Abstract
We present a structure of the mammalian ribosome determined at ∼8.7Å resolution by cryo-electron microscopy. A molecular model was created by docking homology models of the subunit rRNAs and conserved ribosomal proteins into the density map. We also modeled expansion segments in the subunit rRNAs and identified the positions of 20 novel proteins. In general, we find that many ribosomal proteins interact with the expansion segments to form an integrated framework that may stabilize the mature ribosome. Importantly, our structure gives a snapshot of the mammalian ribosome before the binding of an A-site tRNA. The structure also provides additional support for the idea that movements of the small subunit and L1 stalk occur during the translocation of tRNAs. Finally, new details are presented about novel inter-subunit bridges in the eukaryotic ribosome. These bridges may help reset the conformation of the ribosomal subunits in preparation for the next cycle of chain elongation.
Keywords: eukaryotic ribosome, protein translation, tRNA translocation, expansion segments
Introduction
The cytoplasmic ribosome is a signature component of free-living cells. This machine is comprised of small and large subunits, which work together to translate proteins from mRNAs with the aide of cognate amino-acylated tRNAs and additional factors (Green and Noller, 1997; Ramakrishnan, 2002). Recent crystal structures of the bacterial small subunit have provided insights into the decoding center and the basis of translational fidelity (Wimberly et al., 2000; Schluenzen et al., 2000; Pioletti et al., 2001; Carter et al., 2000; Brodersen et al., 2000; Ogle et al. 2001, Ogle and Ramakrishnan, 2005). Similarly, structures of large subunits from bacteria and archaea have revealed the architecture of functional sites including the peptidyl transferase center, the L1 stalk, the exit tunnel and a universal docking surface for factors that interact with the nascent polypeptide chain (Ban et al., 2000; Harms et al., 2001). In the 70S ribosome, the relative positions of the small and large subunits and the role of the inter-subunit bridges have also been documented in crystal structures (Yusupov et al., 2001 2006; Korostelev et al., 2006; Selmer et al., 2006; Shuwirth et al., 2005). In addition, structures of programmed bacterial ribosomes have defined a path for the translocation of mRNA and tRNAs through the inter-subunit space (Yusupova et al., 2001; Korostelev et al., 2006; Selmer et al., 2006; Berk et al., 2006).
Peptide bond formation is at the heart of translation and is catalyzed by rRNA (Schmeing et al., 2005; Jenni and Ban, 2003). During translation, the small subunit moves with a ratchet-like motion relative to the large subunit (Frank and Agrawal, 2000; Valle et al., 2003). In addition, the head of the small subunit undergoes a swiveling motion that may be coupled with lateral movements of the tRNAs (Shuwirth et al., 2005; Berk et al., 2006; Spahn et al., 2004a). The translation cycle also requires the sequential action of many factors that interact with the ribosome. Snapshots of these processes have provided insights into initiation (Allen et al., 2005), revealed the interactions of EF-Tu/amino-acylated tRNA with the ribosome (Frank et al., 2005; Valle et al., 2003a; Stark et al., 2002) and given details of EF-G mediated translocation (Datta et al., 2005; Stark et al., 2000; Diaconu et al., 2005).
The larger yeast ribosome contains many novel proteins that are not found in bacterial ribosomes (Dresios et al., 2006). In addition, numerous hyper-variable inserts are present in the subunit rRNAs that are known as expansion segments (ES) (Gerbi, 1996; Schnare et al., 1996). Parallel studies on ribosomes from the yeast Saccharomyces cerevisiae have also demonstrated a ratchet-like motion of the small subunit (Spahn et al., 2001, 2004a). In general, the translation cycle in eukaryotes is more complex than in prokaryotes, as additional regulatory steps are used to fine tune this process (Ramakrishnan, 2002).
The mammalian ribosome is even larger than the yeast ribosome (Wool et al., 1995; Dresios et al., 2006). This size difference is due primarily to expansion segments in the large subunit rRNA, as both ribosomes contain a similar number of proteins. The functions of the expansion segments are not known, but at least one insert (ES27) is required for cell viability (Sweeney et al., 1994). The energy burden of synthesizing the ES and novel proteins, coupled with their ubiquitous presence in eukaryotes, suggests that these components must play an important role in ribosome biology. The added complexity of the eukaryotic ribosome is reflected by the assembly of the small and large subunits in nucleoli. This process requires 170-200 accessory proteins and numerous snoRNPs (Rudra and Warner, 2004; Fromont-Racine et al., 2003).
In this paper, we present a molecular model of the canine 80S ribosome, based on a density map at ∼8.7Å resolution. This structure reveals the architecture of the eukaryotic ribosome, including expansion segments and α-helical rods within novel proteins. We also provide a model for mammalian RACK1 and show how it interacts with the small subunit. Our model suggests a role for the eukaryotic bridges: they may provide a restoring force to help reset the conformation of the small and large subunits at the end of the subunit ratcheting motion. This may help overcome impediments to subunit movements that arise from expansion segments located near the subunit interface. We further show that our structure with an E-site tRNA is comparable to bacterial ribosomes with bound P- and E-site tRNAs. Thus, our model corresponds to a conformation of the mammalian ribosome at the “start” of the translation cycle. Finally, our structure provides additional insights into existing models for the role of the small subunit and L1 stalk during translation.
Results
The native ribosome-channel complex
In this study, electron cryo-microscopy was used to obtain an improved, 3-dimensional (3D) density map of the canine ribosome-channel complex (RCC) from the endoplasmic reticulum (Ménétret et al., 2005; Ludtke et al., 1999; see Supplemental online material/SOM). In the map, the ribosome and the channel are at ∼8.7Å and ∼24Å resolution, respectively (Figure S1). As documented previously, the lower resolution of the channel may be due to a limited flexibility of this component (Ménétret et al., 2005). A rotational series of the RCC is shown in Figure 1, with the small subunit in gold and the large subunit in blue. Many α-helical rods and RNA A-form helices are evident on the surface of the ribosome. The “native” channel (shown in magenta) binds to a flat docking surface on the bottom of the large subunit and is comprised of Sec61 and also contains the translocon associated protein complex (TRAP) (Ménétret et al., 2005). To further our understanding of protein synthesis, we built a molecular model of the canine ribosome. We then focused our analysis on novel regions in the structure and evaluated models for movements of the L1 stalk and small subunit during translation.
Figure 1.
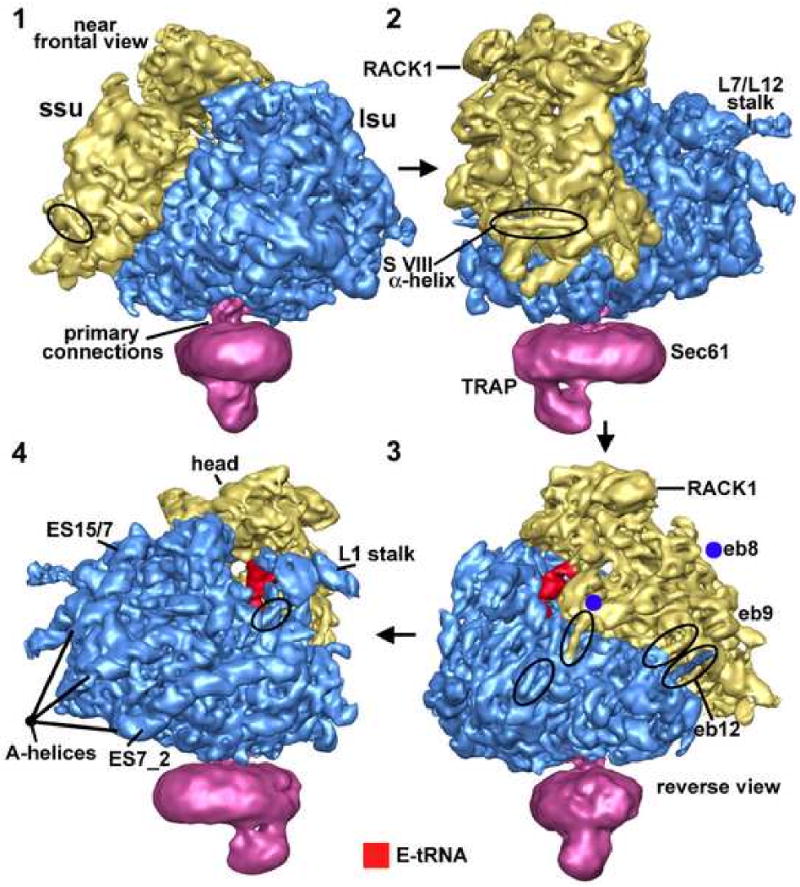
Surface views of the ribosome-channel complex.
In panels 1-4, a rotation series is shown about a vertical axis for the ribosome-channel complex with the ribosome filtered at 8.7Å resolution. The small subunit is shown in gold, the large subunit in blue and the native channel in magenta. Certain key features are indicated. In particular, some surface α-helices are circled and A-form helices of expansion segment RNAs are marked. The E-site tRNA (red) is visible in panels 3 and 4 between the small and large subunits.
Modeling conserved ribosomal RNA and proteins
Our current model of the canine ribosome incorporates crystal structures of ribosomal RNAs from the small subunit of T. thermophilus (Wimberly et al., 2000), the large subunit of H. marismortui (Ban et al. 2000), and certain regions of the 70S ribosome (Korostelev et al., 2006; Selmer et al., 2006). Overall, the fit of the conserved subunit rRNAs within the map was very good (SOM; Figure S2A); indeed only a few regions needed to be repositioned. In the next step, comparative models for conserved proteins in the small and large subunits were simultaneously docked and refined in the map with some manual adjustments (SOM, Figure S2B; Topf et al., 2006; Topf et al., submitted). In total, we modeled 16 chains in the small subunit (including RACK1) and 32 chains in the large subunit (Tables S_1, S_2).
The models and their fit in the map were validated by the correspondence between α-helices and rods in the map. In addition, β-sheets and some hairpins formed flattened features at a suitable threshold, which were used to position these structures in the map. The final cross-correlation values for the small and large subunits were 0.70 and 0.68, respectively, and included all of the modeled components (see SOM). Front and back views of the canine ribosome are shown in Figures 2A, 2C, with the modeled core rRNA and conserved proteins. The full atomic models are shown in Figures 2B and 2D without the map. For completeness, these figures also include expansion segments (red), spherical markers and rods for the novel proteins and the E-site tRNA. The quality of the overall fit can be judged by viewing interface views of the small and large subunits, as these regions are highly conserved (Figures S3A, S3B). Empty density at the periphery of the subunits arises from regions of the expansion segments and novel proteins that were not modeled explicitly. A rotation series of the small (Figure S4) and large subunits (Figure S5) shows the modeled eukaryotic proteins and their fit on the subunit rRNAs. Importantly, sequence comparisons of subunit rRNAs and conserved proteins show that the canine and human ribosomes are virtually identical. Hence, our model should be representative of the human ribosome.
Figure 2.
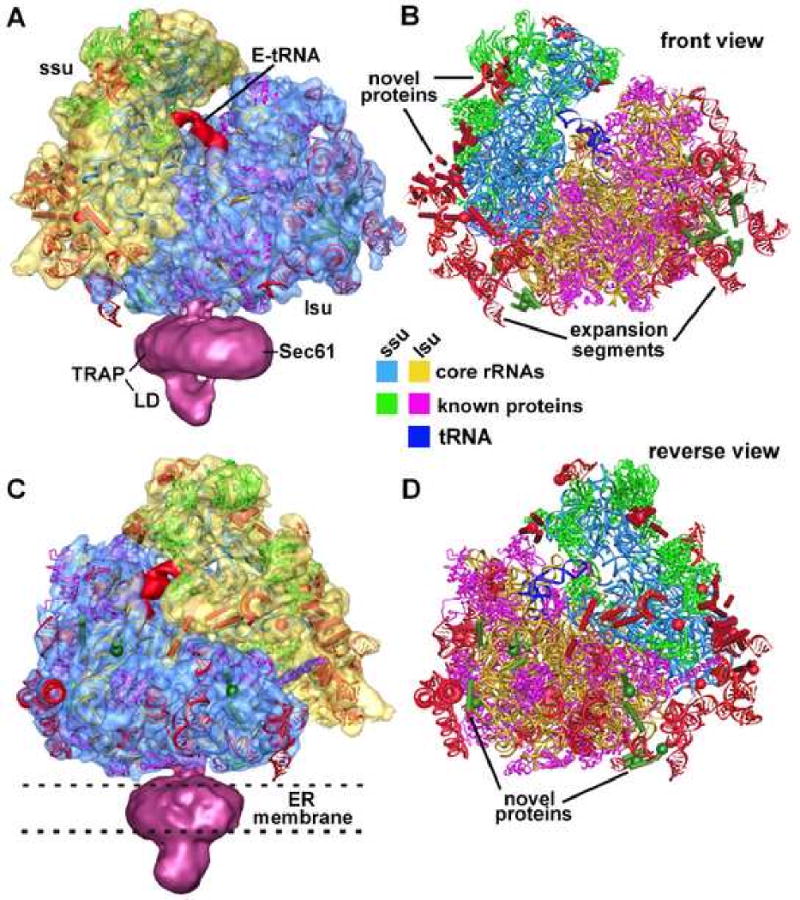
A model of the cytoplasmic 80S ribosome.
A. A model of the canine ribosome is shown within a density map of the ribosome-channel complex. The E-site tRNA is shown in red between the small (ssu) and large subunits (lsu) of the ribosome. This specimen contained a native ER channel (in magenta) comprised of Sec61 and TRAP. The latter has a prominent lumenal domain (LD).
B. The molecular model of the canine ribosome is shown in a front view. The subunit rRNAs and conserved proteins are color coded (see boxes). Novel proteins (spheres and rods) and expansion segments (red helices) are also included.
C. The model of a canine ribosome is shown in a reverse view within the EM density map. The position of the ER membrane is indicated by dashed lines.
D. The molecular model of the canine ribosome is shown in a reverse view.
Expansion segments
Hyper-variable expansion segments in the subunit rRNAs are responsible, in part, for the altered morphology of eukaryotic ribosomes relative to their prokaryotic cousins (Spahn et al., 2001; 2004b; Morgan et al., 2002; Dube et al., 1998). Although a significant fraction of the nucleotides in the expansion segments are disordered (∼50%), we find that many of these inserts are helical. In the small subunit, there are 11 distinct expansion segments (Figure S6). Most of these segments were modeled as A-form helices (Figures 3 and S6), with the exception of the distal half of ES3S and a major part of ES6S, both of which are flexible. In the large subunit, there are 16 notable expansion segments (Figures S7A, S7B); six of these hyper-variable regions are much enlarged relative to their yeast counterparts (Gerbi, 1996; Schnare et al., 1996). Together, these six segments (ES7L, 9L, 15, 27, 30 and 39) account for a mass of ∼0.4 MDa.
Figure 3.

Expansion segments and interacting proteins in the canine ribosome.
A. The model of the canine ribosome is shown in a front view with the ES depicted as red ribbons. The ssu and lsu rRNAs are shown as blue and gold ribbons, respectively. The conserved protein models are shown in green and magenta for the ssu and lsu. Proteins that interact with ES are colored yellow and cyan and are labeled with their partner ES. The novel proteins are marked by spheres and modeled α-helices are depicted as rods.
B. A reverse view is shown of the model.
C. A back view of the lsu reveals ES7L and ES15. The ES7L/ES9L/ES15 pseudo-knot (PK) is marked by a dashed ellipse near the center of the lsu. Note that expansion segments with the same numerical designation in each subunit have a superscript (S or L) to identify them.
D. A bottom view reveals the model as viewed from the ER membrane. The tunnel exit is marked with a circle (TE).
We find that ES3L and ES4L in the 5.8S rRNA form helices within a cluster of expansion segments, which includes ES5L, ES19, ES20 and ES31. This cluster is located on the side and bottom of the large subunit (Figures 3B, 3D). Notably, ES19 and ES31 form 3 helical segments but the insertion points are too close together and do not allow an unambiguous identification of two of these helices (ES19/31 in Figure 3B). Expansion segment 27 (700 nt) originates on the lower surface of the large subunit and faces the small subunit, but is almost entirely disordered. A smaller ES27 in the yeast ribosome extends towards the tunnel exit and is displaced when the Sec61 channel is bound (Beckman et al., 2001). Together, ES27 and ES41 on the large subunit combine with ES3S, ES6S and ES12S on the small subunit to form a dense network of negatively charged RNA located below the subunit interface (Figures 3A, 3B, 3D). This region also contains unmodeled flexible RNA from ES3S, ES6S and ES27. We also found that ES12L forms a small extension from the central protuberance, adjacent to two small helices that arise from ES10 and an unknown insertion (Figure 3C). A short segment (ES30) on the L1 stalk is largely disordered, but its presence could affect the binding of cellular factors to the stalk.
Four major expansion segments on the back of the large subunit form a tubular network comprised of A-form helices and knot-like structure (Figures 3C, S_7). ES7L is the largest expansion segment (813 nt) and one branch, denoted ES7_1, radiates outwards from its insertion point to form a protrusion beneath the L7/L12 stalk. ES7L also “merges” with ES9L and ES15 to form the knot-like structure on the back of the large subunit (see dashed ellipse, labeled PK; Figure 3C). A second branch of ES7L (ES7_2) extends from the ES7L/ES9L/ES15 pseudo-knot, wraps around the back of the large subunit and becomes disordered near the L1 stalk. ES9L starts near the pseudo-knot beneath ES12L and then extends away from the large subunit. A prominent spine runs from the pseudo-knot towards the central protuberance (CP) and may be formed by ES15. A lower branch also extends from the pseudo-knot. Since ES7L is rather large, it could account for one of these ES15 branches; hence, they are labeled ES15/7. Finally, ES39 forms a group of four helices beneath ES7_1 and the L7/L12 stalk (Figure 3C).
Some expansion segments interact with conserved or novel proteins (Table S_3, see next section). In addition, conserved proteins that bind to expansion segments have acquired additional basic residues through which these interactions might be formed. In the case of L35e, a newly modeled, α-helical extension binds to ES5L (Figure 3D). Most of the conserved proteins that bind to expansion segments form links between the subunit rRNA and the ES. These conserved proteins are shown with their expansion segment partners in Figure 3 and are colored yellow and cyan in the small and large subunits. The overall effect of these protein-mediated interactions is to tether proximal regions of expansion segments to the ribosome, while leaving extended and flexible regions in solution. To interact with these extended RNAs, hypothetical cellular proteins may recognize secondary structure and/or the negatively-charged surfaces of the hyper-variable regions.
Novel proteins
During the evolution of eukaryotes, 31 novel proteins have been added to the ribosome, including 17 in the small subunit and 14 in the large subunit. In our map, only 20 unknown protein densities could be identified, with ∼10 in each subunit (Table S_4). A similar number of novel proteins were identified previously in the yeast ribosome (Spahn et al., 2001). In general, the positions of these proteins are conserved, which allowed us to adopt the nomenclature of Spahn et al. (2001). Many novel proteins contain rod-like features that are likely α-helices. In total, 56 of these were modeled as α-helices in the small and large subunits. These helices are shown as rods in the small and large subunits, along with spheres to mark the novel proteins (Figures 4A and 4B). These proteins are generally found on exposed surfaces of the subunits, where they may interact with expansion segments (12 of 20 novel proteins, Table S_3).
Figure 4.

Novel proteins in the canine ribosome.
A) Novel proteins of the ssu are marked with red spheres and possible α-helices are indicated by red cylinders. The core rRNA and ES are shown as blue and red ribbons, respectively, while the conserved proteins are shown in green. The ssu is shown in back, platform and interface views.
B) Novel proteins in the lsu are marked with green spheres and cylinders for α-helices. The core rRNA and ES are shown as gold and red ribbons and the conserved proteins are colored magenta. The lsu is shown in reverse and back views.
C. Novel protein S-VIII is comprised of two long rods between ES3S, ES6S and ES12S. They may also interact with ES1S which forms a bulge.
D. Density for novel protein L-XII reveals two long, intersecting α-helices beneath the ES7L/ES9L/ES15 pseudo-knot on the back of the lsu.
For example, we identified a novel protein (S-VIII) in the small subunit that appears to be comprised of two long α-helical rods. This protein interacts with ES6S and ES12S by spanning the distance between them (Figure 4C) and may also interact with ES3S. Some unmodeled density between the rods (red sphere) may correspond to ES1S, which forms a bulge, and this density may also contain some additional protein. In the large subunit, the L-XII density contains two long α-helices (one of which is ∼90Å long) that intersect to form a “Y” (Figure 4D). These α-helices underlie the ES7L/ES9L/ES15 pseudo-knot and may interact with ES9L, ES15/7L and ES7_1. Hence, long α-helices are used in both subunits to link RNA helices in neighboring expansion segments. These α-helices may stabilize the packing of the ES rRNA against the core. Finally, a domain of the S-IV density contains α-helices and β-strands in an S6p-like configuration and this similarity was verified by a cross-correlation which gave a value of 0.65 with Chimera (Figure 7D). There is no known homolog of S6p in eukaryotes, but this S-IV domain may be related to S6p. The similarity is even more striking because the S6p and S-IV proteins are both located near the platform in the small subunit. The S-IV density links ES7S with the body of the small subunit and also forms an inter-subunit bridge (eb8).
Figure 7.

Inter-subunit bridges in the canine ribosome.
A. Known rotations of the body and head of the small subunit are indicated by arrows on the canine ribosome. Positions of the eukaryotic bridges and b2e are indicated.
B. A reverse view of the ribosome shows that the eukaryotic bridges form a contiguous line along the lateral edge of the small subunit and are also present beneath the subunits (eb11).
C. An un-modeled C-terminal extension of L37ae contacts h22 in bridge 2e.
D. An α-helix originates from S-IV and extends across the subunit interface to form eb8. Additional density from L7ae and ES31 helps to form this bridge near H76 (the L1 stalk helix).
E. Bridge 9 (eb9) involves extensive interactions between L30e and S13e. The N-terminal helix of S13e is flipped out to interact with novel protein S-IX.
F. Novel protein S-VII forms a bridge between ES3S and ES41. An icon view in the lower left shows the proximity of eb11 and eb12.
G. A C-terminal extension of L19e forms a long helix that crosses the inter-subunit gap to interact with one branch of ES6S.
The exit-site and L1 stalk
Transfer RNAs (tRNAs) are the adaptors that translate information encoded in mRNA into protein sequences. The ribosome in our map contains an exit-site tRNA, which may have originated from the displacement of a P-site tRNA after puromycin treatment of RNCs (Morgan et al., 2002). Electron density for the E-site tRNA (Figures 5A-5C, shown in silver) reveals that its conformation is similar in mammalian and bacterial ribosomes (Selmer et al., 2006, Korostelev et al., 2006). Thus, the anti-codon arm interacts with S5e while the acceptor arm is near L44e (Figures 5A, 5B). The L1 stalk is flexible and may play a role in the release of E-site tRNAs (Gomez-Lorenzo et al., 2000; Harms et al., 2001; Valle et al. 2003b). In our structure, the L1 stalk was modeled with H76-H78 from the T. thermophilus ribosome (Selmer et al., 2006). Based on the map, the D- and T-loops of the E-site tRNA contact the N-terminal domain of L10ae and H76, respectively. In a yeast ribosome with bound eEF2 and sordarin (Spahn et al., 2004a), the L1 stalk adopts an inwards conformation that partially blocks the canonical E-site (labeled Y, Figure 5C). However, the L1 stalk is displaced laterally when an E-site tRNA is bound in mammalian (gold ribbon, Figure 5C) and bacterial ribosomes (not shown; see Selmer et al., 2006; Korostelev et al., 2006). Extensive contacts made by the E-site tRNA imply that a significant conformational change of the L1 stalk may be required to release the tRNA. Indeed, an open conformation of the L1 stalk was visualized in the D. radiodurans large subunit (Figure 5C, labeled D; Harms et al., 2001) and the yeast ribosome (Spahn et al., 2004a; Morgan et al., 2002). As proposed previously, the L1 stalk may act as a lateral gate to release the E-site tRNA during translocation of the A- and P-site tRNAs (Spahn et al., 2004a; Morgan et al., 2002; Harms et al., 2001). Interestingly, we observed a tubular density that links the body of the large subunit (near L27ae and protein L-IX) to L10ae on the L1 stalk (Figure 5D, Table S_4). This novel feature was modeled as an α-helix and it may be necessary to disconnect this link when the E-site tRNA is released.
Figure 5.

The E-site tRNA, L1 and L7/L12 stalks.
A. Electron density for the E-site tRNA (in silver) is shown in a reverse view in the canine ribosome model. The anti-codon stem interacts with a β-hairpin of S5e. The tRNA density was zoned out of the 3D map with Chimera.
B. The E-site tRNA (in silver) interacts with L44e, L10ae and the top of H76 as seen in an oblique top view of the ribosome.
C. The L1 stalk may function as a lateral gate for the E-site tRNA. L1 stalks are shown from a yeast ribosome with bound eEF2 and sordarin (Y) and from a D. radiodurans crystal structure (D) in blue and red, respectively. The L1 stalk in the canine ribosome with an E-site tRNA is shown in gold and a possible hinge in H76 is marked.
D. A rod-like density links L27ae on the body of the lsu and L10ae on the L1 stalk.
E. The L7/L12 stalk is anchored by the N-terminal helix of L10p/P0 to the lsu (see asterisk). This flexible stalk may contain two dimers of P1/P2 modeled here by L12p.
The L7/L12 stalk
Currently, little is known about the structure of the L7/L12 stalk in a mammalian ribosome. In bacteria, the L7/L12 stalk forms a remarkable machine that uses motions of flexibly-linked, C-terminal domains of L7-L12 dimers to recruit EF-Tu, EF-G and other factors to the ribosome (Diaconu et al., 2005). The L7/L12 stalk is comprised of either 4 or 6 copies of the L7-L12 homologs in different species, The N-terminal domains of the L7-L12 dimers bind to helix α8 of L10p, to form an extended helix bundle in the stalk. The number of bound L12p dimers is proportional to the length of helix α8 (Diaconu et al., 2005). In eukaryotes, two copies of the L12p homologs, known as P1/P2, may be bound to the P0/L10p protein (Gonzalo & Reboud, 2003).
In our map, the L7/L12 stalk is present when the resolution is truncated to ∼12Å, but at ∼8.7Å resolution the distal region of the stalk is fragmented. This behavior results from the innate flexibility of the stalk. We docked a model of the Thermotoga maritima L10-L12 complex with two L12 dimers into the map (Figure 5E; Diaconu et al., 2005). This docking was guided by the fit of the N-terminal helix of L10p/P0 into a well resolved rod near H42 (not shown). We also introduced a slight bend in the complex to accommodate the density. While the fit is not perfect, the length of complex is consistent with the idea that the mammalian L7/L12 stalk contains two dimers of P1/P2 bound to P0.
RACK1 interactions with the small subunit
RACK1 is a conserved receptor for activated, protein kinase C and forms a β-propeller with 7 WD40 repeats. The RACK1 protein has been identified on the small subunit of eukaryotic ribosomes, where it is bound near the mRNA exit site (Figure 6, central icon; Sengupta et al., 2004). Thus, RACK1 may serve as a binding site for protein kinase C, which in turn regulates the initiation step of protein translation by phosphorylating eIF6 on the large subunit (Sengupta et al., 2004; Ceci et al., 2003). RACK1 may also direct ribosomes to focal adhesions for targeted translation (Nilsson et al., 2004).
Figure 6.
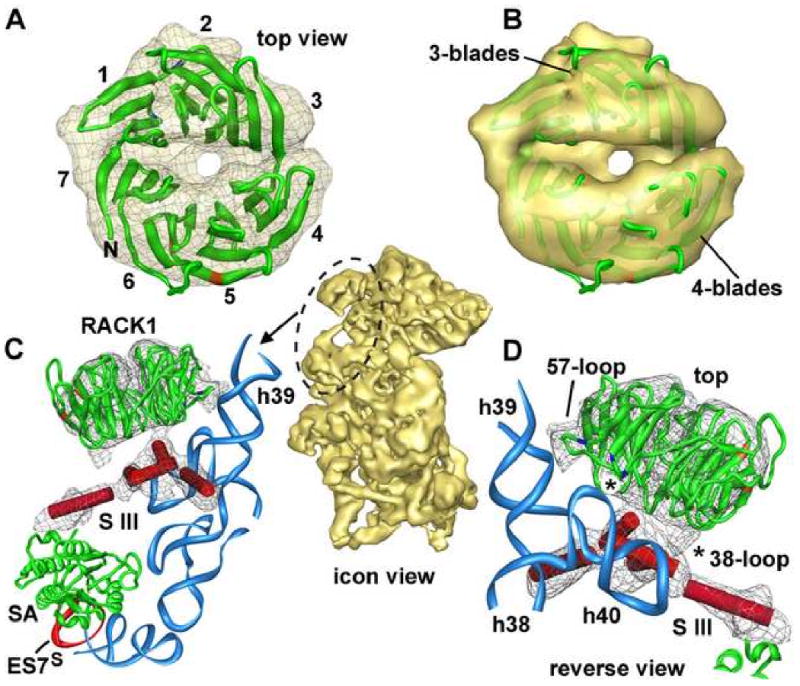
RACK1 binding to the small subunit.
A. A top view of the RACK1 homology model (green ribbon) is shown in the EM density map (grey mesh) along the pseudo 7-fold axis. The blades of the β-propeller are grouped into sets of 3- and 4-blades respectively, due to a distortion of the cylindrical symmetry. Individual blades are visible at higher thresholds (not shown). Tyrosines that may interact with SRC kinase (Y229, Y247) are shown in red.
B. A surface view of RACK1 at a low threshold shows the division into two halves with 3- and 4-blades.
C. RACK1 interacts with h39 and a novel protein density (marked S-III). This view corresponds to the region circled in the central icon of the ssu. The EM map is shown in grey mesh.
D. RACK1 is shown at higher magnification in a reverse view. Interactions are indicated between the 57-loop and h39 and between the 38-loop and h40. The S-III protein density may be an extension of SA/S0 and sits directly below RACK1.
For these studies, an homology model of canine RACK1 was created using a 7-bladed β-propeller from the PDB. When this model was docked into our map, we found that mammalian RACK1 is shifted a bit outwards, relative to its position in the yeast ribosome near h40 (Sengupta et al., 2004). However, the rotational alignment of the β-propellers is similar in the two models (not shown). Intriguingly, the 7-fold rotational symmetry of the RACK1 β-propeller is distorted somewhat by a lateral offset between opposing 3-blade and 4-blade segments (Figures 6A, 6B). This distortion could arise from the binding of the three N-terminal blades to the small subunit. However, other β-propellers display similar distortions in solution (c.f. clathrin, pdb code: 1BPO; ter Haar et al., 1998).
Our docking placed RACK1 next to h39 and showed that this receptor also interacts with a novel protein near h40 (Figure 6C). The interaction with h40 involves the “38-loop” which contains Arg36 and Lys38 (Figure 6D). The canine RACK1 also contains a loop with Arg57 that fits into density and may interact with helix h39 (near nt 1121; Figure 6D). Thus, mammalian RACK1 makes distinct contacts to helices h39 and h40 using adjacent blades of the β-propeller. In contrast, yeast RACK1 appears to make two contacts to h40 that involve the 36- and 280-loops (Sengupta et al., 2004). Yeast and human RACK1 proteins share ∼53% sequence identity, so their global folds should be similar. The different ways in which the yeast and canine RACK1 interact with rRNA may reflect evolutionary changes over ∼1.5 billion years.
RACK1 binding also involves a novel protein domain located beneath the β-propeller near h38 and h40 (Figure 6D). Intriguingly, the adjacent SA/S0 protein contains an un-modeled C-terminal extension (∼90 residues) that is specific to eukaryotes. The C-terminal extension could form a long α-helix in our map (labeled S-III). There are also sufficient residues to form a helical domain at the end of this long α-helix. This small helical domain sits on h40, contacts h38 and binds to RACK1. Importantly, this small domain would not interfere with binding partners, such as the SRC and protein C kinases, which may interact with the top and lateral surfaces of the RACK1 β-propeller (Sengupta et al., 2004).
Inter-subunit bridges
Ribosomes undergo a ratchet-like subunit rearrangement (RSR) during translation that involves a rotation of the body of the small subunit relative to the large subunit (Figure 7A; Frank and Agrawal, 2000; Valle et al., 2003b; Spahn et al., 2004a). This process is coupled with an inwards rotation of the head towards the E-site (Spahn et al., 2004a; Schuwirth et al., 2005; Berk and Cate, 2007). During the RSR, many inter-subunit bridges must rearrange locally or be transiently broken (Gao et al., 2003; Spahn et al., 2004a). The origin for the ratcheting motion of the small subunit is near bridge 2c (b2c). This rotation displaces distal regions of ES3S and ES6S by ∼20Ǻ. In addition, the exact nature of certain bridges is dependent upon the conformational state of the yeast ribosome (Spahn et al., 2004a). To place the mammalian structure in context with previous structures from yeast and bacteria, we identified bridges at the interface between the small and large subunits in the 3D map (Figure S_8; Table S_5). Interestingly, the pattern of bridges in our model is similar to that observed in a vacant yeast ribosome (Spahn et al., 2004a), as exemplified by b6, which forms a link between h14 and L23e (not shown).
We now describe bridges of particular interest in the canine ribosome. First, we observed a novel bridge (b3b) between h44 and H64 with spacing of ∼4.9Å (Figure S_8; Table S_5). Very strong density for this feature suggests the possibility of an unusual RNA conformation in this region, as there are no proteins which could contribute to this bridge. As expected, bridges specific to eukaryotes are located along the platform side of the small subunit and also beneath this subunit (Figures 7A, S_8; Spahn et al., 2004a). This alignment is shown in Figure 7B, where eukaryotic bridges (eb) 9, 12 and 11 are nearly contiguous. Bridge 2e is positioned between eb8 and eb9; this link has an additional feature that is formed by an extension of the short C-terminal α-helix of L37ae, which extends towards h22. This flattened density may correspond to a β-hairpin or an ordered loop (Figure 7C).
Many of the eukaryotic bridges involve expansion segments, novel proteins or extensions of a conserved protein. For example, eb8 is formed by an extended feature in the novel S-IV protein. The actual bridge is comprised of two helices with a flexible kink between them and connects h23 with H79 on the large subunit. In addition, a possible extension of L7ae and ES31 may help form eb8 (Figure 7D). We also found a new bridge (denoted eb8b) between h23 and H76 that is near eb8 (Figure S_8). Another bridge (eb9) is formed by S13e and L30e and is similar to one identified previously in a plant ribosome (Halic et al., 2005). In this bridge, the N-terminal α-helix of S13e is flipped outwards by ∼180° at the subunit interface and interacts with novel protein S-IX (Figure 7E). A weak contact may also be present between h11 and H63 to form eb10 (not shown). Another bridge (eb11) is formed by a novel protein (S-VII), which makes contacts with h9 and ES3S in the small subunit and ES41 in the large subunit (Figure 7F). Finally, we found that eb12 is formed by a remarkable α-helix that extends from the C-terminus of L19e. This α-helix spans a distance of ∼40Å as it crosses the subunit interface and interacts with a branch of ES6S in the small subunit (Figure 7G). The L19e extension is also present in yeast ribosomes and thus, is conserved in eukaryotes (see Schuler et al., 2006).
The actual functions of the novel eukaryotic bridges are not known, but a clue to their role may lie in the distribution of these links on the small subunit (Figures 7A, 7B). The eukaryotic bridges may provide a restoring force to counter the ratcheting motion of the small subunit (Figure 7A; Spahn et al., 2004a, Frank and Agrawal, 2000) and help reset the ribosome for the next round of protein synthesis. This process may be more complicated in higher eukaryotes due to expansion segments near the subunit interface (Figures 3A, 3B, 3D).
Conformation of the small subunit
In the yeast ribosome, a latch is present between the head and shoulder of the small subunit. This latch helps to form the mRNA entrance into the ribosome and may regulate mRNA access into the decoding center (Spahn et al., 2004a). A similar latch is present in our structure. This latch is formed by h34 and a loop of S3e in the head, which are vertically-aligned with h18 in the shoulder (Figures 8A, 8B). Remarkably, a similar latch was observed in a recent crystal structure of a programmed E. coli ribosome (Berk et al., 2006). Remarkably, the positions of the small and large subunits in the programmed E. coli ribosome are similar to the subunits in our model, even though a P-site tRNA is absent in the canine ribosome (not shown).
Figure 8.
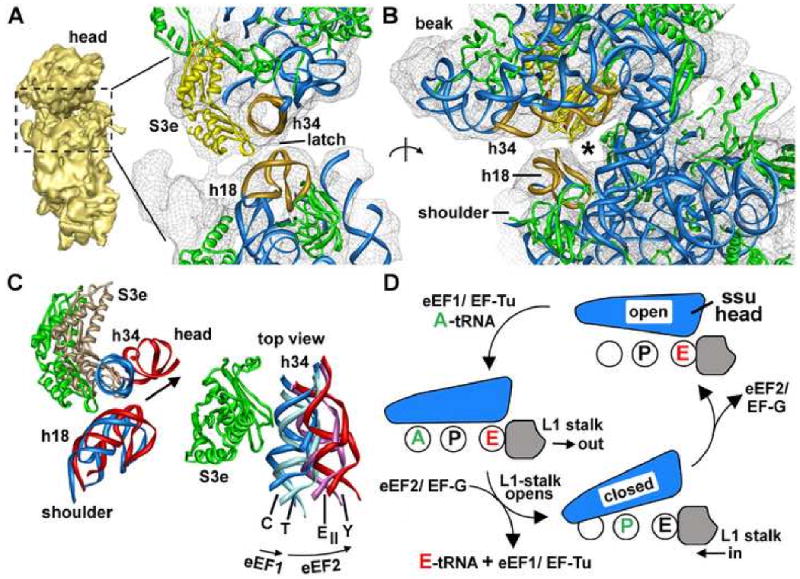
Conformational changes of the small subunit.
A. A latch is present between H34 and S3e in the head of the ssu and h18 in the shoulder. The helices are shown in gold and S3e is in yellow.
B. The head of the ssu is rotated ∼90° clockwise to show the latch and the mRNA entrance channel (asterisk).
C. (left) A comparison between the canine ribosome with an E-site tRNA (rRNA is blue, S3e green) and a yeast model with bound eEF2-sordarin (rRNA is red and S3e is tan) reveals a large rotation of the head relative to h18 in the shoulder. (right) A top view reveals a hypothetical, progressive rotation series for the head of the ssu. The h34 helices are color coded, as above with the cyan h34 from a programmed Thermus ribosome (pdb code: 2j00) and the pink h34 from the second conformational model of an E. coli ribosome (pdb code: 2aw7).
D. A model is shown for head rotation in the ssu and movements of the L1 stalk during the translation cycle.
Our structure supports the idea that core regions of the small and large subunits are structurally similar in all cytoplasmic ribosomes. This suggests that conformational changes which play a role in the translocation of tRNAs may be conserved between prokaryotic and eukaryotic ribosomes. To test this hypothesis, we aligned available structures of 70S and 80S ribosomes using conserved regions of the large subunit. We first looked at the latch region in 80S ribosomes. A close-up of this region is shown for the canine ribosome with an E-site tRNA (shown in blue, Figure 8C, left) and the yeast 80S-eEF2-sordarin complex (shown in red; Spahn et al., 2004a). This comparison shows that the head of the eukaryotic small subunit is quite dynamic, as proposed previously (Spahn et al., 2004a). Remarkably, the head of the small subunit moves from an open position, as seen in our model, to a closed conformation as seen in the yeast ribosome with bound eEF2 and sordarin (Spahn et al., 2004a). During this movement, the head of the small subunit pivots towards the Exit-site through an angle of ∼25° and h34 is displaced by ∼22Å. Similar movements occur for most of the features along the interface of the head, because the pivot point is located near h39 and h28 in the neck region (not shown).
The generality of this head movement is shown in Figure 8C (right) with known structures viewed from the top. These structures are placed in the order: 80S ribosome with E-site tRNA (labeled “C”)/70S ribosome with P-site tRNA (not shown; Berk et al., 2006), 70S ribosome with A-, P- and E-site tRNAs (“T”; Selmer et al., 2006; Korostelev et al., 2006), conformers I (not shown) and II of the E. coli ribosome (“E-II”; Shuwirth et al., 2005) and the yeast ribosome with eEF2 and sordarin (“Y”; Spahn et al., 2004a). This striking progression is consistent with the idea that conformational dynamics of the small subunit are similar in prokaryotes and eukaryotes.
Discussion
In this paper, we have presented a molecular model of the canine 80S ribosome with an E-site tRNA. Our model is based on a structure determined at 8.7Å resolution and provides insights into novel features of the eukaryotic ribosome. Importantly, our structure lends support to idea that conformational changes in the head of the small subunit and L1 stalk may play significant roles during the translocation of tRNAs. In addition, the eukaryotic bridges may help restore the subunits of the ribosome to a conformation with bound P- and E-site tRNAs, in preparation for the next cycle of chain elongation.
The mammalian ribosome
As expected, we found that subunit rRNAs and conserved proteins in the mammalian ribosome are structurally similar to their counterparts in bacterial and archaeal ribosomes, although numerous variations have occurred during evolution. Overall, eukaryotic ribosomes are much larger than their bacterial counterparts, due to the presence of rRNA expansion segments and novel proteins. Moreover, expansion segments and novel proteins are found on solvent exposed regions of the small and large subunits. A large amount of rRNA also lines the subunit interface which faces the ER membrane. In addition, both conserved and novel proteins bind to the expansion segments and stabilize their packing on the ribosome. Thus, expansion segments and novel proteins have evolved as an integrated framework, which envelopes the core bacterial ribosome without impeding its function.
In general, little is known about the functions of the expansion segments and novel proteins. However, the expansion segments could play a role in subunit assembly within the nucleolus or in stabilizing the mature ribosome (Sweeney et al., 1994). In addition, expansion segments may facilitate certain aspects of translation (Gao et al., 2005) or play other roles in eukaryotic cells (Beckmann et al., 2001). Some expansion segments could act as a negatively-charged sink that recruits positively-charged factors to the ribosome to enhance the efficiency of translation. Alternatively, these segments may be involved in targeting the ribosome to cytoplasmic sites within the larger eukaryotic cell.
Novel proteins in the eukaryotic ribosome are positioned to interact with expansion segments, conserved subunit rRNAs, or both RNAs simultaneously. Thus, it is striking that ordered density has been found for only about two-thirds of these proteins (20 of 31; Spahn et al., 2001; 2004a, this work). Some novel proteins may be lost during purification, they may be present normally at low occupancy or alternatively, they could interact with flexible regions of the ribosome. It seems likely that additional processes will be discovered which use the expansion segments and novel proteins.
In this work, we also showed that RACK1 forms a slightly distorted, 7-bladed β-propeller that is intimately associated with the small subunit in the canine ribosome. Indeed, RACK1 has multiple binding sites that involve rRNA and protein contacts on the small subunit. Since RACK1 is also found in plant and yeast ribosomes (Chang et al., 2005; Sengupta et al., 2004), it appears that RACK1 has evolved as an intrinsic component of the eukaryotic ribosome. Hence, RACK1 is perfectly positioned on the head and neck of the small subunit to form a docking platform for factors that regulate initiation and translation (Sengupta et al., 2004; Nilsson et al., 2004).
Structural rearrangements during translation
The sequential actions of eEF1/EF-Tu and eEF2/EF-G introduce a cognate amino-acylated tRNA into the A-site and then power the translocation of this tRNA into the P-site during peptide bond formation. Concomitant with this process, the small subunit undergoes a ratcheting of the main body relative to the large subunit and the head may move towards the E-site (Frank and Agrawal, 2000; Valle et al., 2003; Spahn et al., 2004a). Within this context, our work supports the idea that the head of the small subunit may pivot from an open to a more closed conformation during the translation cycle (Spahn et al., 2004a; Schuwirth et al., 2005). The magnitude of the observed head movements (∼20Å) suggests that this process may facilitate the lateral translocation of tRNAs (Spahn et al., 2004a; Berk and Cate, 2007).
As summarized in Figure 8D, the transition from a ribosome with bound P- and E-site tRNAs and an open head conformation to a ribosome with three occupied tRNA sites is catalyzed by eEF1/EF-Tu. This transition would involve a small rotation of the head of the small subunit towards a less open position and local changes around h18 to accommodate tRNA binding in the A-site. After dissociation of eEF1/EF-Tu, the binding of eEF2/EF-G displaces the A-site tRNA into the P-site, with a further rotation of the head of the small subunit towards the E-site. Lateral movements of the tRNAs during this process would induce the L1 stalk to open laterally with a concomitant release of the E-site tRNA. This release is facilitated by eEF3 in fungi (Andersen et al., 2006). In mammals, this release process may involve a novel α-helical connection between the large subunit and L10ae on the L1 stalk. After the E-site tRNA is released, the L1 stalk may bind a deacylated tRNA as it moves into the vacant E-site. Finally, release of eEF2/EF-G would allow the head of the small subunit to swivel back to its open position (Figure 8D, top). As the head of the small subunit moves back to an open conformation, the body of the small subunit pivots back to its original position.
This “head” cycle may be conserved as part of the ratcheting motion of the small subunit in all cytoplasmic ribosomes. During this cycle, the L1 stalk may adopt at least two different conformations, with the major positional determinant being the presence or absence of an E-site tRNA. However, the relative positions of the L1 stalk and head of the small subunit are probably independent yet correlated, as they would each move during translocation of the tRNAs.
Subunit bridges may play a significant role in modulating movements of the small subunit. In line with this idea, we find that bridges specific to eukaryotes are formed by novel proteins, expansion segments in rRNA and insertions in conserved ribosomal proteins. These links are present along the platform side of the small subunit and two additional bridges are located at the bottom of the small subunit. Remarkably, two of the bridges (eb8 and eb12) use α-helices to span the gap between the subunits. Bridge 12 is particularly striking, as a long helical extension of L19e has evolved to interact with ES6S in the small subunit, thereby stabilizing this region of the subunit interface. These eukaryotic bridges may help restore the small subunit to an initial conformation with bound P- and E-site tRNAs, when the small subunit has completed its ratchet-like movement. This may occur when the elongation factor EF2 dissociates from the mammalian ribosome and would set the stage to add another amino acid residue to the nascent chain.
Supplementary Material
Table S_1. Summary of modeled proteins in the canine ribosome.
Table S_2. Listing of ribosomal protein families in the small and large subunits.
Table S_3. Proteins that interact with the expansion segments.
Table S_4. Novel proteins in the canine ribosome.
Table S_5. Subunit bridges in the canine 80S ribosome with an E-site tRNA.
Figure S_1. FSC curves for the final 3D map of the canine RCC.
The complex has a resolution of 8.7Å based on the FSC0.5, while the channel is less ordered (∼24Å, red curve).
Figure S_2. Examples of modelled rRNAs and ribosomal proteins.
Features are shown in thin slabs in ‘O’ and non-related density has been omitted for clarity.
A. RNA helices in the small subunit (h44 and h2) are shown on the left and middle, respectively. Helix 69 forms a bridge (b2a) from the large subunit (lsu) to the small subunit (ssu). A-form helices are well resolved throughout the map and as also exemplified by density for the E-site tRNA (see silver surface of tRNA in Figures 5A-5C, main text).
B. Examples are shown of conserved ribosomal proteins modelled in the electron density. They include L9e, L23e and L26e from the large subunit and S13e from the small subunit. Proteins L9e and L26e contain β-sheets and regions of extended strands. Two β-hairpins (βH) are in good density in L26e. The α-helices are in rod-like density in all of the proteins. Some local rRNA is shown in gold.
Figure S_3. Overall fit of molecular models in the small and large subunits.
A. The fit of the molecular model (on the right) is shown within the 3D map of the small subunit (left, ssu). This view from the subunit interface is useful for evaluating the fit since this surface is strongly conserved.
B. The molecular model of the lsu (on the right) is shown inside a transparent surface of the subunit calculated from the 3D map.
Figure S_4. Modeled proteins in the small subunit.
A. Conserved proteins in the ssu were modeled using known crystal structures and a quantitative evaluation of the fit of the resulting homology models in the map. Modeled proteins (in green) are shown inside calculated volumes that are superimposed on a density map depiction of the core rRNA (blue) and expansion segments (red). The proteins are labeled. The rRNA density maps were created by filtering atomic models of these features to ∼9Å resolution. Novel proteins are also indicated.
B. The distribution of conserved proteins is shown in a platform view of the ssu.
C. The modeled proteins are shown in an interface view of the ssu.
Figure S_5. Modeled proteins in the large subunit.
A. An interface view is shown of the lsu and the modeled proteins (in magenta) are labelled.
B. A reverse view of the lsu (from the L1-stalk side) is shown with the modeled proteins labeled.
C. The distribution of conserved proteins is shown on a back view of the lsu.
D. A bottom view of the lsu shows conserved proteins on the surface which faces the ER membrane.
Figure S_6. Secondary structure diagram of the small subunit rRNA and modeling information. Hyper-variable expansion segments in the ssu are labeled. Regions highlighted in green are variable regions in the human sequence that were modeled using existing helical features in the crystal structure of the T. thermophilus ssu. Regions in blue were modeled de novo using the EM density map. Regions in yellow are nucleotides that were not modeled as they are in flexible regions, bulges or pseudo-knots.
Figure S_7. Secondary structure diagram of the large subunit rRNA with expansion segments and modeling information. A. A secondary structure diagram is shown for the 5′ half of the lsu rRNA. B. A secondary structure diagram is shown for the 3′ half of the lsu rRNA.
Figure S_8. An overview of the inter-subunit bridges in the canine ribosome.
A. The conserved bridges (in grey) and the eukaryotic bridges (in dark blue) are marked by spherical markers (∼10Å diameter) on interface views of the small and large subunits from the final 3D map. The bridges are also labeled.
B. The bridges are indicated in interface views of the final model of the small (left) and large (right) subunits, respectively.
Acknowledgments
We thank A. Neuhof for preparation of the canine RCC samples, T. Rapoport for his support, F. Fabiola, A. Korostelev and M.S. Chapmann for discussions on CNS-RSREF, B. Webb for help with MODELLER and T.D. Goddard for help with Chimera. This work was supported by grants to R.G. (GM067317), A.S (NIH R01 GM54762, NIH PN2 EY016525, and NSF IIS-0705196) and C.W.A (NIH RO1 GM45377). A.S. also acknowledges support from The Sandler Family Supporting Foundation, Hewlett-Packard, NetApps, IBM, and Intel. M.T. is supported by an MRC Career Development Award. The ribosome map and coordinates for the 80S model have been deposited in the EMD (- XXXXXX) and RCSB (xxxx, xxxx).
References
- Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005;121:703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Andersen CB, Becker T, Blau M, Anand M, Halic M, Balar B, Mielke T, Boesen T, Pedersen JS, Spahn CM, Kinzy TG, Andersen GR, Beckmann R. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature. 2006;443:663–668. doi: 10.1038/nature05126. [DOI] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Beckmann R, Spahn CM, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 2001;107:361–372. doi: 10.1016/s0092-8674(01)00541-4. [DOI] [PubMed] [Google Scholar]
- Berk V, Zhang W, Pai RD, Cate JH. Structural basis for mRNA and tRNA positioning on the ribosome. Proc Natl Acad Sci U S A. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk V, Cate JH. Insights into protein biosynthesis from structures of bacterial ribosomes. Curr Opin Struct Biol. 2007;17:302–309. doi: 10.1016/j.sbi.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Brodersen DE, Clemons WM, Jr, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhauser N, Marchisio PC, Biffo S. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature. 2003;426:579–584. doi: 10.1038/nature02160. [DOI] [PubMed] [Google Scholar]
- Chang IF, Szick-Miranda K, Pan S, Bailey-Serres J. Proteomic characterization of evolutionarily conserved and variable proteins of Arabidopsis cytosolic ribosomes. Plant Physiol. 2005;137:848–862. doi: 10.1104/pp.104.053637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta PP, Sharma MR, Qi L, Frank J, Agrawal RK. Interaction of the G′ domain of elongation factor G and the C-terminal domain of ribosomal protein L7/L12 during translocation as revealed by cryo-EM. Mol Cell. 2005;20:723–731. doi: 10.1016/j.molcel.2005.10.028. [DOI] [PubMed] [Google Scholar]
- Diaconu M, Kothe U, Schlunzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Dresios J, Panopoulos P, Synetos D. Eukaryotic ribosomal proteins lacking a eubacterial counterpart: important players in ribosomal function. Mol Microbiol. 2006;59:1651–1663. doi: 10.1111/j.1365-2958.2006.05054.x. [DOI] [PubMed] [Google Scholar]
- Dube P, Bacher G, Stark H, Mueller F, Zemlin F, van Heel M, Brimacombe R. Correlation of the expansion segments in mammalian rRNA with the fine structure of the 80S ribosome: a cryo-electron microscopic reconstruction of the rabbit reticulocyte ribosome at 21Å resolution. J Mol Biol. 1998;279:403–421. doi: 10.1006/jmbi.1998.1804. [DOI] [PubMed] [Google Scholar]
- Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- Frank J, Sengupta J, Gao H, Li W, Valle M, Zavialov A, Ehrenberg M. The role of tRNA as a molecular spring in decoding, accommodation, and peptidyl transfer. FEBS Lett. 2005;579:959–962. doi: 10.1016/j.febslet.2004.10.105. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- Gao H, Ayub MJ, Levin MJ, Frank J. The structure of the 80S ribosome from Trypanosoma cruzi reveals unique rRNA components. Proc Natl Acad Sci U S A. 2005;102:10206–10210. doi: 10.1073/pnas.0500926102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Sengupta J, Valle M, Korostelev A, Eswar N, Stagg SM, Van Roey P, Agrawal RK, Harvey SC, Sali A, Chapman MS, Frank J. Study of the structural dynamics of the E coli 70S ribosome using real-space refinement. Cell. 2003;113:789–801. doi: 10.1016/s0092-8674(03)00427-6. [DOI] [PubMed] [Google Scholar]
- Gerbi SA. Expansion segments: regions of variable size that interrupt the universal core secondary structure of ribosomal RNA. In: Zimmermann RA, Dahlberg AE, editors. Ribosomal RNA, Structure, Evolution, Processing, and Function in Protein Biosynthesis. CRC Press; New York: 1996. pp. 71–87. [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure (Camb) 2005;13:473–482. doi: 10.1016/j.str.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Green R, Noller HF. Ribosomes and translation. Annu Rev Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- Gomez-Lorenzo MG, Spahn CM, Agrawal RK, Grassucci RA, Penczek P, Chakraburtty K, Ballesta JP, Lavandera JL, Garcia-Bustos JF, Frank J. Three-dimensional cryo-electron microscopy localization of EF2 in the Saccharomyces cerevisiae 80S ribosome at 17.5Å resolution. EMBO J. 2000;19:2710–2718. doi: 10.1093/emboj/19.11.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo P, Reboud JP. The puzzling lateral flexible stalk of the ribosome. Biol Cell. 2003;95:179–93. doi: 10.1016/s0248-4900(03)00034-0. [DOI] [PubMed] [Google Scholar]
- Halic M, Becker T, Frank J, Spahn CM, Beckmann R. Localization and dynamic behavior of ribosomal protein L30e. Nat Struct Mol Biol. 2005;12:467–468. doi: 10.1038/nsmb933. [DOI] [PubMed] [Google Scholar]
- Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- Jenni S, Ban N. The chemistry of protein synthesis and voyage through the ribosomal tunnel. Curr Opin Struct Biol. 2003;13:212–219. doi: 10.1016/s0959-440x(03)00034-4. [DOI] [PubMed] [Google Scholar]
- Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Ménétret JF, Hegde RS, Heinrich S, Chandramouli P, Ludtke SJ, Rapoport TA, Akey CW. Architecture of the ribosome-channel complex derived from native membranes. J Mol Biol. 2005;348:445–457. doi: 10.1016/j.jmb.2005.02.053. [DOI] [PubMed] [Google Scholar]
- Morgan DG, Ménétret JF, Neuhof A, Rapoport TA, Akey CW. Structure of the mammalian ribosome-channel complex at 17Å resolution. J Mol Biol. 2002;324:871–886. doi: 10.1016/s0022-2836(02)01111-7. [DOI] [PubMed] [Google Scholar]
- Nilsson J, Sengupta J, Frank J, Nissen P. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 2004;5:1137–1141. doi: 10.1038/sj.embor.7400291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM, Jr, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Pioletti M, Schlunzen F, Harms J, Zarivach R, Gluhmann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C, Hartsch T, Yonath A, Franceschi F. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 2001;20:1829–1839. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- Rudra D, Warner JR. What better measure than ribosome synthesis? Genes Dev. 2004;18:2431–2436. doi: 10.1101/gad.1256704. [DOI] [PubMed] [Google Scholar]
- Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluhmann M, Janeli D, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A. Structure of functionally activated small ribosomal subunit at 3.3Å resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Huang KS, Strobel SA, Steitz TA. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- Schnare MN, Damberger SH, Gray MW, Gutell RR. Comprehensive comparison of structural characteristics in eukaryotic cytoplasmic large subunit (23S-like) ribosomal RNA. J Mol Biol. 1996;256:701–719. doi: 10.1006/jmbi.1996.0119. [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV, 4th, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Sengupta J, Nilsson J, Gursky R, Spahn CM, Nissen P, Frank J. Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat Struct Mol Biol. 2004;11:957–962. doi: 10.1038/nsmb822. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae-tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004a;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004b;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Wieden HJ, Zemlin F, Wintermeyer W, van Heel M. Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat Struct Biol. 2002;9:849–854. doi: 10.1038/nsb859. [DOI] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Wieden HJ, van Heel M, Wintermeyer W. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell. 2000;100:301–309. doi: 10.1016/s0092-8674(00)80666-2. [DOI] [PubMed] [Google Scholar]
- Sweeney R, Chen L, Yao MC. An rRNA variable region has an evolutionarily conserved essential role despite sequence divergence. Mol Cell Biol. 1994;14:4203–4215. doi: 10.1128/mcb.14.6.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Haar E, Musacchio A, Harrison SC, Kirchhausen T. Atomic structure of clathrin: a beta propeller terminal domain joins an alpha zigzag linker. Cell. 1998;13:563–573. doi: 10.1016/s0092-8674(00)81623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topf M, Baker ML, Marti-Renom MA, Chiu W, Sali A. Refinement of protein structures by iterative comparative modeling and CryoEM density fitting. J Mol Biol. 2006;357:1655–1668. doi: 10.1016/j.jmb.2006.01.062. [DOI] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol. 2003a;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003b;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- Wimberly B, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, von Rhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Wool IG, Chan YL, Gluck A. Structure and evolution of mammalian ribosomal proteins. Biochem Cell Biol. 1995;73:933–947. doi: 10.1139/o95-101. [DOI] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZh, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S_1. Summary of modeled proteins in the canine ribosome.
Table S_2. Listing of ribosomal protein families in the small and large subunits.
Table S_3. Proteins that interact with the expansion segments.
Table S_4. Novel proteins in the canine ribosome.
Table S_5. Subunit bridges in the canine 80S ribosome with an E-site tRNA.
Figure S_1. FSC curves for the final 3D map of the canine RCC.
The complex has a resolution of 8.7Å based on the FSC0.5, while the channel is less ordered (∼24Å, red curve).
Figure S_2. Examples of modelled rRNAs and ribosomal proteins.
Features are shown in thin slabs in ‘O’ and non-related density has been omitted for clarity.
A. RNA helices in the small subunit (h44 and h2) are shown on the left and middle, respectively. Helix 69 forms a bridge (b2a) from the large subunit (lsu) to the small subunit (ssu). A-form helices are well resolved throughout the map and as also exemplified by density for the E-site tRNA (see silver surface of tRNA in Figures 5A-5C, main text).
B. Examples are shown of conserved ribosomal proteins modelled in the electron density. They include L9e, L23e and L26e from the large subunit and S13e from the small subunit. Proteins L9e and L26e contain β-sheets and regions of extended strands. Two β-hairpins (βH) are in good density in L26e. The α-helices are in rod-like density in all of the proteins. Some local rRNA is shown in gold.
Figure S_3. Overall fit of molecular models in the small and large subunits.
A. The fit of the molecular model (on the right) is shown within the 3D map of the small subunit (left, ssu). This view from the subunit interface is useful for evaluating the fit since this surface is strongly conserved.
B. The molecular model of the lsu (on the right) is shown inside a transparent surface of the subunit calculated from the 3D map.
Figure S_4. Modeled proteins in the small subunit.
A. Conserved proteins in the ssu were modeled using known crystal structures and a quantitative evaluation of the fit of the resulting homology models in the map. Modeled proteins (in green) are shown inside calculated volumes that are superimposed on a density map depiction of the core rRNA (blue) and expansion segments (red). The proteins are labeled. The rRNA density maps were created by filtering atomic models of these features to ∼9Å resolution. Novel proteins are also indicated.
B. The distribution of conserved proteins is shown in a platform view of the ssu.
C. The modeled proteins are shown in an interface view of the ssu.
Figure S_5. Modeled proteins in the large subunit.
A. An interface view is shown of the lsu and the modeled proteins (in magenta) are labelled.
B. A reverse view of the lsu (from the L1-stalk side) is shown with the modeled proteins labeled.
C. The distribution of conserved proteins is shown on a back view of the lsu.
D. A bottom view of the lsu shows conserved proteins on the surface which faces the ER membrane.
Figure S_6. Secondary structure diagram of the small subunit rRNA and modeling information. Hyper-variable expansion segments in the ssu are labeled. Regions highlighted in green are variable regions in the human sequence that were modeled using existing helical features in the crystal structure of the T. thermophilus ssu. Regions in blue were modeled de novo using the EM density map. Regions in yellow are nucleotides that were not modeled as they are in flexible regions, bulges or pseudo-knots.
Figure S_7. Secondary structure diagram of the large subunit rRNA with expansion segments and modeling information. A. A secondary structure diagram is shown for the 5′ half of the lsu rRNA. B. A secondary structure diagram is shown for the 3′ half of the lsu rRNA.
Figure S_8. An overview of the inter-subunit bridges in the canine ribosome.
A. The conserved bridges (in grey) and the eukaryotic bridges (in dark blue) are marked by spherical markers (∼10Å diameter) on interface views of the small and large subunits from the final 3D map. The bridges are also labeled.
B. The bridges are indicated in interface views of the final model of the small (left) and large (right) subunits, respectively.


