Abstract
Plasmodium falciparum, the Apicomplexan parasite that is responsible for the most lethal forms of human malaria, is exposed to radically different environments and stress factors during its complex lifecycle. In any organism, Hsp70 chaperones are typically associated with tolerance to stress. We therefore reasoned that inhibition of P. falciparum Hsp70 chaperones would adversely affect parasite homeostasis. To test this hypothesis, we measured whether pyrimidinone-amides, a new class of Hsp70 modulators, could inhibit the replication of the pathogenic P. falciparum stages in human red blood cells. Nine compounds with IC50 values from 30 nM to 1.6 μM were identified. Each compound also altered the ATPase activity of purified P. falciparum Hsp70 in single-turnover assays, although higher concentrations of agents were required than was necessary to inhibit P. falciparum replication. Varying effects of these compounds on Hsp70s from other organisms were also observed. Together, our data indicate that pyrimidinone-amides constitute a novel class of anti-malarial agents.
Keywords: Molecular chaperone, Hsp70, Hsp40, J domain, ATPase, Pyrimidinone
1. Introduction
Plasmodium falciparum malaria kills ~3000 people each day, mostly children in sub-Saharan Africa. Several anti-malarial agents are available that–in principle–could prevent this disease. Unfortunately, the use of some of these agents is compromised by parasite resistance, high cost, and/or dangerous or unpleasant side-effects. Also, some therapies are combinations of existing drugs (e.g., artemether-lumefantrine, dihydroartemisinin-piperaquine, or artesunate-amodiaquine)1,2 that depend on the inability of the parasite to become resistant to artemisinin derivatives. Therefore, continued efforts to identify novel compounds that kill P. falciparum are critical.
The P. falciparum genome encodes six Hsp70 and forty-three Hsp40 molecular chaperones.3,4 Hsp70 molecular chaperones couple the hydrolysis of ATP with the binding and release of polypeptide substrates and play vital roles in protein folding, degradation and transport.5 Hsp40s interact with Hsp70 through a conserved, four helix-bundle known as the ‘J domain’. J domain interactions enhance Hsp70 ATPase activity; similar to Hsp70s, many Hsp40s are also polypeptide-binding proteins.6 Hsp70s and Hsp40s can constitute a significant amount of total cellular protein, a percentage that rises when cells are stressed (hence the ‘heat shock protein’ nomenclature).7
There are several reasons why P. falciparum might require the function of Hsp70 and Hsp40 chaperones. First, this parasite, like other members of the Apicomplexa, contains several endo-membrane systems.8 Because Hsp70-Hsp40 pairs engineer protein transport across membranes and are essential for membrane integrity, 9 each internal membrane might require its own set of chaperones. In support of this hypothesis, it was recently shown that a J domain-containing protein in P. falciparum (PF10_0381) is required for ‘knob’ formation, a structure that helps the presentation of PfEMP1 proteins on the red blood cell surface; this, in turn, leads to the binding of parasitized red blood cells to the vascular endothelium. 10 Second, the parasite is exposed to radically different environments during its life cycle. It is capable of thriving in the mosquito, in the host liver, and in the highly oxidizing environment of the red blood cell.11 Thus, Hsp70s and Hsp40s might be necessary to offset cellular stresses that are encountered during the P. falciparum life cycle. Third, P. falciparum exhibits sudden bursts of protein synthesis as it enters the trophozoite stage that marks the initiation of several rounds of intracellular division. Molecular chaperones help retain newly synthesized polypeptides in soluble conformations and facilitate folding.12 For example, an inhibitor of the Hsp90 chaperone, which is required for folding select cellular proteins, was shown to inhibit the ‘ring’ to trophozoite transition.13 Finally, it appears that the parasite contains extensive chaperone networks that are involved in a multitude of cellular activities.14 Based on these data, P. falciparum viability should be exceptionally sensitive to Hsp70-Hsp40 inhibition. Indeed, parasite growth is inhibited by 15-deoxyspergualin,15,16 a non-specific chaperone modulator that binds to Hsp70 and to Hsp90 with a KD of ~5 μM.17
We previously reported the synthesis and characterization of pyrimidinone-peptoid hybrid molecules that modulate Hsp70 activity in vitro and that, in some cases, prevent cancer cell proliferation. 18–21 These data indicate that specific Hsp70 modulators can be identified and that at least a sub-group of these compounds is membrane-permeable, as evidenced by their activity in a cellular assay. To test whether molecules in this class also compromise P. falciparum replication, we screened a small collection of pyrimidinones and identified nine compounds that exhibited potent effects on parasite metabolism. Some of these compounds inhibited P. falciparum viability with similar potencies to some established anti-malarial drugs.22 We also developed new purification schemes for Hsp70 proteins from Homo sapiens and P. falciparum and compared the effects of these compounds on the ATPase activities of the human, yeast, and parasite chaperones. Together, our data support the continued investigation of pyrimidinones as antimalarial agents.
2. Results and discussion
To assess whether pyrimidinones inhibit P. falciparum growth, we examined the effects of 157 compounds in this class and related Biginelli and Ugi multicomponent condensation-derived compounds on the uptake of [3H]hypoxanthine into infected human erythrocytes. The hypoxanthine assay provides a rapid, quantifiable read-out of parasite viability, and the compounds assayed included several recently described agents,18 as well as precursors and structurally related analogs.
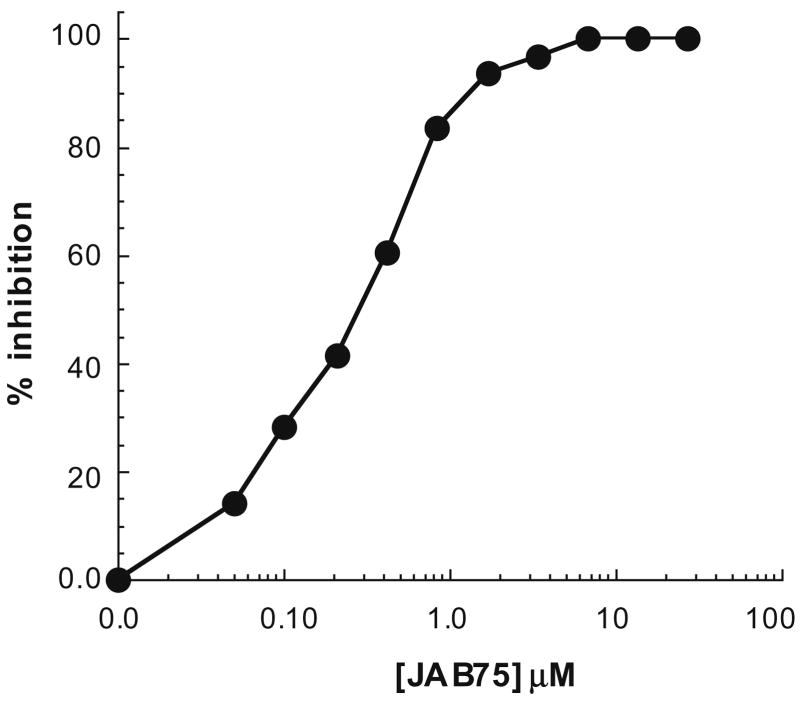
The impact of JAB75 (see Section 3) on [3H]hypoxanthine up-take is shown in Figure 1. In this and all other assays, we used the chloroquine (CQ)-resistant Dd2 clone and employed CQ as an internal control because CQ is known to inhibit Dd2 with an IC50 value of ~0.2 μM.23,24 In this experiment, the IC50 for JAB75 was calculated to be ~0.3 μM (Table 1, second column) and the IC50 for CQ was 0.19 μM (data not shown). From our initial analysis of 157 compounds, we identified nine molecules (Fig. 2; see Section 3) with IC50 values between 30 nM and 1.6 μM (Table 1, second column).
Figure 1.
JAB75 inhibits [3H]hypoxanthine uptake into red blood cells infected with P. falciparum. The uptake of [3H]hypoxanthine into infected human erythrocytes and the IC50 value were determined as described in Section 3. The raw data from a single assay were standardized such that the inhibition of uptake in the absence of JAB75 was set to 0%.
Table 1.
Effects of most potent P. falciparum inhibitors on the steady-state ATPase activities of P. falciparum, yeast (Ssa1), and human (HsHsp70)
| Compound | P. falciparum IC50 (μM) | PfHsp70 (ATPase) | Ssal (ATPase) | HsHsp70 (ATPase) |
|---|---|---|---|---|
| DMSO | 1 | 1 | 1 | |
| DMT3024 | 0.2 | 0.73 ± 0.003 | 1.0 ± 0.004 | 0.74 ± 0.003 |
| DMT2264 | 1.1 | 0.74 ± 0.002 | 0.97 ± 0.004 | 0.95 ± 0.002 |
| MAL2-29 | 1.6 | 1.01 ± 0.1 | 0.93 ± 0.007 | 0.85 ± 0.06 |
| MAL2-39 | 0.2 | 0.92 ± 0.05 | 0.95 ± 0.004 | 0.80 ± 0.08 |
| MAL2-61 | 0.1 | 0.99 ± 0.07 | 1.1 ± 0.003 | 0.74 ± 0.07 |
| MAL2-213 | 0.03 | 1.02 ± 0.11 | 0.89 ± 0.016 | 0.80 ± 0.06 |
| MAL2-215 | 0.05 | 1.01 ± 0.08 | 1.2 | 0.72 ± 0.09 |
| MAL3-39 | 0.8 | 0.72 ± 0.08 | 0.75 | 0.82 ± 0.05 |
| JAB75 | 0.3 | 1.0 ± 0.06 | 1.2 | 0.70 ± 0.06 |
The IC50 value for inhibition of [3H]hypoxanthine uptake is shown in the second column. The three columns on the right depict the relative activities of each enzyme (after standardization to the activity with an equal volume of DMSO, which was set to ‘1’) in the presence of a final concentration of 300 μM of the designated compound. The turnover numbers of each enzyme in the presence of DMSO were: P. falciparum Hsp70: 0.020 min−1; Ssa1: 0.032 min−1; HsHsp70: 0.025 min−1. Stained gels showing the enriched proteins used in this analysis are provided in a Supplementary Figure (Fig. S1). Data represent the means of 6 independent experiments, and where shown, ±SD.
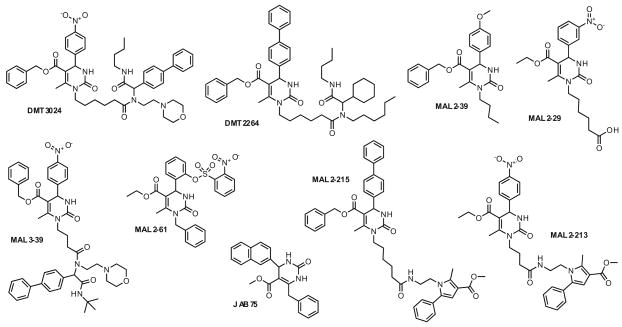
Figure 2.
Select pyrimidinones inhibit hypoxanthine uptake into P. falciparum-infected red blood cells. The depicted compounds inhibited P. falciparum replication with IC50 values of 30 nM–1.6 μM (see Table 1). The structures were drawn to maximize chemical similarity.
To ensure that the compounds were not generally cytotoxic, we also determined the 50% growth inhibitory concentrations (GI50) for each of these nine agents in two human cell lines, HepG2 hepatocellular carcinoma cells and WI-38 embryonic diploid lung cells, as previously described.18 Based on this analysis, all GI50 values in these cells were >10 μM, which is well above the concentration needed to inhibit P. falciparum growth (Table 1). As a control for this experiment, we found that the GI50 values for paclitaxel in HepG2 and WI-38 cells were 1.0 ± 0.6 nM and 13.7 ± 0.2 nM, respectively (data not shown).
We previously showed that a subset of pyrimidinones inhibit the activity of Hsp70.18,19 Therefore, we next assessed the effects of the nine compounds on Hsp70 ATPase activity. We first examined the ability of each pyrimidinone to modulate the ATP hydrolytic rate of a purified yeast Hsp70, Ssa1, as previously published.18–20 In addition, we wished to compare the effects of these agents on human and P. falciparum Hsp70. The three chaperones are 71–74% identical to one another at the amino acid level and, not surprisingly, Hsp70s from different species have been reported to substitute functionally for one another. For example, the growth of bacteria containing a mutation in the gene encoding DnaK, the Hsp70 homolog, can be rescued at high temperature by expression of a P. falciparum Hsp70.25 One might also envision, however, that a specific inhibitor could selectively target the P. falciparum chaperone but have no effect on Hsp70s from different species.
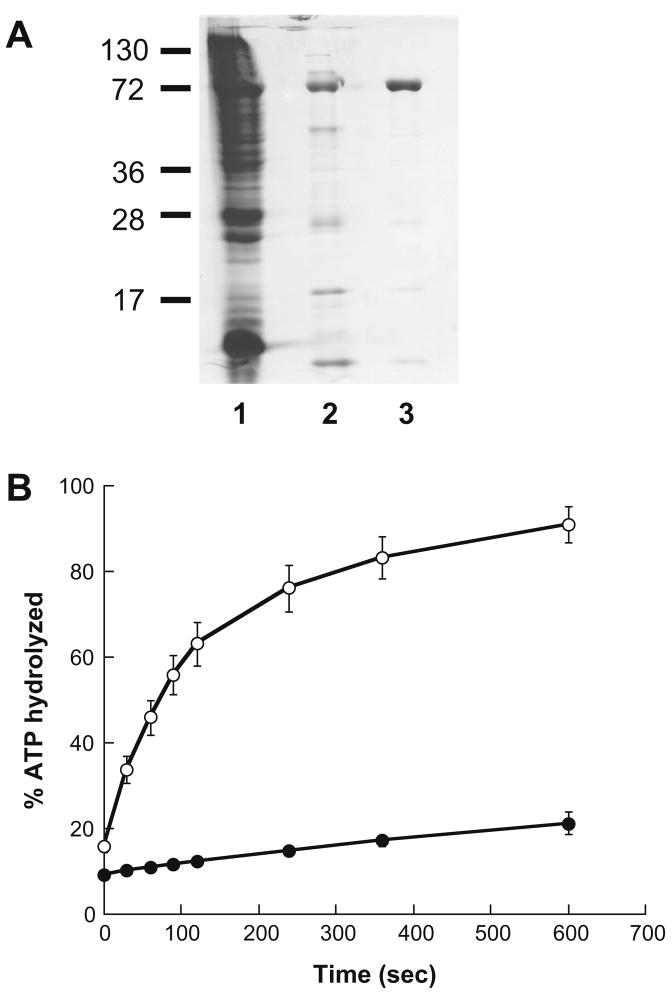
To compare the effects of the lead compounds on the ATPase activities of the Hsp70s, we first modified purification schemes for both the H. sapiens and P. falciparum chaperones (see Section 3). The peak fractions from the P. falciparum Hsp70 purification are shown Figure 3A, and the single-turnover ATPase activities of the enzyme in the absence and presence of a J domain chimera (see below) are shown in Figure 3B.
Figure 3.
Purification and analysis of P. falciparum Hsp70. (A) A Coomassie Brilliant blue-stained gel summarizing individual steps during the P. falciparum Hsp70 purification is shown: lane 1: crude E. coli lysate from the P. falciparum Hsp70 over-expressing strain; lane 2: pooled peak fractions after nickel-affinity resin chromatography; lane 3: pooled peak fractions after Q-sepharose column chromatography. The positions of molecular weight markers (×10−3) are indicated to the left. Densitometry analysis indicates >95% purity. (B) The single-turnover ATPase activity of P. falciparum Hsp70 is stimulated by a J domain-containing hybrid protein. ATPase assays were performed as described in Section 3 in the presence of 3 μg of the Hlj1 chimeric protein. Data represent the means of 6 independent determinations, ±SD.
Next, each of the nine compounds was incubated with the P. falciparum, yeast and human chaperones, and steady-state ATPase assays were performed. The results presented in Table 1, columns 3–5, indicate that the compounds display a range of activities and alter the activities of each chaperone distinctly. For example, JAB75, MAL2-61, and MAL2-215 reduced the rate of ATP hydrolysis by the human enzyme by 30%, but had no effect or stimulated the activities of the parasite and yeast enzymes. In contrast, MAL2-39 compromised the ATPase activities of all three enzymes, but mainly HsHsp70. MAL3-39 inhibited all enzymes to a very similar extent. Perhaps most intriguing, DMT2264 affected the ATPase activity of P. falciparum Hsp70 significantly more than Ssa1 and HsHsp70. These data indicate that the pyrimidinones can be classified based on their in vitro effects on Hsp70s from different species. Nevertheless, it is important to note that these analyses were performed at a final concentration of 300 μM in order to maximize the effects. We also selected this concentration because the action of pyrimidinones was first studied under these conditions;18 however, when used at concentrations that inhibited P. falciparum replication (Table 1), the steady-state ATPase activity of Hsp70 was unaltered, suggesting the possibility of secondary targets or other issues related to compound metabolism or accumulation (see below).
The cellular activity of Hsp70 most often requires interaction with J domain-containing Hsp40 partners.5,6 In fact, one of our most potent pyrimidinone-based inhibitors of breast cancer cell proliferation has no effect on endogenous ATPase activity but compromises the ability of a J domain-containing co-chaperone to enhance ATP hydrolysis.19,21 Therefore, it was possible that the compounds might have greater effects on the J domain-stimulated ATPase activity of the Hsp70s. Because the purification of a P. falciparum Hsp40 has not been reported, we instead chose to use a chimeric protein that contains the J domain from Hlj1, a yeast Hsp40, fused to glutathione-S-transferase.26 Recent work indicates that the chimera interacts promiscuously with a variety of Hsp70s and augments their ATPase activities.36 For comparison, we also used a full-length ‘type-I’ Hsp40 co-chaperone from yeast, Ydj1.
We added each of the nine P. falciparum inhibitors in single-turnover ATPase reactions containing either the parasite (PfHsp70) or yeast Hsp70 (Ssa1) in the presence or absence of the Hlj1 chimera and Ydj1 (see Table 2). Furthermore, the activity of human Hsp70 in the presence or absence of Ydj1 and one of its known Hsp40 partners, Hdj1, was examined in the presence or absence of each pyrimidinone. We chose to use single-turnover conditions for this analysis because the level of J domain-mediated stimulation of Hsp70 ATPase activity is significantly greater for single-turnover assays as compared to steady-state assays (see, e.g., Fig. 3B). Furthermore, the effects of compounds on enzyme kCAT values can specifically be monitored and the fold change measured. As shown in Table 2, these nine compounds exhibited a range of effects. First, relatively subtle changes were observed when the effects of the compounds were examined in the presence of the J domain protein. For example, a <50% decrease (in the presence of MAL2-213 or DMT2264) and a ~40% increase (in the presence of MAL2-61 or MAL3-39) was measured when the J domain was added into the assay in the presence of these chemicals (Table 2, ‘fold change PfHsp70 kCAT + Hlj1’). In contrast, two of the compounds induced a potent ‘burst’ of ATP hydrolysis in the single-turnover assay in the absence of a J domain (‘fold change PfHsp70 kCAT’): Based on a fit of the data to a single exponential, MAL2-39 and MAL2-61 enhanced P. falciparum Hsp70 ATPase activity by 7.2 and 6.5-fold, respectively. Each of the other compounds also enhanced ATP hydrolysis to varying extents, an effect that has been previously noted for certain other pyrimidinones.18,19 Because the cycle of Hsp70 ATP binding/hydrolysis is coupled with substrate binding and release, altered rates of endogenous or J domain-stimulated ATP hydrolysis will correspondingly alter the efficacy of substrate binding. Therefore, enhanced rates of ATP hydrolysis can lead to defects in the ability of Hsp70s to act as a molecular chaperone, especially under stress conditions.27,28 Moreover, recent data confirm that the individual chaperone cycles and conformations have been tailored to match the folding of specific substrates.29 In the future, it will be important to examine whether these agents affect the binding of known Hsp70 substrates. Moreover, it is critical to note that effects on ATPase activity in steady-state assays may result from alterations in any one of a number of steps in the hydrolytic cycle, including the kCAT, ATP binding, and inorganic phosphate and/or ADP release.
Table 2.
Effects of the P. falciparum inhibitors on the single-turnover ATPase activities of a malarial parasite, yeast, and human Hsp70s
| Compound | Fold change PfHsp70 kCAT | Fold change PfHsp70 kCAT + Hlj1 | Fold change PfHsp70 kCAT + Ydj1 | Fold change Ssal kCAT | Fold change Ssal kCAT + Hlj1 | Fold change Ssal kCAT + Ydj1 | Fold change hHsp70 kCAT | Fold change hHsp70 kCAT + Hdj1 | Fold change hHsp70kCAT + Ydj1 |
|---|---|---|---|---|---|---|---|---|---|
| DMSO | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| DMT3024 | 2.3 | −1.3 | −1.1 | −1.6 | −1.1 | −2.6 | 1 | 4.7 | 1 |
| DMT2264 | 2.9 | −1.5 | 1.1 | −1.7 | 1.2 | −1.3 | 2.3 | 2.8 | 1.1 |
| MAL2-29 | 1.8 | −1.1 | −1.2 | 1.3 | 1 | 1.1 | 1.4 | 1.1 | −1.3 |
| MAL2-39 | 7.2 | −1.1 | 1.1 | 2.3 | 1 | −1.1 | 4.5 | 4.5 | 1.1 |
| MAL2-61 | 6.5 | 1.4 | 1 | 2.1 | 1 | −1.3 | 3.6 | 3.5 | −1.2 |
| MAL2-213 | 3.7 | 1.5 | 1.1 | −1.3 | −1.6 | −1.6 | 2 | 4.3 | −1.2 |
| MAL2-215 | 2.7 | −1.1 | 1.1 | 13.3 | −6.5 | −1.7 | 3.5 | 5.3 | 1.5 |
| MAL3-39 | 2.2 | 1.4 | 2.4 | 1.2 | 1 | −1.5 | 2 | 2.9 | −1.1 |
| JAB75 | 1.4 | −1.2 | 1 | 2 | −1.2 | −1.3 | 1.3 | 2.5 | −1.2 |
The indicated compounds were supplemented into single-turnover reactions, as described,18,19 either using the P. falciparum Hsp70, yeast (Ssa1) enzyme, or human Hsp70. Where indicated, the kCAT values were also measured in the presence of the following concentrations of distinct J domain-containing proteins: Hlj1 (1.1 μM), Ydj1 (0.045 μM when used with Ssa1 and 0.9 μM when used with P. falciparum and human Hsp70), and Hdj2 (1.0 μM). Compounds were added at a final concentration of 100 μM, and all data were standardized to the level of activity determined in the absence of compound but in the presence of an equal volume of DMSO. For example, the activity of the P. falciparum Hsp70 in the presence of DMSO and in the presence or absence of Hlj1 was set to ‘1’, and all other reactions under the same conditions were calculated and are shown as a fraction of this value. The data represent the means of 2–4 independent experiments, and where shown, a negative sign indicates an inhibitory effect of the agent. The average stimulation by Ydj1 (in the absence of compound) for P. falciparum Hsp70, for Ssa1, and for human Hsp70 in this experiment was 8.2, 5.5, and 7.2-fold, respectively.
An examination of the endogenous and J domain-stimulated ATPase activities of the yeast enzyme in the presence or absence of the compounds yielded quite different results (Table 2, ‘fold change Ssa1 kCAT’ and ‘fold change Ssa1 kCAT + Hlj1’). We found that MAL2-215 exerted significant effects on both the endogenous and J domain-stimulated activity of Ssa1, but the compounds with the greatest impact on the P. falciparum enzyme (e.g., MAL2-39, MAL2-61, and MAL2-213) only modestly altered the activity of the yeast enzyme. Notably, the strongest effects of each pyrimidinone were observed when the activity of the human enzyme was examined in the presence of its partner, Hdj1. At this point, the substructure features that mediate these distinct phenomena remain to be elucidated. Our data nevertheless indicate that the continued examination of pyrimidinones with diverse Hsp70s will prove worthwhile, especially if a specific binding site on the chaperone for this class of modulators can be identified. Efforts toward this goal are underway.
It is important to note that the relative effects of the compounds on the endogenous or J domain-stimulated P. falciparum ATPase activities do not correlate with the IC50 values in the [3H]hypoxanthine uptake assay (compare Table 1 and Table 2). There are several explanations for this fact. First, it is possible that some compounds have secondary cellular targets, which may enhance the antimalarial effects. Second, some of the compounds may be metabolized when added to the infected erythrocytes to produce derivatives that may be more or less potent, depending on the nature of the modification. Third, the less potent agents in the [3H]hypoxanthine uptake assay might be actively excluded from cells due to the action of multi-drug transporters or other gene products that are known to mediate drug resistance in this parasite. 30 Assays with radiolabeled or fluorescently labeled compounds will help clarify this possibility by enabling measurements of compound accumulation in the parasite; however, these results are not yet available. Fourth, the bonafide Hsp70 that is a target of the active compounds may be any one of the other five Hsp70s that are encoded by the P. falciparum genome. It is striking how distinctly some compounds affect the activities of the yeast Ssa1 and human proteins and the P. falciparum Hsp70 utilized in this study (PfHsp70-1; PF08_0054) even though these proteins are >70% identical. Because Hsp70-1 and the mitochondrial Hsp70 in P. falciparum (PfHsp70-3; PF11_0351) are only 48% identical, the compounds are also expected to exhibit distinct effects on parasitic Hsp70s. The problem of identifying the target(s) of these compounds could, in principle, be rectified by the purification of each of the six P. falciparum Hsp70s and 43 P. falciparum J proteins so that each combination could be tested in ATPase assays in the presence of the modulators. A streamlined approach would be to prepare activated, affinity tagged derivatives of our novel P. falciparum inhibitors and then identify potential cellular target(s) using an unbiased screen. This experimental regimen would ideally isolate the ‘correct’ Hsp70 and/or Hsp40 chaperone target(s).
We note structural trends that relate to potency in the P. falciparum replication assay. Specifically, there are several common and distinct features amongst the identified inhibitors depicted in Figure 2. For example, all nine compounds share an ester pyrimidine core, substituted at C-4, and eight of the nine are alkylated at N1. Among the compounds, DMT3024, DMT2264, and MAL3-39 are structurally closely related. They share a benzyl ester pyrimidine core that is substituted at C-4 with an arene moiety, and an N-alkylated amide side chain that is attached via a 3–5 carbon linker. They differ in their side chain lipophilicity and in the presence of the morpholine moiety on DMT3024 and MAL3-39, which is absent in DMT2264. The most potent compounds, MAL2-215 and MAL2-213, have a slightly different ester substitution on the pyrimidine core, as well as a distinct tetrasubstituted pyrrole side chain. This tetrasubstituted pyrrole appears to be an important determinant of potency because modifications in ester identity (Bn vs Et), N1 linker length (C-4 vs C-6), and C-4 aryl substitution (Ph vs NO2) do not appear to affect activity. Finally, MAL2-61, MAL2-39, and MAL2-29 are truncated derivatives that still have the signature pyrimidine heterocycles but a minimal N1 side chain substituent lacking an amide function, i.e. H or Bn (JAB75), butyl (MAL2-39), and hexanoic acid (MAL2-29).
In summary, we describe the discovery of a novel class of anti-malarial agents. In the future, second-generation chemical libraries of these pyrimidinone sub-classes should be analyzed for their effects on P. falciparum, but already we have identified compounds that have equal or greater potency in the [3H]hypoxanthine uptake assay as established antimalarial agents and that are synthetically readily modified.31 Based on the success of our initial efforts, reported herein, we are confident that compounds with greater potencies and improved pharmacological properties are within reach.
3. Experimental
3.1. Synthesis of compounds using in this study
The synthesis of MAL3-39 and MAL2-215,19 DMT3024,18 and DMT226431 has previously been reported. The following compounds were synthesized as described.
3.1.1. Synthesis of MAL2-29
TFA (50 μL) was added to a mixture of 6-ureiadohexanoic acid (100 mg, 0.57 mmol), 3-nitrobenzaldehyde (73 mg, 0.48 mmol), and ethylacetate (61 μL, 0.48 mmol) in dichloroethane (3 mL), and the resulting mixture was heated to reflux under N2 for 12 h. Next, dichloroethane was removed in vacuo, and the residue was stirred in Et2O, filtered, and washed with hexane/acetone to afford UPCMLD00WMAL2-29 (128.0 mg, 53%) as a white solid. 1H NMR (300 MHz, DMSO-d6) δ 11.96 (s, 1H), 8.12 (d, 1H, J = 3.8 Hz), 8.06 (s, 1H), 7.65–7.63 (m, 2H), 5.26 (d, 1H, J = 3.6 Hz), 4.02 (q, 2H, J = 7.0 Hz), 3.87–3.83 (m, 1H), 3.48–3.42 (m, 1H), 3.33 (s, 1H), 2.49 (s, 3H), 2.09 (t, 2H, J = 7.2 Hz), 1.48–1.29 (m, 6H), 1.10 (t, 3H, J = 7.1 Hz); MS (API-ES) m/z (rel. intensity) 402.1 (MH+, 100); purity determined by LC–UV at 254 nm: 94.9%.
3.1.2. Synthesis of MAL2-39
Benzyl acetoacetate (248 μL, 1.43 mmol) and para-anisaldehyde (175 μL, 1.43 mmol) were added to a solution of butylurea acid (200 mg, 1.72 mmol) in THF (3 mL). The solution was stirred for 10 min followed by the addition of concentrated HCl (two drops) and the resulting solution was stirred at ambient temperature for 48 h. The reaction mixture was concentrated to an oil, which was then purified by chromatography on SiO2 with hexane/EtOAc (2:1) and eluted with CHCl3 to afford UP-CMLD00WMAL2-39 (502.9 mg, 86%). 1H NMR (300 MHz, CDCl3) δ 7.22–7.17 (m, 3H), 7.08–7-05 (m, 2H), 7.01 (dd, 2H, J = 8.7, 2.0 Hz), 6.69 (dd, 2H, J = 8.8, 2.1 Hz), 5.40 (d, 1H, J = 2.7 Hz), 5.22 (d, 1H, J = 2.7 Hz), 4.98 (d, 2H, J = 2.6 Hz), 3.82–3.75 (m, 1H), 3.70 (s, 3H), 3.53–3.47 (m, 1H), 2.45 (s, 3H), 1.49–1.43 (m, 2H), 1.26–1.19 (m, 2H), 0.84 (t, 3H, J = 7.2 Hz); MS (API-ES) m/z (rel. intensity) 409.1 (MH+, 100); purity determined by LC–UV at 254 nm: 92.2%.
3.1.3. Synthesis of MAL2-61
Ethyl acetoacetate (141 μL, 1.11 mmol) and 2-formylphenyl-2-nitrobenzenesulfonate (341 mg, 1.11 mmol) were added to a solution of benzylurea acid (200 mg, 1.33 mmol) in THF (3 mL). The solution was stirred for 10 min followed by the addition of concentrated HCl (two drops) and the resulting solution was stirred at ambient temperature for 48 h. The reaction mixture was concentrated to a viscous oil, which was then purified by chromatography on SiO2 and eluted with hexane/EtOAc (2:1) to afford UP-CMLD00WMAL2-61 (488.3 mg, 80%) as a fine solid. 1H NMR (300 MHz, CDCl3) δ 8.07 (d, 1H, J = 7.5 Hz), 7.96–7.88 (m, 2H), 7.78 (ddd, 1H, J = 14.6, 7.2, 2.2 Hz), 7.39–7.17 (m, 8H), 6.97 (dd, 1H, J = 7.1, 2.2 Hz), 5.90 (s, 2H), 5.22 (d, 1H, J = 16.3 Hz), 4.94 (d, 1H, J = 16.3 Hz), 4.00–3.90 (m, 2H), 2.56 (s, 3H), 1.04 (t, 3H, J = 7.1 Hz); 13C NMR (300 MHz, CDCl3) δ 165.5, 152.7, 150.4, 148.7, 145.5, 137.9, 137.2, 135.8, 132.3, 132.1, 129.2, 129.1, 128.7, 128.6, 128.4, 127.4, 126.7, 125.2, 122.3, 102.8, 60.2, 48.3, 45.9, 16.3, 13.6; MS (API-ES) m/z (rel. intensity) 552.2 (MH+, 100); purity determined by LC–UV at 254 nm: 83.8%.
3.1.4. Synthesis of MAL2-213
A stirred solution of dihydropyrimidinone (20 mg, 0.051 mmol) in CH2Cl2 (1 mL) containing DMAP (10 mg, 0.082 mmol) was treated with EDCI (11 mg, 0.056 mmol) followed by methyl 1-(2-aminoethyl)- 2-methyl-5-phenyl-1H-pyrrole-3-carboxylate (15 mg, 0.051 mmol). The resulting solution was stirred/shaken at room temperature for 18 h. The mixture was washed with water, dried over MgSO4, and then purified by chromatography on SiO2 and eluted with hexane/EtOAc (3:1) and CH3Cl/EtOAc (6:1) to afford UPCMLD00WMAL2-213 (26 mg, 81%). 1H NMR (300 MHz, CDCl3) δ 8.18 (d, 2H, J = 6.8 Hz), 7.45–7.34 (m, 7H), 6.55 (s, 1H), 5.74 (br s, 1H), 5.54 (br s, 1H), 5.47 (d, 1H, J = 3.1 Hz), 4.16–4.09 (m, 4H), 3.80 (s, 3H), 3.79–3.73 (m, 1H), 3.63–3.60 (m, 1H), 3.30–3.23 (m, 2H), 2.62 (s, 3H), 2.55 (s, 3H), 1.99 (t, 2H, J = 6.5 Hz), 1.83–1.72 (m, 2H), 1.20 (t, 3H, J = 7.1 Hz); MS (APCI) m/z (rel. intensity) 1285 (2MNa+, 60), 686 (100), 664 (40), 654 (MNa+, 40), 632 (MH+, 10); purity determined by 1H NMR: >95%.
3.1.5. Synthesis of JAB75
In the presence of polyphosphate ester (PPE, 300.0 mg), a solution of methyl 3-oxo-4-phenylbutanoate (384.0 mg, 2.0 mmol), 2-naphthaldehyde (312.0 mg, 2,0 mmol) and urea (180.0 mg, 3.0 mmol) in THF (40 mL) was heated at reflux for 15 h. The product was extracted into EtOAc, and the combined EtOAc extracts were dried (Na2SO4), concentrated, and purified by chromatography on SiO2 with hexane/EtOAc (3:1) to afford 330 mg (44%) of UP-CMLD00WJAB75. 1H NMR (300 MHz, DMSO-d6) δ 9.43 (s, 1H), 7.91–7.75 (m, 4H), 7.58–7.24 (m, 8H), 5.35 (d, 1H, J = 3.3 Hz), 4.12, 4.08 (AB, 2H, J = 13.7 Hz), 3.52 (s, 3H); MS (API-ES) m/z (rel. intensity) 373 ([M+H]+, 100), 395 ([M+Na]+, 15); HRMS (EI) m/z calcd for C23H20N2O3 372.1474, found 372.1477; purity determined by LC–UV at 254 nm: 95%.
3.2. Protein purifications
The yeast Hsp70, Ssa1, and a J domain-containing Hlj1-glutathi-one-S-transferase protein chimera were purified as previously described. 26,32 Ydj1 was kindly provided by Dr. D. Cyr (University of North Carolina School of Medicine), and Hdj1 was purchased from StressGen/Assay Designs. The purification of a hexahistidine-tagged version of P. falciparum Hsp70 (PF08_0054) was performed as described33 with several modifications. The plasmid pQE30-P. falciparum Hsp70 was introduced into chemically competent Escherichia coli Rosetta 2 cells (Novagen), and transformed colonies were selected on LB with ampicillin and chloramphenicol. Over-night cultures were grown at 26 °C and diluted 1:10 into 1 L of the same media, the cells were grown to mid-log phase, and IPTG was added before growth was continued for another 5 h at 26 °C. Cells were harvested and lysed, and the extract was loaded on a nickel-chelating Sepharose column in lysis buffer (8 M urea, 300 mM NaCl, 10 mM Tris, pH 8.0, 10 mM imidazole, 1 mM PMSF, 2 mM leupeptin, 0.7 mM pepstatin A). The column was washed with 10 mL of low imidazole buffer (10 mM Tris, pH 8.0, containing 300 mM NaCl and 10 mM imidazole), followed by 20 mM imidazole buffer, and the bound proteins were eluted using a 10 mL × 10 mL linear gradient of 25 mM to 500 mM imidazole. Samples from 1 mL fractions were analyzed by SDS–PAGE and peak fractions were pooled and dialyzed against 50 mM Tris, pH 7.4, containing 50 mM NaCl, 0.8 mM DTT, 2 mM MgCl2, and 5% glycerol. The dialysate was then loaded onto a Q-Sepharose column and washed, and the purified protein was eluted with 50 mM Tris, pH 7.4, containing 200 mM NaCl, 0.8 mM DTT, 2 mM MgCl2, and 5% glycerol. Peak fractions were identified and were dialyzed as above. Protein aliquots were frozen in liquid nitrogen and stored at −80 °C. The activity of the enzyme was comparable to that isolated using previously described methods.33
Human Hsp70 (HSPA1A) was purified in one of two manners. First, an untagged version was purified based on a protocol provided by R. Morimoto (Northwestern University). A 25 mL culture of E. coli containing plasmid pMSHSP BL21 FS in LB-ampicillin was used to inoculate a 1 L culture, and after 4 h at 37 °C protein expression was induced with IPTG. After another 6 h, the cells were harvested and washed, and the pellet was frozen in 50 mL of TEK50 buffer (20 mM Tris, pH 8.0, containing 0.1 mM EDTA and 50 mM KCl). The thawed pellets were resuspended in TEK50 containing 1 mg/ml lysozyme, 0.5 mM PMSF, 1 μg/ml leupeptin, and 1 μg/ml pepstatin A, and the cells were subjected to three freeze-thaw cycles and lysed by sonication. Cell debris was removed by low-speed centrifugation, and the supernatant was cleared by ultracentrifugation and chromatographed on a DEAE column (Sigma-Aldrich) equilibrated with TMC50 buffer (20 mM Tris, pH 7.2, containing 5 mM MgCl2 and 50 mM NaCl). Bound proteins were eluted with a step gradient of TMC50 and TMC100 (20 mM Tris, pH 7.2, containing 5 mM MgCl2 and 100 mM NaCl), and fractions containing Hsp70, as assessed by SDS-PAGE, were pooled and loaded onto an ATP-agarose column (Sigma-Aldrich), which was recirculated for 4 h. The column was washed with TMC100 and the bound Hsp70 was eluted with TMC100 containing 10% glycerol and 25 mM ATP, and then TMC100 containing glycerol, ATP, and 4 M NaCl. Fractions containing the majority of the Hsp70 protein, as assessed above, were pooled and dialyzed against 20 mM Tris, pH 7.2, containing 50 mM NaCl and 1 mM EDTA. The final dialysate was loaded onto a Q-Sepharose column (Sigma–Aldrich) equilibrated with TMC100, and the column was washed with TMC100 and the purified Hsp70 was eluted using a linear gradient of TMC100 to TMC400. The purified Hsp70 was dialyzed against 50 mM Tris, pH 7.4, containing 50 mM NaCl, 0.8 mM DTT, 2 mM MgCl2, and 5% glycerol, snap-frozen in liquid nitrogen, and stored at −80 °C.
Second, a hexastidine (His6)-tagged form of human Hsp70 was constructed and purified, and significantly higher yields of protein were obtained. To this end, plasmid DNA pMSHSP (see above) was used as template in a polymerase chain reaction using oligonucleotide primers with the following sequences: ACGATCGAAGAAGTGG ACCACCATCACCATCACCATTAG (forward) and GTCCACTTCTTCGA TCGTGGGGCCTGACCCAGACCCTCC (reverse), which replaced the six C-terminal amino acids with a His6 polypeptide tag at the C-terminus of the Hsp70 protein coding sequence. A ligation-independent mutagenesis protocol was used to design these primers and carry out the mutagenesis cloning.34 The sequence of the protein coding region of the recombinant plasmid was confirmed by DNA sequencing. For expression of recombinant Hsp70–His6, E. coli cultures containing the recombinant plasmid (pMSHSP- His6) were grown in LB containing 100 μg/ml ampicillin to O.D.600 = 0.8 at 37 °C, and induced at 25 °C for 18 h with 0.4 mM IPTG. Following centrifugation, the cell pellets were lysed by a combination of sonication (model W-220F, Branson) and passing the cells through a pressurized cell disruptor (Microfluidics) in 50 mM Tris, pH 8.0, containing 500 mM NaCl, 10% glycerol, and 20 mM imidazole. The Hsp70 was then purified at 4 °C using a standard purification protocol recommended for use with Ni-NTA resin (QIAGEN), and the recombinant Hsp70–His6 was eluted from the resin with 100 mM Tris, pH 8.0, containing 200 mM imidazole and 500 mM NaCl,. Next, the protein was further purified by size exclusion chromatography using a preparative Superdex-200 gel filtration column equilibrated in 50 mM Tris, pH 8.0, containing 500 mM NaCl. Fractions containing the purified protein were dialyzed over-night in 10 mm Tris, pH 8.0, at 4 °C, and the dialyzed protein was pooled, aliquoted, and stored at −70 °C.
3.3. In vitro and cell-based assays
Steady state and single turnover ATPase assays with the indicated Hsp70 preparations were performed as previously described.18,19
Compounds were assessed for their ability to inhibit the growth of P. falciparum asexual blood stages using [3H]hypoxanthine up-take assays, involving exposure of parasites to drug for 72 h and adding [3H]hypoxanthine for the last 24 h, as previously described. 35 Each compound was screened at final concentrations of 1 μM and 5 μM. Compounds that initially appeared to inhibit [3H]hypoxanthine uptake by >50% at 5 μM and displayed at least as much inhibition at 1 μM were selected and re-screened. The group of nine compounds that passed this second test (Fig. 2) was then subjected to dose-response analyses. IC50 values were calculated using a linear regression analysis based on the linear portions of plotted values. The IC50 value for CQ was assessed as described.23
The effect of the compounds on the growth of HepG2 and WI-38 cells was assessed as previously described.18 Data are reported as GI50 values and were calculated relative to a DMSO (negative) control.
Supplementary Material
Acknowledgments
This work was supported in part by a pilot grant from the University of Pittsburgh Drug Discovery Institute to J.L.B. R.J.C. was supported by a fellowship from the Beckman Foundation, and S.B. received funding from a Howard Hughes Medical Institute undergraduate research program. P.W. and D.M.H. thank the NIH-NIGMS for Grant P50-GM067082 for support of the CMLD program at the University of Pittsburgh. We also wish to thank Greg Blatch, James Burden, Pete Chambers, Doug Cyr, Myriam Gorospe, Michael Lyon, Richard Morimoto, Stephanie Nicolay, Bill Strieff, David Turner, and Stefan Werner for technical assistance and/or for providing reagents.
Footnotes
Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bmc.2009.01.024.
References and notes
- 1.McKerrow JH. PLoS Med. 2005;2:e210. doi: 10.1371/journal.pmed.0020210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pecoul B. PLoS Med. 2004;1:e6. doi: 10.1371/journal.pmed.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Botha M, Pesce ER, Blatch GL. Int J Biochem Cell Biol. 2007;39:1781. doi: 10.1016/j.biocel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 4.Shonhai A, Boshoff A, Blatch GL. Protein Sci. 2007;16:1803. doi: 10.1110/ps.072918107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayer MP, Bukau B. Cell Mol Life Sci. 2005;62:670. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig EA, Huang P, Aron R, Andrew A. Rev Physiol Biochem Pharmacol. 2006;156:1. doi: 10.1007/s10254-005-0001-0. [DOI] [PubMed] [Google Scholar]
- 7.Hartl FU. Nature. 1996;381:571. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 8.Tonkin CJ, Pearce JA, McFadden GI, Cowman AF. Curr Opin Microbiol. 2006;9:381. doi: 10.1016/j.mib.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 9.Fewell SW, Travers KJ, Weissman JS, Brodsky JL. Annu Rev Genet. 2001;35:149. doi: 10.1146/annurev.genet.35.102401.090313. [DOI] [PubMed] [Google Scholar]
- 10.Maier AG, Rug M, O’Neill MT, Brown M, Chakravorty S, Szestak T, Chesson J, Wu Y, Hughes K, Coppel RL, Newbold C, Beeson JG, Craig A, Crabb BS, Cowman AF. Cell. 2008;134:48. doi: 10.1016/j.cell.2008.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunt NH, Stocker R. Blood Cells. 1990;16:499. [PubMed] [Google Scholar]
- 12.Frydman J, Nimmesgern E, Ohtsuka K, Hartl FU. Nature. 1994;370:111. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- 13.Banumathy G, Singh V, Pavithra SR, Tatu U. J Biol Chem. 2003;278:18336. doi: 10.1074/jbc.M211309200. [DOI] [PubMed] [Google Scholar]
- 14.Pavithra SR, Kumar R, Tatu U. PLoS Comput Biol. 2007;3:1701. doi: 10.1371/journal.pcbi.0030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Midorikawa Y, Haque QM, Nakazawa S. Chemotherapy. 1998;44:409. doi: 10.1159/000007152. [DOI] [PubMed] [Google Scholar]
- 16.Ramya TN, Karmodiya K, Surolia A, Surolia N. J Biol Chem. 2007;282:6388. doi: 10.1074/jbc.M610251200. [DOI] [PubMed] [Google Scholar]
- 17.Nadeau K, Nadler SG, Saulnier M, Tepper MA, Walsh CT. Biochemistry. 1994;33:2561. doi: 10.1021/bi00175a027. [DOI] [PubMed] [Google Scholar]
- 18.Wright CM, Chovatiya RJ, Jameson NE, Turner DM, Zhu G, Werner S, Huryn DM, Pipas JM, Day BW, Wipf P, Brodsky JL. Bioorg Med Chem. 2008;16:3291. doi: 10.1016/j.bmc.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fewell SW, Smith CM, Lyon MA, Dumitrescu TP, Wipf P, Day BW, Brodsky JL. J Biol Chem. 2004;279:51131. doi: 10.1074/jbc.M404857200. [DOI] [PubMed] [Google Scholar]
- 20.Fewell SW, Day BW, Brodsky JL. J Biol Chem. 2001;276:910. doi: 10.1074/jbc.M008535200. [DOI] [PubMed] [Google Scholar]
- 21.Rodina A, Vilenchik M, Moulick K, Aguirre J, Kim J, Chiang A, Litz J, Clement CC, Kang Y, She Y, Wu N, Felts S, Wipf P, Massague J, Jiang X, Brodsky JL, Krystal GW, Chiosis G. Nat Chem Biol. 2007;3:498. doi: 10.1038/nchembio.2007.10. [DOI] [PubMed] [Google Scholar]
- 22.Noedl H, Krudsood S, Leowattana W, Tangpukdee N, Thanachartwet W, Looareesuwan S, Miller RS, Fukuda M, Jongsakul K, Yingyuen K, Sriwichai S, Ohrt C, Knirsch C. Antimicrob Agents Chemother. 2007;51:651. doi: 10.1128/AAC.01023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sidhu AB, Verdier-Pinard D, Fidock DA. Science. 2002;298:210. doi: 10.1126/science.1074045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wellems TE, Panton LJ, Gluzman IY, do Rosario VE, Gwadz RW, Walker-Jonah A, Krogstad DJ. Nature. 1990;345:253. doi: 10.1038/345253a0. [DOI] [PubMed] [Google Scholar]
- 25.Shonhai A, Boshoff A, Blatch GL. Mol Genet Genom. 2005;274:70. doi: 10.1007/s00438-005-1150-9. [DOI] [PubMed] [Google Scholar]
- 26.Youker RT, Walsh P, Beilharz T, Lithgow T, Brodsky JL. Mol Biol Cell. 2004;15:4787. doi: 10.1091/mbc.E04-07-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandez-Saiz V, Moro F, Arizmendi JM, Acebron SP, Muga A. J Biol Chem. 2006;281:7479. doi: 10.1074/jbc.M512744200. [DOI] [PubMed] [Google Scholar]
- 28.Jiang J, Prasad K, Lafer EM, Sousa R. Mol Cell. 2005;20:513. doi: 10.1016/j.molcel.2005.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang YC, Chang HC, Chakraborty K, Hartl FU, Hayer-Hartl M. EMBO J. 2008;27:1458. doi: 10.1038/emboj.2008.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ekland EH, Fidock DA. Curr Opin Microbiol. 2007;10:363. doi: 10.1016/j.mib.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Werner S, Turner DM, Lyon MA, Huryn DM, Wipf P. Synlett. 2006:2334. [Google Scholar]
- 32.McClellan AJ, Brodsky JL. Genetics. 2000;156:501. doi: 10.1093/genetics/156.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matambo TS, Odunuga OO, Boshoff A, Blatch GL. Protein Exp Purif. 2004;33:214. doi: 10.1016/j.pep.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 34.Chiu J, March PE, Lee R, Tillett D. Nucleic Acids Res. 2004;32:e174. doi: 10.1093/nar/gnh172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fidock DA, Nomura T, Wellems TE. Mol Pharmacol. 1998;54:1140. doi: 10.1124/mol.54.6.1140. [DOI] [PubMed] [Google Scholar]
- 36.Vembar, S.S.; Jin, Y.; Brodsky, J.L.; Hendershot, L.M., in preparation.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.