Summary
Addison’s disease, an immune-mediated disorder caused by destruction of the adrenal glands, is a rare disorder of Western European populations. Studies indicate that the disorder is polygenic in nature, involving specific alleles of the CTLA-4, DRB1*04 and DQ, Cyp27B1, VDR and MIC-A and -B loci. A similar immune form of Addison’s disease occurs in several breeds of domestic dog, with frequencies ranging from 1.5 to 9.0%. The high frequency of the disease in domestic dog breeds likely reflects the small number of founders associated with many breeds, subsequent inbreeding, and the frequent use of popular sires.
The Portuguese Water Dog (PWD) is a significantly affected breed. An analysis of 11 384 PWDs surveyed between 1985 and 1996 suggests a breed-specific disease incidence of 1.5%. As with humans, the disease is typically of late onset.
This study involves a genetic comparison of Addison’s disease in the PWD to the analogous disease in humans. The study is facilitated by the existence of complete pedigrees and a relatively high degree of inbreeding among PWDs. The breed originated from 31 founders, with 10 animals responsible for 90% of the current gene pool. We describe, specifically, the identification of two disease-associated loci, on Canis familiaris (CFA) chromosomes CFA12 and 37, which are syntenic with the human DRB1 histocompatibility locus alleles HLA-DRB1* 04 and DRB1*0301, and to a locus for immunosuppression syntenic with CTLA-4. Strong similarities exist therefore in the complex genetic background of Addison’s disease in humans and in the PWD. With the completion of the canine and human genome sequence, the purebred dog is set to become an important comparative model for Addison’s as well as other human immune disorders.
Introduction
Genetic mapping of complex disease traits and, in particular, those associated with autoimmune disease, remains a challenge for researchers interested in both human medicine and companion animal health. One autoimmune disorder of particular interest is Addison’s disease (primary adrenocortical insufficiency) that is characterized by destruction of the adrenal cortex, resulting in the inability to produce cortisone when stimulated with adrenocorticotrophic hormone (ACTH). While the disease is relatively rare in the Western European human population, studies suggest a polygenic aetiology involving many loci including alleles of CTLA-4 (Vaidya et al., 2000; Blomhoff et al., 2004), DRB1*04 and DQ (Yu et al., 1999; Bilbao et al., 2003), Cyp27B1 (Lopez et al., 2004), VDR (Pani et al., 2002), as well as MIC-A and -B (Gambelunghe et al., 1999).
The domestic dog (Canis familiaris) is well established as a clinical model for Addison’s disease, with a similar presentation, age at onset and response to clinical interventions. The disease occurs in several breeds (Pedersen, 1999), with frequencies ranging from 1.5% to as high as 9% for Standard Poodles and Bearded Collies (Pedersen, 1999; Oberbauer et al., 2002). As with humans, low serum cortisol levels, following ACTH stimulation, are used to establish a clinical diagnosis.
Because diseases like Addison’s are likely to involve many interacting loci, genetic analyses are typically restricted to populations where the variant alleles of interest are sufficiently concentrated, resulting in high disease frequencies. The comparatively higher frequency of Addison’s disease in some dog breeds compared to others may reflect founder effects, inbreeding and/or frequent use of ‘popular sires’. A comprehensive list of affected breeds would be difficult to establish, but would primarily include Standard Poodles, Duck Tolling Retrievers, Leon-bergers, Portuguese Water Dogs (PWD), Bearded Collies (Oberbauer et al., 2002; Famula et al., 2003) and West Highland White Terriers (AKC Canine Health Foundation, personal communication). There is some suggestion that the disease is more common in purebred domestic dog than mixed-breed dogs (Melian et al., 1999). Genetic, rather than environmental, factors are likely to be the most important predisposing factor associated with observed breed specificity (Pedersen, 1999). It is unclear if the same genes are responsible for the disease in different breeds, however, as we demonstrate here, breeds with the appropriate population structure can be use to identify quantitative trait loci (QTLs) associated with the disease.
Prior to any disease gene mapping, population genetic studies of the disease are often used to develop statistical models for subsequent linkage analysis. Towards this end, Famula and colleagues (2003) undertook a complex segregation analysis of the disease in Standard Poodles, evaluating 778 Standard Poodles of known Addisonian phenotypes. Disease was confirmed clinically by ACTH challenge for 8.6% of dogs. Heritability was estimated to be 0.75 and the segregation analyses suggested that the disease is influenced by at least one autosomal recessive locus (Famula et al., 2003). The same researchers also examined the incidence of the disease in the Bearded Collie using a large data set of 635 dogs, of which 9.4% were affected (Oberbauer et al., 2002). However, the analysis did not provide support for a simple autosomal dominant mode of inheritance; indeed, the analysis did not support claims for a single locus of large effect assuming any mode of inheritance.
The PWD breed offers some specific advantages for the study of complex traits like Addison’s disease over other breeds. The breed is relatively small, and all dogs registered today can trace their lineage to two founding kennels, termed Algarbiorum and Alvalade. The breed club is highly motivated to improve the health of their breed, and collaboration is facilitated through a program of interaction between owners and scientists at the University of Utah called the Georgie Project (www.georgieproject.com). PWD owners in the USA have provided DNA samples and phenotypic information on over 1000 dogs. Finally, the breed has been the subject of intense study for identification of QTLs association with morphological variation (Chase et al., 1999, 2002, 2005a). Thus, a large amount of molecular data has already been collected on an extensive and well-characterized population of PWDs.
Analysis of data from 11 384 PWDs born between 1985 and 1996 suggests that the frequency of Addison’s in the PWD is about 1.5% (K. Chase, K. Miller and K.G. Lark et al., unpublished data). This estimate may not be representative of the breed as a whole because it derives solely from dogs participating in the Georgie Project. Nevertheless, Addison’s disease is clearly a significant health problem for PWDs.
In this report, we examine the genetic basis for delayed onset Addison’s disease in the PWD. We describe, specifically, the identification of two loci, located on chromosomes CFA12 and 37, syntenic with alleles of the human major histocompatability complex (such as HLA-DRB1*04 and HLA-DQA1 *0301) and a locus for immunosuppression (CTLA-4), respectively, that contribute to Addison’s disease in the PWD.
Materials and methods
The present US population of approximately 20 000 American Kennel Club (AKC)-registered PWDs is derived from 31 founders, of which 10 are responsible for more than 90% of the current gene pool (Molinari, 1993; Chase et al., 1999). Pedigree records are complete and their accuracy has been validated by comparing experimentally determined consanguinities based on alleles of microsatellite DNA markers with consanguinities derived from the pedigree records (Chase et al., 1999). Consanguinities in the current population range from 0.0 to 0.6 with a mean of 0.2 (Chase et al., 1999).
Previously, to facilitate research on skeletal structure, we adapted statistical and computational tools to analyse the genetic and phenotypic relationships within the PWD population and have used these to identify QTLs regulating skeletal size and shape (Chase et al., 2002, 2004; 2005a, b; Carrier et al., 2005; Lark et al., 2005). This necessitated completing a genome-wide scan on approximately 800 PWDs. The work reported here is based on the initial genome-wide scans done for the above-mentioned morphology study. Specifically, the data reported here are based on a total of 682 largely tetranucleotide-based microsatellite markers (simple sequence repeats or SSRs) that were selected from publicly available resources (Breen et al., 2004). A total of 806 AKC-registered PWDs had been previously genotyped using these markers, including 57 Addisonians. An additional 148 dogs in the pedigree have confirmed Addison’s disease, but their samples were unavailable for genotyping, primarily due to the death of the dog.
A genetic map detailing the associated marker coverage of the genome has been published elsewhere (Carrier et al., 2005). Details regarding polymerase chain reaction conditions, primers and microsatellite variants can be found at web sites describing the canine map http://idefix.Univ-rennes1.fr:8080/Dogs/RH10K-SOM.html. The complete canine genome map can be accessed at http://www.genome.ucsc.edu; http://www.ncbi.nih.gov; and http://www.ensembl.org
We tested for associations between Addison’s disease and a marker allele using Fisher’s exact test (Agresti, 1990). All alleles with a population frequency greater than 0.05 for 682 markers across the genome were included in the analysis. In all, 2223 marker alleles were tested. Data from 57 affected genotyped dogs and 749 unaffected genotyped dogs were considered.
Narrow sense heritability is estimated as one half of the slope of the regression line of phenotypic similarity (Ritland, 1996) on to the pedigree estimation of consanguinity between each pair of dogs (Falconer & Mackay, 1996). This is the ratio of the additive variance to the total variance. A total of 5000 permutation tests were used to establish the significance of the heritability estimate.
Results and discussion
To date, 205 cases of Addison’s disease have been documented involving dogs within the extensive PWD pedigree assembled at the University of Utah. Of the 205, 136 are female and 69 are male. Age at diagnosis was available for 124 of these dogs. Based on these data, a minimum estimate of disease incidence is 1.5%; however, incorrect diagnoses, as well as unreported cases, could increase that figure appreciably. Comparison of the consanguinity of Addisonian with non-Addisonian dogs demonstrated a significant genetic component to the PWD disease. However, the additive heritability was low (narrow sense heritability is 0.039, P < 0.001), suggesting that environmental effects may act on allele penetrance. In addition, the disease may be caused by interacting loci. The late onset, together with the different incidence in males and females, is consistent with an autoimmune aetiology.
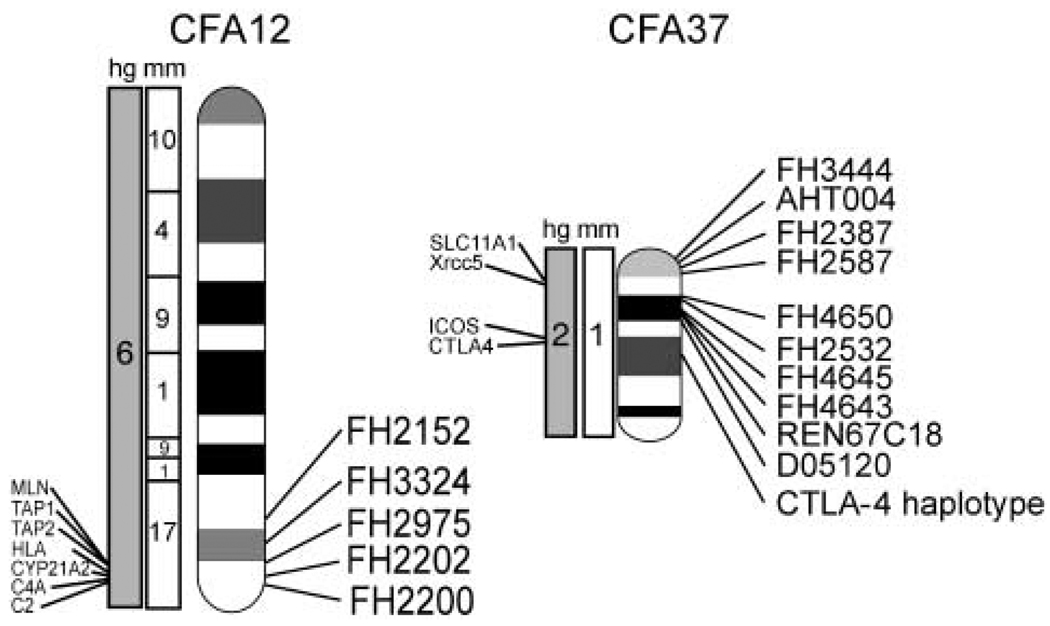
In this study, association between marker and disease state was sought across the entire canine genome using 682 microsatellite-based markers. Data from 57 affected genotyped dogs and 749 unaffected genotyped dogs were considered. QTLs were identified on CFA12 and CFA37 (Fig. 1). Markers on CFA12 are associated significantly with an increase in the frequency of the disease. By comparison, a marker on CFA37 was associated with a decrease in the frequency of the disease. The parameters of these associations and all markers in the corresponding regions are presented in Table 1 and Table 2.
Figure 1.
Diagrams of canine autosomes CFA12 and CFA37 and markers associated with the quantitative trait loci are discussed in the text. Details of their association are presented in Table 1. Syntenic portions of the mouse (mm) and human (hg) genomes are noted, as well as the candidate gene positions.
Table 1.
Association of markers on canine chromosomes 12 and 37 with increased and decreased Addisonian frequency
| Marker name | Dog chromosome |
Base pair positiona |
Allele size |
Frequency total |
Frequency in affectedsb |
Raw P value |
Adjusted P value |
|---|---|---|---|---|---|---|---|
| FH2200 | 12 | 3943089 | 402 | 0.32 | 0.44 | 0.005901 | 1 |
| FH2200 | 12 | 3943089 | 406 | 0.51 | 0.38 | 0.010242 | 1 |
| FH2200 | 12 | 3943089 | 430 | 0.10 | 0.10 | 1 | 1 |
| FH2202 | 12 | 5281978 | 402 | 0.07 | 0.04 | 0.237749 | 1 |
| FH2202 | 12 | 5281978 | 446 | 0.49 | 0.39 | 0.032355 | 1 |
| FH2202 | 12 | 5281978 | 450 | 0.11 | 0.05 | 0.058462 | 1 |
| FH2202 | 12 | 5281978 | 462 | 0.28 | 0.51 | 1.02E-07 | 0.000226 |
| FH2975 | 12 | 7347139 | 280 | 0.15 | 0.10 | 0.130924 | 1 |
| FH2975 | 12 | 7347139 | 284 | 0.34 | 0.27 | 0.145558 | 1 |
| FH2975 | 12 | 7347139 | 300 | 0.29 | 0.51 | 3.58E-07 | 0.000795 |
| FH2975 | 12 | 7347139 | 336 | 0.05 | 0.03 | 0.478362 | 1 |
| FH2975 | 12 | 7347139 | 352 | 0.11 | 0.05 | 0.057436 | 1 |
| FH3324 | 12 | 10396440 | 330 | 0.26 | 0.44 | 1.48E-05 | 0.032816 |
| FH3324 | 12 | 10396440 | 338 | 0.12 | 0.12 | 1 | 1 |
| FH3324 | 12 | 10396440 | 426 | 0.20 | 0.15 | 0.208464 | 1 |
| FH3324 | 12 | 10396440 | 430 | 0.37 | 0.26 | 0.021096 | 1 |
| FH2152 | 12 | 14019604 | 374 | 0.05 | 0.03 | 0.271026 | 1 |
| FH2152 | 12 | 14019604 | 414 | 0.45 | 0.31 | 0.003446 | 1 |
| FH2152 | 12 | 14019604 | 418 | 0.10 | 0.09 | 0.869032 | 1 |
| FH2152 | 12 | 14019604 | 422 | 0.06 | 0.09 | 0.210427 | 1 |
| FH2152 | 12 | 14019604 | 434 | 0.26 | 0.40 | 0.000797 | 1 |
| D05120 | 37 | 21315646 | 105 | 0.51 | 0.57 | 0.16189 | 1 |
| D05120 | 37 | 21315646 | 107 | 0.49 | 0.43 | 0.16189 | 1 |
| REN67C18 | 37 | 22361181 | 134 | 0.08 | 0.06 | 0.693665 | 1 |
| REN67C18 | 37 | 22361181 | 136 | 0.39 | 0.29 | 0.042277 | 1 |
| REN67C18 | 37 | 22361181 | 138 | 0.18 | 0.18 | 1 | 1 |
| REN67C18 | 37 | 22361181 | 140 | 0.35 | 0.45 | 0.037226 | 1 |
| FH4643 | 37 | 22849867 | 190 | 0.34 | 0.29 | 0.328812 | 1 |
| FH4643 | 37 | 22849867 | 224 | 0.16 | 0.14 | 0.576227 | 1 |
| FH4643 | 37 | 22849867 | 226 | 0.32 | 0.45 | 0.005667 | 1 |
| FH4643 | 37 | 22849867 | 230 | 0.16 | 0.12 | 0.261422 | 1 |
| FH4645 | 37 | 23392170 | 314 | 0.19 | 0.11 | 0.019324 | 1 |
| FH4645 | 37 | 23392170 | 326 | 0.25 | 0.29 | 0.345437 | 1 |
| FH4645 | 37 | 23392170 | 330 | 0.32 | 0.35 | 0.511924 | 1 |
| FH4645 | 37 | 23392170 | 334 | 0.12 | 0.16 | 0.273219 | 1 |
| FH2532 | 37 | 25197477 | 338 | 0.24 | 0.09 | 2.07E-05 | 0.045953 |
| FH2532 | 37 | 25197477 | 346 | 0.31 | 0.37 | 0.139822 | 1 |
| FH2532 | 37 | 25197477 | 354 | 0.05 | 0.08 | 0.109296 | 1 |
| FH2532 | 37 | 25197477 | 398 | 0.30 | 0.36 | 0.168622 | 1 |
| FH4650 | 37 | 25596157 | 348 | 0.05 | 0.06 | 0.62649 | 1 |
| FH4650 | 37 | 25596157 | 352 | 0.16 | 0.25 | 0.0279 | 1 |
| FH4650 | 37 | 25596157 | 364 | 0.32 | 0.36 | 0.450064 | 1 |
| FH4650 | 37 | 25596157 | 372 | 0.16 | 0.19 | 0.406419 | 1 |
| FH4650 | 37 | 25596157 | 380 | 0.23 | 0.12 | 0.005589 | 1 |
| FH2587 | 37 | 29726009 | 192 | 0.25 | 0.23 | 0.641298 | 1 |
| FH2587 | 37 | 29726009 | 196 | 0.30 | 0.28 | 0.825373 | 1 |
| FH2587 | 37 | 29726009 | 200 | 0.41 | 0.45 | 0.412508 | 1 |
| FH2387 | 37 | 30837793 | 384 | 0.14 | 0.11 | 0.376054 | 1 |
| FH2387 | 37 | 30837793 | 404 | 0.18 | 0.17 | 1 | 1 |
| FH2387 | 37 | 30837793 | 408 | 0.59 | 0.68 | 0.048651 | 1 |
| FH2387 | 37 | 30837793 | 416 | 0.05 | 0.03 | 0.622066 | 1 |
| AHT004 | 37 | 31321462 | 100 | 0.07 | 0.05 | 0.542715 | 1 |
| AHT004 | 37 | 31321462 | 112 | 0.42 | 0.27 | 0.001237 | 1 |
| AHT004 | 37 | 31321462 | 114 | 0.15 | 0.13 | 0.478817 | 1 |
| AHT004 | 37 | 31321462 | 124 | 0.36 | 0.56 | 2.58E-05 | 0.057385 |
| FH3444 | 37 | 32143327 | 270 | 0.59 | 0.77 | 6.3E-05 | 0.140142 |
| FH3444 | 37 | 32143327 | 318 | 0.16 | 0.10 | 0.100776 | 1 |
| FH3444 | 37 | 32143327 | 378 | 0.22 | 0.11 | 0.002545 | 1 |
The dog chromosome position of each marker is given in Mb.
The significant marker allele associated with the phenotype is given followed by its frequency in the total population, as well as its frequency in the affected population. Significance of association between marker and QTL (raw P values) is corrected by a factor of 2223 for the number of trials (marker alleles tested).
Table 2.
Marker name, position, base pair, primer sequences, and repeat motif for relevant markers on CFA12 and 37
| Marker namea | Dog chromosome | Base pair position | Forward primer | Reverse primer | Repeat |
|---|---|---|---|---|---|
| FH2200 | 12 | 3943089 | CATGATCCTGGAGTCCCG | GAAAGCTGCTTCAGTGGACC | GAAA |
| FH2202 | 12 | 5281978 | GTTGAGTGGTTGCCTTTAGC | CAGGATCTTCATATGTCACC | AAGA |
| FH2975 | 12 | 7347139 | TGAACCAATTGAGGAACACA | TGGGGTCTTGAGTTTGAGTC | CTTT |
| FH3324 | 12 | 10396440 | TGCATATCAGACTGCCAACC | TCAGTGGTTTAGAGCCTTCG | CTTT |
| FH2152 | 12 | 14019604 | ACCCCTCTTCATGCTCTCTG | AGCATATCAGTAGGGAAGGGAG | GAAA |
| D05120 | 37 | 21315646 | ACTCTGCTGTATAGACATCTTGT | AGCAGAGGACTATGGGAAATAAC | TG |
| REN67C18 | 37 | 22361181 | TCTGTGCGTTTCCGTTTATG | TTAGTACCTGTTTGTTATCC | CA |
| FH4643 | 37 | 22849867 | GGTAGCTGGAAGGAATCTGG | GCATCTGTTTCAGCCTTTTT | TG |
| FH4645 | 37 | 23392170 | CAAAGACATTGAGGCGTCTT | CTGCAAAAGGAGAGACTGGA | AAAG |
| FH2532 | 37 | 25197477 | CACGCAGAAAGGCAGAAAG | TTTCCATAGTGGCTGCATCA | AAAG |
| FH4650 | 37 | 25596157 | AGCTCCCGGCTTAGGATCT | CCTTGGCAATTGGAAAAAGA | CTTT |
| FH2587 | 37 | 29726009 | GGCATGAACAAATCAGTGGA | TTTGCTGTTTAATCCATCTGG | AAAG |
| FH2387 | 37 | 30837793 | TTGTTCACTCAGCTAGGAGACG | TTTTTATTCAACAGCAGCTAGGC | TTTC |
| AHT004 | 37 | 31321462 | CATCATGCATCAAGCAGAGC | TCATGTAAGCAGAGACTGAC | TG |
| FH3444 | 37 | 32143327 | GCAGTCCCTTCTGATCACACT | AGATACCACTCTGGGCACTG | CTTT |
Crucial information for a subset of markers listed in Table 1.
With regard to CFA12, of particular interest on Table 1 are two markers, FH2202 and FH2975, that have adjusted P = 0.00022, 0.00079, respectively, for alleles 462 and 300. These two markers are positioned only about 2 million base pairs apart. Supportive data are also provided by maker FH3324, which is another 3 million base pairs closer to the centromere, but has a corrected P = 0.03 for allele 330. The lower P value suggests that the linkage signal begins to drop off at this position. These markers are within the canine DLA region. Thus, the locus on CFA12 is associated with increased frequency of disease, supporting our assumption that late onset Addison’s in the PWD is an autoimmune disease. The canine counterparts of human HLA-DRB1*04 and HLA-DQ (Yu et al., 1999; Bilbao et al., 2003) are primary candidate genes and are currently under investigation.
For CFA37, we note a single marker, FH2532, at position 25 Mb on CFA37 that shows an adjusted P = 0.045 for allele 338. This marker is about 10 million bases from the CTLA-4 locus, which is an excellent candidate gene. More markers need to be tested in both this region on CFA37 and on CFA12 to select the best candidate genes for detailed study.
Table 3 lists each candidate in the linkage regions for both CFA12 and 37, as well as their position on their respective chromosome based on the CANFam1.0 assembly (Lindblad-Toh et al., 2005).
Table 3.
Names and positions for candidate genes in the 20 Mb region around the quantitative trait loci listed in Table 1
| Candidate genesa | Dog chromosome | Base pair position |
|---|---|---|
| CTLA-4 | 37 | 15617156 |
| ICOS | 37 | 15682406 |
| Xrcc5 | 37 | 26085873 |
| SLC11A1 | 37 | 28017155 |
| C2 | 12 | 4311579 |
| C4A | 12 | 4356840 |
| CYP21A2 | 12 | 4373931 |
| HLA-DRA | 12 | 5054036 |
| HLA-DRB3 | 12 | 5072536 |
| HLA-DRB1 | 12 | 5072536 |
| HLA-DRB5 | 12 | 5072561 |
| HLA-DRB4 | 12 | 5072562 |
| HLA-DPB1 | 12 | 5073680 |
| HLA-DQA1 | 12 | 5141465 |
| HLA-DQB1 | 12 | 5165247 |
| HLA-DQB1 | 12 | 5225549 |
| TAP2 | 12 | 5319648 |
| Tap1 | 12 | 5345683 |
| MLN | 12 | 6057634 |
Column 1 lists candidate genes of interest, column 2 lists the canine chromosome on which each gene is located based on the CANFam1.0 assembly. The base pair position on each canine chromosome is listed in column 3.
Recent studies in humans have implicated the cytotoxic T lymphocyte antigen-4 (CTLA-4) gene region as having a role in Addison’s disease in human populations (Blomhoff et al., 2004), as well as a role in other immune-mediated diseases (Ide et al ., 2004; Zhernakova et al., 2005). This gene is located about 10 Mb from the most closely linked CFA37 marker (FH2532) that we found to be associated with Addison’s disease in the PWD population in the initial genome-wide screen. Consequently, we analysed the CTLA-4 sequence directly. We sequenced all exons and splice junctions associated with CTLA-4, together with a limited amount of intron 1, all of short intron 2 and half of intron 3, plus ~1.5kb of 5′ promoter regions out to 10 kb upstream and 5 kb downstream. Sequence comparison between 10 affected and 10 unaffected dogs revealed three different haplotypes in the breed. Further analysis of 48 affected and 181 unaffected dogs showed a weak non-significant association between the disease state and the rarest haplotype. Specifically, of the three CTLA-4 haplotypes, two are shared approximately equally between affected and unaffected dogs. The third and more rare haplotype is seen more frequently in unaffected dogs (one chromosome from 48 affected dogs and 18 from the 181 unaffected animals). Markers making up haplotypes were largely from non-coding regions or encompassed alternations that did not change the encoded amino acid. Thus, no conclusions can be drawn about any of the SNPs and potential disease association.
The significance of the association between the CTLA-4 region and Addison’s disease (P = 0.09) is much less than the significance of the FH2532 marker (2.7 × 10−5). Given these results, we hypothesize that the CTLA-4 gene is unlikely to be the underlying cause of the CFA37 QTL that decreases the frequency of the disease. Thus, our data implicate a second gene in this region that may act as an immune suppressor. Possible candidate genes are listed in Table 3.
As more Addisonian PWDs are diagnosed within the Georgie Project, both the sequence and the haplotype analyses will be expanded in an attempt to find the relevant disease variants, particularly on CFA37, and, ultimately, to determine if the orthologous human gene is contributing to Addison’s in humans. This work is particularly facilitated by the canine system for several reasons (Parker & Ostrander, 2005). First, Parker et al.(2004) have established phylogeny of domestic dog breeds using cluster analysis. These data suggest that closely related breeds may be particularly susceptible to a disease because they share a common ancestral mutation. This fact can be used to facilitate the identification of a disease gene, as a common haplotype is certain to exist around the disease allele in breeds that share the same ancient mutation. Consequently, we are in the process of collecting Addisonian dogs from related breeds. In an unstructured cluster analysis using the program structure, the PWD falls into a large cluster of breeds that includes what we have historically called the Hunting group, as it contains all the Spaniel, Pointer and Retriever breeds (Parker & Ostrander, 2005). Breeds with significant levels of Addison’s like the Poodle do not fall in this group, and likely have an independent origin for the disease.
We are therefore focusing on breeds like the Retrievers. We will use the available SNP data, generated from the 6.5× Boxer and 1.5× Poodle sequences (Kirkness et al., 2003 and Lindblad-Toh et al., 2005) to select and genotype SNPs across the regions of linkage in affected and unaffected dogs from multiple breeds. Because of meiotic recombination, the region of shared haplotype among dogs with Addison’s is likely to be smaller than the haplotype observed in any single breed. This will permit more judicious selection of candidate genes for further study.
Acknowledgements
This research was supported by major gifts from the Judith L. Chiara Foundation and the Nestle Purina Co. as well as gifts from > 100 PWD owners, in part by an extramural grant (GM 063056) from the NIH to KGL, and in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health. We thank Dr Nathan Sutter for useful suggestions during the course of this research.
References
- Agresti A. Categorical Data Analysis. New York: John Wiley; 1990. [Google Scholar]
- Bilbao JR, Martin-Pagola A, Perez de Nanclares G, Calvo B, Vitoria JC, Vazquez F, Castano L. HLA-DRB1 and MICA in autoimmunity: common associated alleles in autoimmune disorders. Annals of New York Academy of Sciences. 2003;1005:314. doi: 10.1196/annals.1288.049. [DOI] [PubMed] [Google Scholar]
- Blomhoff A, Lie BA, Myhre AG, Kemp EH, Weetman AP, Akselsen HE, Huseby ES, Undlien DE. Polymorphisms in the cytotoxic T lymphocyte antigen-4 gene region confer susceptibility to Addison’s disease. Journal of Clinical Endocrinology & Metabolism. 2004;89:3474. doi: 10.1210/jc.2003-031854. [DOI] [PubMed] [Google Scholar]
- Breen M, Hitte C, Lorentzen TD, Thomas R, Cadieu E, Sabacan L, et al. An integrated 4249 marker FISH/RH map of the canine genome. BMC Genomics. 2004;5:1. doi: 10.1186/1471-2164-5-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrier DR, Chase K, Lark KG. Genetics of canid skeletal variation: size and shape of the pelvis. Genome Research. 2005;15:1825. doi: 10.1101/gr.3800005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase K, Adler FR, Miller-Stebbings K, Lark KG. Teaching a new dog old tricks: identifying quantitative trait loci using lessons from plants. Journal of Heredity. 1999;90:43. doi: 10.1093/jhered/90.1.43. [DOI] [PubMed] [Google Scholar]
- Chase K, Carrier DR, Adler FR, Jarvik T, Ostrander EA, Lorentzen TD, Lark KG. Genetic basis for systems of skeletal quantitative traits: Principal component analysis of the canid skeleton. Proceedings of the National Academy of Sciences of the USA. 2002;99:9930. doi: 10.1073/pnas.152333099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase K, Lawler DF, Adler FR, Ostrander EA, Lark KG. Bilaterally asymmetric effects of quantitative trait loci (QTLs): QTLs that affect laxity in the right versus left coxofemoral (hip) joints of the dog (Canis familiaris) American Journal of Medical Genetics. 2004;124:239. doi: 10.1002/ajmg.a.20363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase K, Carrier DF, Adler FR, Ostrander EA, Lark KG. Size sexual dimorphism in Portuguese Water Dogs: interaction between an autosome and the X chromosome. Genome Research. 2005a;15:1825. doi: 10.1101/gr.3712705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase K, Lawler DF, Carrier DR, Lark KG. Genetic regulation of osteoarthritis: a QTL regulating cranial and caudal acetabular osteophyte formation in the hip joint of the dog (Canis familiaris) American Journal of Human Genetics. 2005b;135:334. doi: 10.1002/ajmg.a.30719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer DS, Mackay T. Introduction to Quantitative Genetics. New York: Longman; 1996. [Google Scholar]
- Famula TR, Belanger JM, Oberbauer AM. Heritability and complex segregation analysis of hypoadrenocorticism in the standard poodle. Journal of Small Animal Practice. 2003;44:8. doi: 10.1111/j.1748-5827.2003.tb00096.x. [DOI] [PubMed] [Google Scholar]
- Gambelunghe G, Falorni A, Ghaderi M, Laureti S, Tortoioli C, Santeusanio F, Brunetti P, Sanjeevi CB. Microsatellite polymorphism of the MHC class I chain-related (MIC-A and MIC-B) genes marks the risk for autoimmune Addison’s disease. Journal of Clinical Endocrinology & Metabolism. 1999;84:3701. doi: 10.1210/jcem.84.10.6069. [DOI] [PubMed] [Google Scholar]
- Ide A, Kawasaki E, Abiru N, Sun F, Kobayashi M, Fukushima T, et al. Association between IL-18 gene promoter polymorphisms and CTLA-4 gene 49A/G polymorphism in Japanese patients with type 1 diabetes. Journal of Autoimmunity. 2004;22:73. doi: 10.1016/j.jaut.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Kirkness EF, Bafna V, Halpern AL, Levy S, Remington K, Rusch DB, et al. The dog genome: survey sequencing and comparative analysis. Science. 2003;301:1898. doi: 10.1126/science.1086432. [DOI] [PubMed] [Google Scholar]
- Lark KG, Chase K, Carrier DR, Adler FR. Genetic analysis of the canid skeleton: morphological loci in the Portuguese Water Dog population. In: Ostrander EA, Giger U, Lindblad-Toh K, editors. The Dog and its Genome. Vol Monograph 44. New York: Cold Spring Harbor Laboratory Press; 2005. p. 67. [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Lopez ER, Zwermann O, Segni M, Meyer G, Reincke M, Seissler J, Herwig J, Usadel KH, Badenhoop K. A promoter polymorphism of the CYP27B1 gene is associated with Addison’s disease, Hashimoto’s thyroiditis, Graves’ disease and type 1 diabetes mellitus in Germans. European Journal of Endocrinology. 2004;151:193. doi: 10.1530/eje.0.1510193. [DOI] [PubMed] [Google Scholar]
- Melian C, Stefanacci J, Peterson ME, Kintzer PP. Radiographic findings in dogs with naturally-occurring primary hypoadrenocorticism. Journal of the American Animal Hospital Association. 1999;35:208. doi: 10.5326/15473317-35-3-208. [DOI] [PubMed] [Google Scholar]
- Molinari C. The Portuguese Water Dog. Lisbon, Portugal: ELO-Publicidade; 1993. [Google Scholar]
- Oberbauer AM, Benemann KS, Belanger JM, Wagner DR, Ward JH, Famula TR. Inheritance of hypoadrenocorticism in bearded collies. American Journal of Veterinary Research. 2002;63:643. doi: 10.2460/ajvr.2002.63.643. [DOI] [PubMed] [Google Scholar]
- Pani MA, Seissler J, Usadel KH, Badenhoop K. Vitamin D receptor genotype is associated with Addison’s disease. European Journal of Endocrinology. 2002;147:635. doi: 10.1530/eje.0.1470635. [DOI] [PubMed] [Google Scholar]
- Parker HG, Kim LV, Sutter NB, Carlson S, Lorentzen TD, Malek TB, Johnson GS, DeFrance HB, Ostrander EA, Kruglyak L. Genetic structure of the purebred domestic dog. Science. 2004;304:1160. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- Parker HG, Ostrander EA. Canine genomics and genetics: running with the pack. PLOS Genetics. 2005;1:0507. doi: 10.1371/journal.pgen.0010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen NC. A review of immunologic diseases of the dog. Veterinary Immunology and Immunopathology. 1999;69:251. doi: 10.1016/S0165-2427(99)00059-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritland K. A marker-based method for inferences about quantitative inheritance in natural populations. Evolution. 1996;50:1062. doi: 10.1111/j.1558-5646.1996.tb02347.x. [DOI] [PubMed] [Google Scholar]
- Vaidya B, Imrie H, Geatch DR, Perros P, Ball SG, Baylis PH, et al. Association analysis of the cytotoxic T lymphocyte antigen-4 (CTLA-4) and autoimmune regulator-1 (AIRE-1) genes in sporadic autoimmune Addison’s disease. Journal of Clinical Endocrinological Metabolism. 2000;85:688. doi: 10.1210/jcem.85.2.6369. [DOI] [PubMed] [Google Scholar]
- Yu L, Brewer KW, Gates S, Wu A, Wang T, Babu SR, et al. DRB1*04 and DQ alleles: expression of 21-hydroxylase autoantibodies and risk of progression to Addison’s disease. Journal of Clinical Endocrinology & Metabolism. 1999;84:328. doi: 10.1210/jcem.84.1.5414. [DOI] [PubMed] [Google Scholar]
- Zhernakova A, Eerligh P, Barrera P, Weseloy JZ, Huizinga TW, Roep BO, Wijmenga C, Koeleman BP. CTLA4 is differentially associated with autoimmune diseases in the Dutch population. Human Genetics. 2005;118:58. doi: 10.1007/s00439-005-0006-z. [DOI] [PubMed] [Google Scholar]