solution and using the extinction coefficient of 91300 M-1cm-1 for Alexa-Fluor-568 and ε0.1% = 0.18 at A280nm for calmodulin The labelled calmodulin (molar ratio of 20 per MyoV-HMM) was exchanged with endogenous calmodulin as previously described22. This resulted in an average of six Alexa-Fluor-568 dyes per MyoV-HMM as determined spectrophotometrically. This value was confirmed by comparing the intensity of single Alexa-Fluor-568 molecules bound nonspecifically to a surface (2000 AU) with that of the average intensity of Alexa-Fluor-568-MyoV-HMM (12,000 AU). Biotinylated actin and biotinylated BSA were prepared22 and deac-aminoATP and deac-aminoADP were synthesized as described previously25
Emission and excitation spectra of 0.5 μM deac-aminoADP in the presence and absence of 1 μM MyoV-HMM were taken with a spectrofluorimeter using 2 nm slits (model: Fluoromax-3, HORIBA Jobin Yvon Inc, NJ).
Two line Total Internal Reflection Fluorescence Microscopy
Alexa-Fluor-568-labelled MyoV-HMM and deac-aminoATP were observed with an objective-type TIRF microscopy using an OLYMPUS IX81 microscope and a 60X, 1.45 N.A. PlanApo objective lens with two magnifying (relay) lenses (1.6X in the microscope, and 2.5X in front of the camera). Temperature was kept at 25°C with an environmental box (Precision plastics, MD). To visualize two colors of fluorescence simultaneously, we used the 568 nm line from an Ar-Kr LASER (I70C, Spectra physics) for Alexa568 and the 442 nm line from a He-Cd LASER (model IK41711-G, KIMMON) for deac-aminoATP. Both laser lines were combined by an acousto-Optical Tunable Filter (AOTF, Prairie Technologies, Inc.), which also controlled the laser power. After the AOTF, the two laser lines were separated by a dichroic mirror onto optical fibers. This allows both wavelengths to be in focus at the same time. The two fibers are guided to individual TIRF illuminators located at the rear end of the microscope. Illumination at 442 nm was via the Olympus TIRF apparatus and the illumination at 568 nm was via the position usually occupied by the mercury arc lamp housing. The two laser lines from the two illuminators were combined with a dichroic mirror and introduced into the objective lens. The power of both the 442 and 568 nm beams were 10 to 20 mW in front of the objective lenses. The emitted light was passed through a dual line dichroic mirror (442/568, Chroma) and split by a dichroic mirror (552dcr, Chroma) in the Dual-View™ system (Optical Insights). Fluorescence was detected by an EMCCD camera (DV897, 512BV, Andor), at -90°C with a gain of either 400 or 1000. Images were digitized by using Metamorph (MSD/Molecular Device ver.7.1).
Intensity measurement of deac-aminoADP bound to either the surface or to MyoV-HMM
To test whether the fluorescence of deac-aminoADP increased in intensity upon binding to MyoV-HMM in the microscope, we directly observed single molecules of deac-aminoADP in the presence and absence of MyoV-HMM. First, 10 pM deac-aminoADP was added into a flowcell coated with 0.1% nitrocellulose and incubated 2 min at room temperature. Free deac-aminoADP was washed out using motility assay buffer (see Fig.2 legend for composition) and imaged at 442 nm using the TIRF microscopy setup described above at EM-CCD camera gains of 1000 (Fig.1a and e) and 400 (Fig. 1b and f). Images were acquired in 330 msec windows. On a second slide 10 pM MyoV-HMM was added into the flowcell and incubated 2 min at room temperature. Free MyoV-HMM was washed out with motility assay buffer. Deac-aminoADP (10 nM) was added into the flowcell and the sample imaged at 442 nm and 568 nm at the same two camera gains. At the single molecule level in the microscope, under these conditions, the intensity of deac-aminoADP increased 4-fold upon binding to myosin.
Single molecule motility assay and data analysis
Single molecule motility assays were performed as previously described26. Position data for Alexa-Fluor-568-MyoV-HMM and deac-aminoATP were analyzed by FIONA18. The integrated intensities of 15 x 15 pixel areas were measured at the indicated concentrations using Metamorph. To observe single molecule movements of MyoV-HMM and deac-aminoATP, simultaneously, we changed the concentration of Alexa-Fluor-568-MyoV-HMM to reduce background. At 100 and 200 nM deac-aminoATP concentrations, 200 pM Alexa-Fluor-568-MyoV-HMM was used, while at 400 nM deac-aminoATP, 4 pM Alexa-Fluor-568-MyoV-HMM was used. Steps were identified by eye and marked by hand.
Run lengths of Alexa-Fluor-568-MyoV-HMM were measured with either 1 mM ATP or 1 mM deac-aminoATP. Actin filaments were labelled with 10% AlexaFluor488-phalloidin and 90% phalloidin. The determination of run length was performed as described previously22, except only myosin molecules that dissociated before reaching the end of actin filaments were scored. The average length of an actin filament was 12.5 μm. Velocities with various deac-aminoATP concentrations were measured with time-lapse where data was taken at 10 second intervals with a 300 millisecond exposure time. Sequential images were taken to analyze velocity.
Determination of number of photons
We determined the number of photons from the integrated intensity of a 10 x 10 pixel image of each chosen spot. Deac-aminoADP was bound non-specifically to the surface or to MyoV-HMM, which was bound on a nitrocellulose-coated surface. The estimated total number of photons in the spot at various camera gains was calculated as described in Thompson et al.27. Alternatively the number of photons was calculated using an equation provided by Andor (data not shown).
| (Eq-1) |
Where 60.14 is the camera sensitivity at 10 MHz, EM amplifier, 1.0X preamp setting (electron per A/D count), a is the percentage of quantum efficiency of the camera at the appropriate wavelength, and I is the detected integrated intensity. The two methods gave reasonable agreement. At least 10,000 photons are required to obtain 2.5 nm localization19. Determination of localization accuracy from single molecule fluorophors has been calculated by theoretical equations27 and measured experimentally.18
Supplementary Material
Acknowledgments
We thank F. Zhang, A. Smith and G. Reid for technical assistance; E. Yokoi for TIRF illuminator to combine two optical cables; C. Fanghella for technical help in the calculation of number of photons; and C. Weaver for help with Metamorph. We are appreciative of the critical comments on the manuscript made by P.J. Knight and E. Homsher. M.R.W. was supported by the Medical Research Council, UK. J.R.S. and T.S. were supported by the National Heart, Lung and Blood Intramural Program. H.D.W. and E.F were support by NIH EB00209 and a postdoctoral fellowship from the AHA.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Author contributions Single molecule motility experiments and data analysis were performed by T.S. and kinetic experiments data by E.F. Deac-aminoATP/ADP were provided by M.R.W. T.S., J.R.S and H.D.W participated in the conception of the experiment. T.S. wrote the first draft of the manuscript and all authors participated in producing the final version. All authors participated in discussion and interpretation of the data.
Author Information Reprints and permission information is available at www.nature.com/nature. The authors declare no competing financial interests.
Reference List
- 1.Sellers JR, Weisman LS. Myosins: A superfamily of molecular motors. In: Coluccio L, editor. Proteins and Cell Regulation. Vol. 7. Springer; Dordrecht: 2008. pp. 289–324. [Google Scholar]
- 2.Vale RD. Myosin V motor proteins: marching stepwise towards a mechanism. J Cell Biol. 2003;163:445–450. doi: 10.1083/jcb.200308093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakamoto T, Yildiz A, Selvin PR, Sellers JR. Step-size is determined by neck length in myosin V. Biochemistry. 2005;44:16203–16210. doi: 10.1021/bi0512086. [DOI] [PubMed] [Google Scholar]
- 4.Mehta AD, et al. Myosin-V is a processive actin-based motor. Nature. 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 5.Rief M, et al. Myosin-V stepping kinetics: a molecular model for processivity. Proc Natl Acad Sci U S A. 2000;97:9482–9486. doi: 10.1073/pnas.97.17.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De La Cruz EM, Wells AL, Rosenfeld SS, Ostap EM, Sweeney HL. The kinetic mechanism of myosin V. Proc Natl Acad Sci U S A. 1999;96:13726–13731. doi: 10.1073/pnas.96.24.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenfeld SS, Sweeney HL. A model of myosin V processivity. J Biol Chem. 2004;279:40111. doi: 10.1074/jbc.M402583200. [DOI] [PubMed] [Google Scholar]
- 8.Purcell TJ, Sweeney HL, Spudich JA. A force-dependent state controls the coordination of processive myosin V. Proc Natl Acad Sci U S A. 2005;102:13873–13878. doi: 10.1073/pnas.0506441102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veigel C, Schmitz S, Wang F, Sellers JR. Load-dependent kinetics of myosin-V can explain its high processivity. Nat Cell Biol. 2005;7:861–869. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- 10.Forgacs E, et al. Kinetics of ADP dissociation from the trail and lead heads of actomyosin V following the power stroke. J Biol Chem. 2008;283:766–773. doi: 10.1074/jbc.M704313200. [DOI] [PubMed] [Google Scholar]
- 11.Veigel C, Wang F, Bartoo ML, Sellers JR, Molloy JE. The gated gait of the precessive molecular motor, myosin V. Nat Cell Biol. 2001;4:59–65. doi: 10.1038/ncb732. [DOI] [PubMed] [Google Scholar]
- 12.Funatsu T, Harada Y, Tokunaga M, Saito K, Yanagida T. Imaging of single fluorescent molecules and individual ATP turnovers by single myosin molecules in aqueous solution. Nature. 1995;374:555–559. doi: 10.1038/374555a0. [DOI] [PubMed] [Google Scholar]
- 13.Ishijima A, et al. Multiple- and single-molecule analysis of the actomyosin motor by nanometer piconewton manipulation with a microneedle: Unitary steps and forces. Biophys J. 1996;70:383–400. doi: 10.1016/S0006-3495(96)79582-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tokunaga M, Kitamura K, Saito K, Iwane AH, Yanagida T. Single molecule imaging of fluorophores and enzymatic reactions achieved by objective-type total internal reflection fluorescence microscopy. Biochem Biophys Res Commun. 1997;235:47–53. doi: 10.1006/bbrc.1997.6732. [DOI] [PubMed] [Google Scholar]
- 15.Oiwa K, et al. Comparative single-molecule and ensemble myosin enzymology: sulfoindocyanine ATP and ADP derivatives. Biophys J. 2000;78:3048–3071. doi: 10.1016/S0006-3495(00)76843-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webb MR, Reid GP, Munasinghe VR, Corrie JE. A series of related nucleotide analogues that aids optimization of fluorescence signals in probing the mechanism of P-loop ATPases, such as actomyosin. Biochemistry. 2004;43:14463–14471. doi: 10.1021/bi0486334. [DOI] [PubMed] [Google Scholar]
- 17.Forgacs E, et al. Kinetic mechanism of myosinV-S1 using a new fluorescent ATP analogue. Biochemistry. 2006;45:13035–13045. doi: 10.1021/bi060712n. [DOI] [PubMed] [Google Scholar]
- 18.Yildiz A, et al. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 19.Dunn AR, Spudich JA. Dynamics of the unbound head during myosin V processive translocation. Nat Struct Mol Biol. 2007;14:246–248. doi: 10.1038/nsmb1206. [DOI] [PubMed] [Google Scholar]
- 20.Burgess S, et al. The prepower stroke conformation of myosin V. J Cell Biol. 2002;159:983–991. doi: 10.1083/jcb.200208172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenfeld SS, Houdusse A, Sweeney HL. Magnesium regulates ADP dissociation from myosin V. J Biol Chem. 2005;280:6072–6079. doi: 10.1074/jbc.M412717200. [DOI] [PubMed] [Google Scholar]
- 22.Sakamoto T, et al. Neck length and processivity of myosin V. J Biol Chem. 2003;278:29201–29207. doi: 10.1074/jbc.M303662200. [DOI] [PubMed] [Google Scholar]
- 23.Thirumurugan K, Sakamoto T, Hammer JA, III, Sellers JR, Knight PJ. The cargo-binding domain regulates structure activity of myosin 5. Nature. 2006;442:212–215. doi: 10.1038/nature04865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang F, et al. Effect of ADP and ionic strength on the kinetic and motile properties of recombinant mouse myosin V. J Biol Chem. 2000;275:4329–4335. doi: 10.1074/jbc.275.6.4329. [DOI] [PubMed] [Google Scholar]
- 25.Webb MR, Corrie JE. Fluorescent coumarin-labeled nucleotides to measure ADP release from actomyosin. Biophys J. 2001;81:1562–1569. doi: 10.1016/S0006-3495(01)75810-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakamoto T, Amitani I, Yokota E, Ando T. Direct observation of processive movement by individual myosin V molecules. Biochem Biophys Res Commun. 2000;272:586–590. doi: 10.1006/bbrc.2000.2819. [DOI] [PubMed] [Google Scholar]
- 27.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.