Abstract
Although classical protein tyrosine phosphatase (PTP) superfamily members are cysteine-dependent, emerging evidence shows that many acid phosphatases (AcPs) function as histidine-dependent PTPs in vivo. These AcPs dephosphorylate phospho-tyrosine substrates intracellularly and could have roles in development and disease. In contrast to cysteine-dependent PTPs, they utilize histidine, rather than cysteine, for substrate dephosphorylation. Structural analyses reveal that active site histidine, but not cysteine, faces towards the substrate and functions as the phosphate acceptor. Nonetheless, during dephosphorylation, both histidine-dependent and cysteine-dependent PTPs use their active site arginine and aspartate for substrate binding and proton donation, respectively. Thus, we propose that they should be referred to as a distinct group of ‘histidine-dependent PTPs’ within the PTP superfamily.
Protein tyrosine phosphatase superfamily
Cellular protein tyrosine phosphorylation is regulated by a dynamic equilibrium between the activity of protein tyrosine kinases (PTKs) and protein tyrosine phosphatases (PTPs). Most PTP superfamily members are structurally conserved and depend on the cysteine residue in their active site for phosphotyrosine (pTyr) dephosphorylation [1]. These phosphatases are broadly categorized into classical PTPs, including the receptor and the non-receptor PTPs, the dual-specificity phosphatases (DSPs), cell division cycle 25 (cdc25) phosphatases and the low molecular weight (LMW) PTPs [2]. The classical PTPs, which regulate diverse cellular signals, are marked by the presence of a conserved active site signature sequence, C(X)5RS/T (where X is any amino acid) [1,2]. Structural comparisons between the classical PTPs and DSPs reveal a conserved architecture in their active sites: a conserved active site cysteine residue that serves as the phosphate group acceptor is crucial for their PTP activity, and a substitution in the cysteine residue results in their inactivation [2–4]. Studies reveal that a distinct group of closely related acid phosphatases (AcPs) are histidine-dependent, but cysteine-independent, PTPs that dephosphorylate various intracellular pTyr protein substrates. Similar to the cysteine-dependent PTPs, several reports indicate that these AcPs, which use histidine as their phosphate acceptor, are expressed in specialized, differentiated cells and regulate a variety of cellular processes, including proliferation and differentiation. Based on their intracellular PTP activity, we propose that these histidine-dependent AcPs should be designated as histidine-dependent PTPs and as a distinct group within the PTP superfamily.
Histidine-dependent AcPs: in vivo PTPs
A closer look at the AcPs that exhibit PTP activity shows that they are expressed in diverse organisms (Figure 1). Such histidine-dependent AcPs from plants, for example Lens esculenta (lentils) and Solanum tuberosom (potato tuber) [5,6], prokaryotes, for example Escherichia coli, Staphylococcus aureus and Streptomyces coelicolor [7–10], lower eukaryotes, for example Entamoeba histolytica [11], and specialized cells in higher eukaryotes [12–19] exhibit both AcP and PTP activity. Amino acid sequence analyses reveal that histidine-dependent AcPs have a conserved amino acid sequence motif, RHGXRXP (where X is any amino acid), within their active domain [20]. The conservation of the PTP activity of histidine-dependent AcPs from bacteria to humans suggests the preservation of a specialized function of these members.
Figure 1.
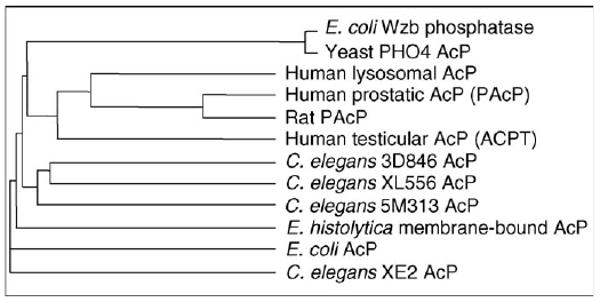
Relatedness of histidine-dependent acid phosphatases (AcPs) in prokaryotic and eukaryotic cells. A phylogenetic tree based on the amino acid sequences of different prokaryotic and eukaryotic AcPs that use active site histidine as their phosphate acceptor. The tree was constructed using the Vector NTI program (Invitrogen, USA).
It should be noted that the nomenclature of these phosphatases as ‘AcPs’ is essentially based on the in vitro biochemical measurement of their optimal phosphatase activity at an acidic pH over small organic compounds containing phosphates, such as paranitrophenyl phosphate (pNPP). It is noteworthy that most of the classical cysteine-dependent PTPs, including PTP1A and PTP1B, also hydrolyze pNPP in vitro with acidic pH optima [21]. Similarly, Wzb, an E. coli histidine-dependent phosphatase containing the AcP signature sequence, dephosphorylates pTyr residues of the Wzc kinase in vivo, whereas it biochemically hydrolyzes pNPP at an acidic pH of 6.5 [8]. Thus, the nomenclature of these phosphatases, which exhibit PTP activity in vivo, as AcPs based on their biochemical activity at acidic pH towards anomalous artificial substrates, such as pNPP, is misleading [22,23]. Indeed, like the cysteine-dependent PTPs, histidine-dependent phosphatases can dephosphorylate pTyr proteins optimally under a neutral pH, that is, at a pH level similar to intracellular pH [17,19,22]. Thus, we propose that these phosphatases should be renamed as PTPs based on their in vivo activity towards intracellular pTyr substrates. Previous suggestions to rename cellular prostatic AcP (cPAcP) as prostate-specific PTP and the LMW AcPs as LMW PTPs were based, in part, on their PTP activity at neutral pH [22,24]. Recent studies have characterized PTP-dependent roles for these histidine-dependent AcPs within key cellular signaling processes in various differentiated cells [8,19,25–28]. Thus, the in vivo role of histidine-dependent phosphatases in the dephosphorylation of pTyr proteins indicates that they represent a distinct group of PTPs under the PTP superfamily.
Structural features of histidine-dependent versus cysteine-dependent PTPs
Structural analyses of classical PTPs, such as PTP1B and CD45 (cluster of differentiation 45 phosphatase), reveal that these PTPs have deep active sites (9 Å deep), making them ‘reachable’ only by longer residues, for example tyrosine, but not shorter serine or threonine residues. By contrast, DSPs, such as human vaccinia h1-related phosphatases (VHRs) and phosphatase and tensin homolog phosphatases (PTENs), have active sites shallow enough (∼6–8 Å) to be ‘reached’ by shorter serine or threonine residues or phospholipids (Figure 2a) [29]. Human PAcP and E. coli AcP subunits exhibit large, open active site conformations similar to the DSPs (Figure 2a) [30–33].
Figure 2.
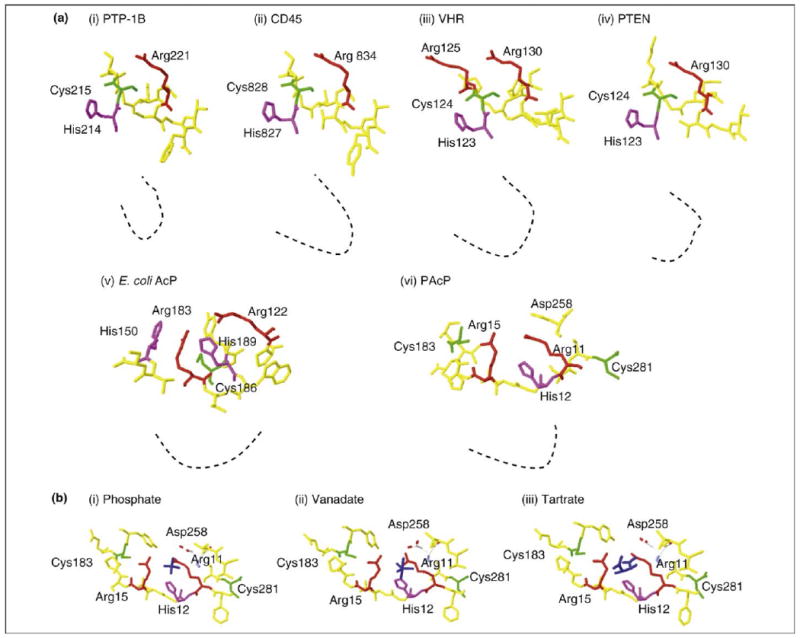
Structural comparison of the active sites of classical cysteine-dependent and histidine-dependent PTPs. The structures of the active sites of cysteine-dependent and histidine-dependent PTPs are compared using Swiss-PDB viewer software. Key amino acids are labeled. Color codes for amino acids: histidine – magenta; cysteine – green; arginine – red; structural amino acids encompassing active site – yellow; substrate or inhibitors – dark blue. (a) Comparison of X-ray crystallographic structures of both cysteine-dependent and histidine-dependent PTPs with their substrates/inhibitors; these comparisons reveal striking differences in their active site conformations. Classical cysteine-dependent PTPs, for example human PTP-1B (i) and CD45 phosphatases (ii), possess a deep active site that can be reached by tyrosine residues, which determines their substrate specificity. By contrast, both dual specificity phosphatases, such as human VHR (iii) and PTEN (iv), and histidine-dependent AcPs, for example E. coli AcP (v) and human cPAcP (vi), have relatively shallow active sites. The dotted black line depicts the depth of the active site cleft of the PTPs. (b) In histidine-dependent PTPs, here PAcP, the phosphate acceptor His12 is positioned towards phosphate (i), vanadate (ii) and tartrate (iii). A comparison with Figure 2a indicates that in E. coli AcP, another histidine-dependent PTP, histidine faces towards the substrate. By contrast, in cysteine-dependent PTPs (Figure 2a, i–iv), the active site cysteine is positioned towards the substrate, whereas the histidine residue faces away from the substrate during a dephosphorylation reaction.
Histidine-dependent PTPs, for example PAcP and ACPT (testicular AcP), contain the conserved histidine, arginine and aspartic acid residues in their active sites. The analysis of phospho-enzyme intermediates revealed that PAcP His12 functions as the phosphate acceptor for its AcP and PTP activities; Asp258 apparently serves as the proton donor for the substrate-leaving group [34]. Further analyses using chemical modification, site-directed mutagenesis and X-ray crystallographic approaches have confirmed the importance of His12 and Asp258 in PAcP AcP and PTP activity [30–32,34,35]. The sequence homology among the AcPs exhibiting PTP activity in their active sites suggests that the histidine corresponding to the His12 of PAcP is a crucial residue for their PTP activity.
A closer examination of histidine-dependent phosphatase crystal structures indicates that the nucleophilic His12 residue is located spatially closer to the substrate, whereas active site cysteine residues face away from the substrate (Figure 2). A comparison of the PAcP active site bound to a phosphate ion with that of the PAcP holoenzyme reveals the outward movement of Cys183 from phosphate (Figure 2a and Figure 2b). Similarly, in E. coli AcP, substrate binding causes His150 to move closer to the substrate, which forms the hydrogen bonds with the substrate when the substrate forms covalent bonds with His189 [33]. The nucleophilic cysteine in cysteine-dependent phosphatases, for example PTP-1B, VHR and PTEN, is spatially closer to the substrate than the histidine residues are to the substrate during the dephosphorylation reaction, even though the active site histidine residue is located next to the cysteine residue in these cysteine-dependent PTPs (Figure 2a). Further studies are needed to understand the requirement of differential utilization of cysteine and histidine in pTyr dephosphorylation.
Interestingly, the cysteine residues in the active sites of histidine-dependent PTPs, corresponding to Cys183 and Cys281 of PAcP, are not essential for their PTP activity [35]. Although these residues are conserved in many eukaryotes, they are not conserved in lower prokaryotes, implying that these cysteinyl residues might be used to dephosphorylate substrates other than pTyr proteins in higher eukaryotes. For example, in vitro biochemical studies indicate that human PAcP hydrolyzes lysophosphatidic acid and phosphatidyl inositol 3 phosphate (PI3P) and thus could function as a phospholipid phosphatase, similar to PTEN [36,37]. However, the potential involvement and biological relevance of the active cysteine in this context requires further validation.
Based on the available data for cPAcP, a model for the histidine-dependent PTP-mediated dephosphorylation reaction can be generated (Figure 3). cPAcP binds its substrate noncovalently using side chains of three arginine residues (Arg11, Arg15 and Arg79) [31,34]. The His12 imidazole ring provides a pair of electrons for nucleophilic attack to the phosphate group. This utilization of histidine by histidine-dependent PTPs parallels the use of cysteine for nucleophilic attack of phosphate groups by cysteine-dependent PTPs. The function of Asp258 in dephosphorylation is apparently conserved between histidine-dependent and cysteine-dependent PTP activity. Cooperatively, Asp258 donates a proton from its carboxyl group to the substrate resulting in the formation of the phosphoenzyme intermediate and the liberation of dephosphorylated substrate. Additionally, Asp258 might also stabilize the phospho-His12 intermediate. Subsequently, nucleophilic attack of the phosphoenzyme intermediate occurs through a water molecule to release the phosphate group and to return a proton to the Asp258 carboxyl group [31,34]. Based on X-ray crystallographic studies, Asp258 is proposed to polarize the water molecule to facilitate the nucleophilic attack of water on the phospho-histidine intermediate [32]. Alternatively, solvent isotope studies indicate that Asp258 functions in positioning the water molecule during the dephosphorylation reaction [31]. Further studies will be required to address the biochemical role of this aspartic acid in histidine-dependent PTPs.
Figure 3.
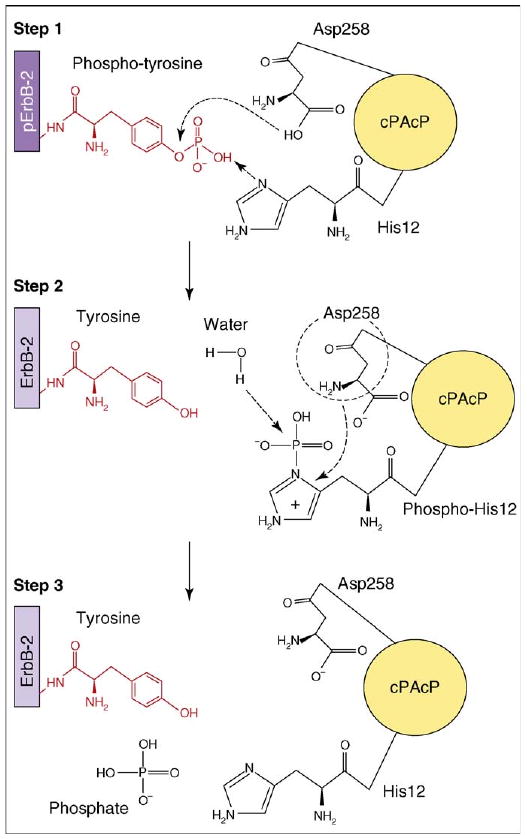
Schematic representation of substrate dephosphorylation by a histidine-dependent phosphatase. The dephosphorylation reaction is summarized using cPAcP as a model histidine-dependent PTP and tyrosine-phosphorylated ErbB-2 as the model substrate. cPAcP His12 serves as the phosphate acceptor in Step 1 of the reaction. Asp258 functions as the proton donor (Steps 1 and 2). Alternatively, Asp258 might also stabilize the phospho-His12 intermediate and position the water molecule to favor the nucleophilic attack on the phosphate (Step 2).
Studies of cPAcP histidine-dependent PTP activity have been paralleled by research on other AcPs that are expressed in a variety of tissues. For example, ACPT, which is expressed in human testicular tissue and brain cells, shows 60% amino acid identity to PAcP and exhibits PTP activity at physiological pH [18,19]. Importantly, the crucial histidine (His12) and the aspartate (Asp259) residues in the ACPT active site, which are homologous to PAcP His12 and Asp258, are conserved [19]. Other AcPs with a conserved active site histidine residue, corresponding to His12 of PAcP, that either exhibit demonstrable PTP activity or are sensitive to PTP inhibitors, for example orthovanadate, include those expressed in E. coli, yeast (Schizosaccharomyces pombe), Caenorhabditis elegans and in human lysosomes [7,38–40]. Although detailed kinetic mechanisms are not yet available, we propose that these histidine-dependent phosphatases represent a discrete group of PTPs, distinct from the ubiquitously expressed classical cysteine-dependent PTPs.
Although cysteine-dependent PTPs and histidine-dependent PTPs share common modes of regulation, for example reversible inactivation by oxidation of cysteine or histidine residues in respective PTPs by physiological reactive oxygen species (ROS) levels, differences in activation exist. In particular, subunit dimerization might significantly enhance the specific activity of the histidine-dependent phosphatases in a similar manner to the action of dimerization on the receptor tyrosine kinases, such as the epidermal growth factor receptor (EGFR) family [41–43]. Indeed, Trp106 and His112 residues of mature rat PAcP, which are thought to regulate its dimerization and subsequent activation [44], are conserved in AcPs, for example ACPT, C. elegans AcP and human lysosomal AcPs, thus indicating that they might also be activated by dimerization [19]. By contrast, dimerization of classical receptor PTPs, such as receptor protein tyrosine phosphatase α (RPTPα), leucocyte antigen-related phosphatase (LAR) and CD45, results in their inactivation [45]. Thus, the use of subunit dimerization could differentiate the regulation of histidine-dependent PTP activity from the regulation of cysteine-dependent PTP activity. Future studies will be necessary to determine the role of dimerization in regulating PTP activity of other AcPs.
Intracellular substrates of histidine-dependent PTPs
Evidence that has accumulated over the past decade has shed light on the biological relevance of histidine-dependent PTPs. In terms of their substrates and in regulating key cellular signaling events, these PTPs function in a manner similar to cysteine-dependent PTPs. Here, we discuss the biological functions of representative histidine-dependent PTPs and their respective in vivo substrates.
ErbB-2: a cPAcP substrate in prostate epithelia
In differentiated prostate epithelial cells, PAcP is expressed in two forms – a secreted form, the physiological functions of which remain unidentified, and an intracellular form, which is the focus of this discussion. cPAcP, the intracellular form, regulates growth in prostate epithelial cells. For example, differentiated prostate epithelial cells express high cPAcP levels and exhibit slow cell proliferation, whereas in prostate carcinomas, cPAcP expression is diminished and cell proliferation is increased [25]. Biochemical and in vivo studies indicate that ErbB-2 (erythroblastosis virus oncogene, also called Her-2 or Neu), an EGFR family member and a transmembrane receptor kinase, is an in vivo cPAcP substrate [26,28,46,47]. Ectopic cPAcP expression in aggressive prostate cancer cells promotes decreased ErbB-2 tyrosine phosphorylation that results in decreased growth rates and tumorigenicity [25–28,47,48]. Importantly, intratumoral injection of ACPP cDNA, but not a mutant version that lacks PTP activity, into xenograft tumors leads to tumor regression, at least in part via downregulating ErbB-2 phosphorylation [28]. Conversely, small interfering RNA (siRNA) or antisense-mediated PAcP knockdown in the LNCaP human prostate adenocarcinoma cell line results in increased ErbB-2 tyrosine phosphorylation and subsequent cell growth [47]. Importantly, knockout animal studies validate cPAcP as a tumor suppressor because prostate adenocarcinomas spontaneously develop in male mice that lack exon 3 in the PAcP gene (AcppΔ3/Δ3) [37]. Thus, cPAcP is a histidine-dependent PTP that regulates prostate epithelial cell growth, at least in part, through ErbB-2 dephosphorylation.
ErbB-4: an ACPT substrate in neuronal cells
ACPT, originally identified in testicular epithelial cells, is also highly expressed in neuronal tissues and exhibits more than 60% amino acid identity with PAcP. In differentiated neuronal cells, ACPT colocalizes and directly interacts with ErbB-4, an EGFR family member that regulates neuronal differentiation [19]. Importantly, ACPT dephosphorylates ErbB-4 in the synaptic sites of differentiated neuronal cells, which indicates that ACPT might have a crucial role in neuronal differentiation [19]. In fact, ACPT expression inhibits ErbB-4-mediated neuregulin (NRG)-induced neurite outgrowth in pheochromocytoma (PC)-12 cells, a differentiation phenotype [19]. Thus, the interaction between ACPT and ErbB-4 could participate in neuronal development and differentiation. Further studies are needed to identify the key neuronal signaling pathways that are regulated by the ACPT–ErbB-4 interaction.
Histidine-dependent PTPs in plants
Phosphatases have been linked to several tyrosine phosphorylation signaling pathways in plants, for example seed germination. In lentil seeds, histidine-dependent AcP PTP activity increases at the onset of seed germination, thus indicating its possible involvement in seed germination [5]. The expression of maize (Zea mays) AcP, which has high activity towards pTyr substrates, also correlates with seed germination [49]. In particular, its protein levels negatively correlate with the phosphorylation of a 14-kDa pTyr protein, a putative intracellular substrate for this AcP [49]. Similarly, AcPs exhibiting significant phosphatase activity towards pTyr substrates have also been reported in dormant hazel (Corylus avellana) seeds [50]. Further studies will be required to elucidate the biological significance of these AcPs in plant signaling pathways.
Wzc: a Wzb substrate in prokaryotes
Wzc, a transmembrane kinase, participates in the regulation of prokaryotic extracellular polysaccharide synthesis, for example in E. coli and Lactobacillus rhamnosus, and is dephosphorylated intracellularly by the Wzb phosphatase [8,51]. Wzb phosphatase activity is histidine-dependent. Whereas specific serine-substituted cysteine mutants, for example C78S and C6S, retain more than 60% of phosphatase activity, alanine-substituted histidine mutants, for example H42A, H43A, H5A and H7A, result in the complete loss of phosphatase activity towards pNPP [51]. Further studies will be needed to address the role of histidine in Wzc-mediated Wzb dephosphorylation in vivo. Similarly, phosphatase 1 from psychrophilic bacteria, for example Shewanella spp., exhibits histidine-dependent phosphatase activity towards pTyr substrates [52]. Future studies should be directed at identifying other bacterial AcPs that function as histidine-dependent PTPs, as well as the physiological substrates of these AcPs.
Concluding remarks and future perspectives
Several lines of evidence support the notion that diverse histidine-dependent AcPs from various species function as histidine-dependent PTPs, representing a distinct subgroup of the PTP superfamily. Significantly, the active site histidine is conserved in a wide variety of organisms. Interestingly, histidine-dependent PTPs share some common modes of regulation with cysteine-dependent PTPs, such as ROS-mediated inactivation by reversible oxidation of cysteine or histidine residues [53,54]. Nonetheless, their unique regulatory mechanism(s), including dimerization, could differentiate histidine-dependent PTPs from classical cysteine-dependent PTPs. Additional biochemical studies will be needed to elucidate this point.
Current studies are focused on analyzing the molecular events governed by histidine-dependent PTPs. These phosphatases could function as regulators of specific biological events, for example proliferation and differentiation in specialized cell types. Although they function as AcPs when small organic phospho-compounds are used as substrates, their physiological role as neutral PTPs on cellular pTyr proteins suggests several possibilities. For example, the differential pH optima could allow histidine-dependent phosphatases to interact with different in vivo substrate molecules in different sub-cellular compartments. cPAcP might function both as a PTP and a phospholipid phosphatase: it can dephosphorylate both pTyr and PI3P and regulate ErbB-2-phosphatidylinositol 3-OH kinase signaling [26,27,35,37,47]. Future studies should be directed at identifying additional AcPs that function as histidine-dependent PTPs in vivo. These studies might provide answers regarding the evolutionary relationship of these histidine-dependent phosphatases in comparison to the cysteine-dependent PTPs. In addition, exploring the conditions under which the cells employ two different kinetic mechanisms for dephosphorylation will help us to understand more about the evolutionary adaptation of organisms. Development of novel inhibitors that react with the active site histidine should be useful for mechanism-based studies and would help us to identify the biological relevance of these histidine-dependent PTPs. Importantly, it is necessary to explore their clinical significance in human diseases, for example cancer, and their potential role in therapy against human disease.
Acknowledgments
Work in our laboratory is supported in part by the National Institutes of Health (grant R01 CA88184), the US Department of Defense (grant PC074289), a Nebraska Research Initiative grant, the Nebraska Cancer and Smoking Disease Research Program (LB506 # 2008–20) and a University of Nebraska Medical Center Graduate Student Fellowship. We thank Fen-Fen Lin for her tremendous contributions toward our studies on the functional role of cPAcP.
References
- 1.Fauman EB, Saper MA. Structure and function of the protein tyrosine phosphatases. Trends Biochem Sci. 1996;21:413–417. doi: 10.1016/s0968-0004(96)10059-1. [DOI] [PubMed] [Google Scholar]
- 2.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 3.Stuckey JA, et al. Crystal structure of Yersinia protein tyrosine phosphatase at 2.5 Å and the complex with tungstate. Nature. 1994;370:571–575. doi: 10.1038/370571a0. [DOI] [PubMed] [Google Scholar]
- 4.Jackson MD, Denu JM. Molecular reactions of protein phosphatases – insights from structure and chemistry. Chem Rev. 2001;101:2313–2340. doi: 10.1021/cr000247e. [DOI] [PubMed] [Google Scholar]
- 5.Bose SK, Taneja V. Induction of a germination specific, low molecular weight, acid phosphatase isozyme with specific phosphotyrosine phosphatase activity in lentil (Lens esculenta) seeds. Biochem Biophys Res Commun. 1998;250:629–634. doi: 10.1006/bbrc.1998.9364. [DOI] [PubMed] [Google Scholar]
- 6.Gellatly KS. Purification and characterization of a potato tuber acid phosphatase having significant phosphotyrosine phosphatase activity. Plant Physiol. 1994;106:223–232. doi: 10.1104/pp.106.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ostanin K, et al. Overexpression, site-directed mutagenesis, and mechanism of Escherichia coli acid phosphatase. J Biol Chem. 1992;267:22830–22836. [PubMed] [Google Scholar]
- 8.Vincent C, et al. Cells of Escherichia coli contain a protein-tyrosine kinase, Wzc, and a phosphotyrosine-protein phosphatase, Wzb. J Bacteriol. 1999;181:3472–3477. doi: 10.1128/jb.181.11.3472-3477.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soulat D. Staphylococcus aureus contains two low-molecular-mass phosphotyrosine protein phosphatases. J Bacteriol. 2002;184:5194–5199. doi: 10.1128/JB.184.18.5194-5199.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Strohl WR. Cloning, purification, and properties of a phosphotyrosine protein phosphatase from Streptomyces coelicolor A3(2) J Bacteriol. 1996;178:136–142. doi: 10.1128/jb.178.1.136-142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aguirre-García MM, et al. Membrane-bound acid phosphatase (MAP) from Entamoeba histolytica has phosphotyrosine phosphatase activity and disrupts the actin cytoskeleton of host cells. Parasitology. 2003;126:195–202. doi: 10.1017/s0031182002002767. [DOI] [PubMed] [Google Scholar]
- 12.Raugei G, et al. Chemical synthesis and expression of a gene coding for bovine liver phosphotyrosine-protein phosphatase. Biochem Int. 1991;23:317–326. [PubMed] [Google Scholar]
- 13.Lau KH, et al. Phosphotyrosyl-specific protein phosphatase activity of a bovine skeletal acid phosphatase isoenzyme. Comparison with the phosphotyrosyl protein phosphatase activity of skeletal alkaline phosphatase. J Biol Chem. 1984;260:4653–4660. [PubMed] [Google Scholar]
- 14.Lau KH, et al. Purification and characterization of an acid phosphatase that displays phosphotyrosyl-protein phosphatase activity from bovine cortical bone matrix. J Biol Chem. 1987;262:1389–1397. [PubMed] [Google Scholar]
- 15.Shekels LL. Identification of the adipocyte acid phosphatase as a PAO-sensitive tyrosyl phosphatase. Protein Sci. 1992;1:710–721. doi: 10.1002/pro.5560010603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li HC, et al. A phosphotyrosyl-protein phosphatase activity associated with acid phosphatase from human prostate gland. Eur J Biochem. 1984;138:45–51. doi: 10.1111/j.1432-1033.1984.tb07879.x. [DOI] [PubMed] [Google Scholar]
- 17.Lin MF, Clinton GM. Human prostatic acid phosphatase has phosphotyrosyl protein phosphatase activity. Biochem J. 1986;235:351–357. doi: 10.1042/bj2350351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yousef GM, et al. Molecular cloning of a novel human acid phosphatase gene (ACPT) that is highly expressed in the testis. Genomics. 2001;74:385–395. doi: 10.1006/geno.2001.6556. [DOI] [PubMed] [Google Scholar]
- 19.Fleisig H, et al. Regulation of ErbB4 phosphorylation and cleavage by a novel histidine acid phosphatase. Neuroscience. 2004;127:91–100. doi: 10.1016/j.neuroscience.2004.04.060. [DOI] [PubMed] [Google Scholar]
- 20.Van Etten RL, et al. Covalent structure, disulfide bonding, and identification of reactive surface and active site residues of human prostatic acid phosphatase. J Biol Chem. 1991;266:2313–2319. [PubMed] [Google Scholar]
- 21.Tonks NK, et al. Characterization of the major protein-tyrosine-phosphatases of human placenta. J Biol Chem. 1988;263:6731–6737. [PubMed] [Google Scholar]
- 22.Lin MF, Clinton GM. The epidermal growth factor receptor from prostate cells is dephosphorylated by a prostate-specific phosphotyrosyl phosphatase. Mol Cell Biol. 1988;8:5477–5485. doi: 10.1128/mcb.8.12.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Etten RL. Phosphomonoesterase enzymes that utilize histidine or cysteine as nucleophiles in SN2P reactions. Phosphorus Sulfur. 1993;76:107–110. [Google Scholar]
- 24.Wo YY, et al. Cloning, expression, and catalytic mechanism of the low molecular weight phosphotyrosyl protein phosphatase from bovine heart. Biochemistry. 1992;31:1712–1721. doi: 10.1021/bi00121a019. [DOI] [PubMed] [Google Scholar]
- 25.Lin MF, et al. The cellular level of prostatic acid phosphatase and the growth of human prostate carcinoma cells. Differentiation. 1994;57:143–149. doi: 10.1046/j.1432-0436.1994.5720143.x. [DOI] [PubMed] [Google Scholar]
- 26.Meng TC, Lin MF. Tyrosine phosphorylation of c-ErbB-2 is regulated by the cellular form of prostatic acid phosphatase in human prostate cancer cells. J Biol Chem. 1998;273:22096–22104. doi: 10.1074/jbc.273.34.22096. [DOI] [PubMed] [Google Scholar]
- 27.Meng TC, et al. Interaction between protein tyrosine phosphatase and protein tyrosine kinase is involved in androgen-promoted growth of human prostate cancer cells. Oncogene. 2000;19:2664–2677. doi: 10.1038/sj.onc.1203576. [DOI] [PubMed] [Google Scholar]
- 28.Igawa T, et al. Suppression of LNCaP prostate cancer xenograft tumors by a prostate-specific protein tyrosine phosphatase, prostatic acid phosphatase. Prostate. 2003;55:247–258. doi: 10.1002/pros.10240. [DOI] [PubMed] [Google Scholar]
- 29.Alonso A, et al. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 30.Jakob CG, et al. Crystal structure of human prostatic acid phosphatase. Prostate. 2000;42:211–218. doi: 10.1002/(sici)1097-0045(20000215)42:3<211::aid-pros7>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 31.Ostanin K, et al. Heterologous expression of human prostatic acid phosphatase and site-directed mutagenesis of the enzyme active site. J Biol Chem. 1994;269:8971–8978. [PubMed] [Google Scholar]
- 32.Ortlund E, et al. Crystal structures of human prostatic acid phosphatase in complex with a phosphate ion and α-benzylaminobenzylphosphonic acid update the mechanistic picture and offer new insights into inhibitor design. Biochemistry. 2003;42:383–389. doi: 10.1021/bi0265067. [DOI] [PubMed] [Google Scholar]
- 33.Ishikawa K, et al. X-ray structures of a novel acid phosphatase from Escherichia blattae and its complex with the transition-state analog molybdate. EMBO J. 2000;19:2412–2423. doi: 10.1093/emboj/19.11.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma S, et al. A DFT study on the formation of a phosphohistidine intermediate in prostatic acid phosphatase. J Am Chem Soc. 2008;130:9708–9716. doi: 10.1021/ja710047a. [DOI] [PubMed] [Google Scholar]
- 35.Zhang XQ, et al. Characterization of a prostate-specific tyrosine phosphatase by mutagenesis and expression in human prostate cancer cells. J Biol Chem. 2001;276:2544–2550. doi: 10.1074/jbc.M006661200. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka M, et al. Prostatic acid phosphatase degrades lysophosphatidic acid in seminal plasma. FEBS Lett. 2004;571:197–204. doi: 10.1016/j.febslet.2004.06.083. [DOI] [PubMed] [Google Scholar]
- 37.Zylka MJ, et al. Prostatic acid phosphatase is an ectonucleotidase and suppresses pain by generating adenosine. Neuron. 2008;60:111–122. doi: 10.1016/j.neuron.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schweingruber ME, et al. Identification and characterization of thiamin repressible acid phosphatase in yeast. J Biol Chem. 1986;261:15877–15882. [PubMed] [Google Scholar]
- 39.Beh CT, et al. An acid phosphatase as a biochemical marker for intestinal development in the nematode Caenorhabditis elegans. Dev Biol. 1991;147:133–143. doi: 10.1016/s0012-1606(05)80013-2. [DOI] [PubMed] [Google Scholar]
- 40.Pohlmann R, et al. Human lysosomal acid phosphatase: cloning, expression and chromosomal assignment. EMBO J. 1988;7:2343–2350. doi: 10.1002/j.1460-2075.1988.tb03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luchter-Wasylewska E, et al. Concentration-dependent dissociation/association of human prostatic acid phosphatase. J Protein Chem. 2003;22:243–247. doi: 10.1023/a:1025016402860. [DOI] [PubMed] [Google Scholar]
- 42.Lang P, et al. Monomeric tartrate resistant acid phosphatase induces insulin sensitive obesity. PLoS ONE. 2008;3:e1713. doi: 10.1371/journal.pone.0001713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Normanno N, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366:2–16. doi: 10.1016/j.gene.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 44.Porvari KS, et al. Site-directed mutagenesis of prostatic acid phosphatase. Catalytically important aspartic acid 258, substrate specificity, and oligomerization. J Biol Chem. 1994;269:22642–22646. [PubMed] [Google Scholar]
- 45.Groen A, et al. Redox regulation of dimerization of the receptor protein-tyrosine phosphatases RPTPα, LAR, RPTPμ and CD45. FEBS J. 2008;275:2597–2604. doi: 10.1111/j.1742-4658.2008.06407.x. [DOI] [PubMed] [Google Scholar]
- 46.Sharma S, et al. Theoretical investigations of prostatic acid phosphatase. Proteins. 2005;58:295–308. doi: 10.1002/prot.20335. [DOI] [PubMed] [Google Scholar]
- 47.Veeramani S, et al. Cellular prostatic acid phosphatase: a protein tyrosine phosphatase involved in androgen-independent proliferation of prostate cancer. Endocr Relat Cancer. 2005;12:805–822. doi: 10.1677/erc.1.00950. [DOI] [PubMed] [Google Scholar]
- 48.Lin MF, et al. Decreased expression of cellular prostatic acid phosphatase increases tumorigenicity of human prostate cancer cells. J Urol. 2001;166:1943–1950. [PubMed] [Google Scholar]
- 49.Senna R, et al. Induction of acid phosphatase activity during germination of maize (Zea mays) seeds. Plant Physiol Biochem. 2006;44:467–473. doi: 10.1016/j.plaphy.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 50.Andriotis VM, Ross JD. Isolation and partial characterization of acid phosphatase isozymes from dormant oilseed of Corylus avellana L. Planta. 2004;219:346–358. doi: 10.1007/s00425-004-1229-2. [DOI] [PubMed] [Google Scholar]
- 51.LaPointe G, et al. Characterization and site-directed mutagenesis of Wzb, an O-phosphatase from Lactobacillus rhamnosus. BMC Biochem. 2008;9:10. doi: 10.1186/1471-2091-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsuruta H, et al. Crystal structure of cold-active protein-tyrosine phosphatase from a psychrophile, Shewanella sp. J Biochem. 2005;137:69–77. doi: 10.1093/jb/mvi010. [DOI] [PubMed] [Google Scholar]
- 53.Meng TC, et al. Regulation of insulin signaling through reversible oxidation of the protein-tyrosine phosphatases TC45 and PTP1B. J Biol Chem. 2004;279:37716–37725. doi: 10.1074/jbc.M404606200. [DOI] [PubMed] [Google Scholar]
- 54.Juarez JC, et al. Superoxide dismutase 1 (SOD1) is essential for H2O2-mediated oxidation and inactivation of phosphatases in growth factor signaling. Proc Natl Acad Sci U S A. 2008;105:7147–7152. doi: 10.1073/pnas.0709451105. [DOI] [PMC free article] [PubMed] [Google Scholar]


