Abstract
The present investigation identifies the molecular basis for the well-documented inhibition of the mitochondrial inner membrane citrate transport protein (CTP) function by the lysine-selective reagent pyridoxal 5′-phosphate. Kinetic analysis indicates that PLP is a linear mixed inhibitor of the Cys-less CTP, with a predominantly competitive component. We have previously concluded that the CTP contains at least two substrate binding sites which are located at increasing depths within the substrate translocation pathway and which contain key lysine residues. In the present investigation, the roles of Lys-83 in substrate binding site one, Lys-37 and Lys-239 in substrate binding site two, and four other off-pathway lysines in conferring PLP-inhibition of transport was determined by functional characterization of seven lysine to cysteine substitution mutants. We observed that replacement of Lys-83 with cysteine resulted in a 78% loss of the PLP-mediated inhibition of CTP function. In contrast replacement of either Lys-37 or Lys-239 with cysteine, caused a modest reduction in the inhibition caused by PLP (i.e., 31% and 20% loss of inhibition, respectively). Interestingly, these losses of PLP-mediated inhibition could be rescued by covalent modification of each cysteine with MTSEA, a reagent that adds a lysine-like moiety (i.e. SCH2CH2 NH3+) to the cysteine sulfhydryl group. Importantly the replacement of non-binding site lysines (i.e., Lys-45, Lys-48, Lys-134, Lys-141) with cysteine, resulted in little change in the PLP inhibition. Based upon these results, we conducted docking calculations with the CTP structural model leading to the development of a physical binding model for PLP. In combination, our data support the conclusion that PLP exerts its main inhibitory effect by binding to residues located within the two substrate binding sites of the CTP, with Lys-83 being the primary determinant of the total PLP effect since the replacement of this single lysine abolishes nearly all of the observed inhibition by PLP.
Keywords: citrate transporter, liposomes, membrane proteins, mitochondria, pyridoxal 5′-phosphate, substrate binding site
INTRODUCTION
The mitochondrial citrate transport protein (CTP)1 catalyzes the efflux of citrate across the mitochondrial inner membrane via an obligatory exchange mechanism. In higher eukaryotes the coupled exchange anion can be either external tricarboxylate (i.e., citrate, isocitrate), dicarboxylate (malate), or phosphoenolpyruvate (Palmieri et al., 1972). In yeast, the exchange partner is thought to be external isocitrate (Kaplan et al., 1995). Following passive diffusion across the outer mitochondrial membrane through a voltage-dependent anion channel (Mannella, 1997), cytosolic citrate serves as the main carbon source supporting fatty acid, triacylglycerol, and sterol biosyntheses (Watson and Lowenstein, 1970; Brunengraber and Lowenstein, 1973; Endemann et al., 1982, Conover,1987). Consequently, the CTP occupies a prominent position within eukaryotic intermediary metabolism.
Due to both the importance of the mitochondrial CTP in cellular bioenergetics, as well as its altered function in certain diseases (Kaplan et al., 1982, 1990b), we have extensively studied this transporter from multiple sources. Thus the CTP has been purified in reconstitutively active form (Kaplan et al., 1990a), cloned (Kaplan et al., 1993), and overexpressed (Kaplan et al., 1995; Xu et al., 1995). We then developed a homology modeled structure of the yeast mitochondrial CTP based on the x-ray structure of the mitochondrial ADP/ATP carrier (Walters and Kaplan, 2004). By superposition of functional data, obtained via chemical modification of site-directed CTP mutants, onto this model we have been able to clearly identify substantial portions of the substrate translocation pathway through the CTP (Kaplan et al., 2000; Ma et al., 2004; Ma et al., 2005). Furthermore, transport kinetic studies conducted with a panel of single-Cys CTP mutants in combination with molecular modeling studies, has permitted the clear-cut identification of two substrate binding sites within the CTP that reside at increasing depths in the bilayer (Ma et al., 2007). The composition of these binding sites highlights the importance of lysine and arginine residues in substrate (i.e., citrate) binding.
With this background in mind, the present investigations were launched to identify the lysine(s) within the CTP that is responsible for the inhibition caused by pyridoxal 5′-phosphate, a lysine-selective reagent (Lundblad, 1991), and to glean new insight regarding the functioning of the transporter’s substrate binding sites. We observed that: i) PLP is a linear-mixed inhibitor of the Cys-less CTP, with a predominantly competitive component; ii) replacement of substrate binding site lysines, one at a time with cysteine, caused varying degrees of loss of the PLP inhibition, with Lys-83 being the primary determinant of the total PLP effect; iii) replacement of non-binding site lysines with cysteine, resulted in little change in the PLP-mediated inhibition; and iv) restoration of a positively charged lysine-like side-chain within a substrate binding site, by chemical modification of a given cysteine substitution mutant with MTSEA, restored the observed PLP inhibition. These findings, combined with docking calculations, have permitted the development of a PLP binding model, which provides new insight into the roles of the CTP binding sites in the PLP-mediated inhibition of CTP function.
EXPERIMENTAL PROCEDURES
Construction, Overexpression, Isolation, and Incorporation of Cys-less and Single-Cys CTP Variants into Liposomal Vesicles
Single-Cys CTP mutants were constructed using the QuikChange site-directed mutagenesis kit (Stratagene) as previously described (Ma et al., 2004). The Cys-less yeast mitochondrial CTP gene in pET-21a(+) was utilized as the starting template (Xu et al., 2000). Mutations were confirmed by sequencing both strands of the entire CTP open reading frame. BL21(DE3) growth and induction of CTP expression with IPTG were conducted as previously detailed (Kaplan et al., 1995; Xu et al., 1995). Two hours following induction, cells were harvested, lysed, and the inclusion body fraction was purified by centrifugation in a sucrose gradient (Kaplan et al., 1995; Xu et al., 1995). CTP mutants were then solubilized from the inclusion body pellet with 6 ml of ice-cold 1.2% (w/v) sarkosyl. Following centrifugation at 314,000 × g for 45 min, the supernatant contained a given solubilized CTP mutant. Each mutant was then incorporated into liposomal vesicles, in the presence of 48 - 75 mM citrate, via the freeze-thaw sonication procedure as previously described (Kaplan et al., 1990a, b; Kaplan et al., 1995). It should be noted that, with single-Cys mutants K83C, K37C and K239C, whose Km values were substantially greater than that for the Cys-less CTP, the higher concentration of citrate was included in the reconstitution incubation (i.e., 75 mM). Immediately prior to the transport assay, a given sample was thawed, sonicated on ice, and the extraliposomal citrate was removed via chromatography on a Dowex column (Ma et al., 2007). The sample was immediately assayed for transport. Protein was quantified via the method of Kaplan and Pedersen, 1985.
Characterization of the Type of PLP-mediated Inhibition of the Cys-less CTP and Determination of Inhibition Constants
The type of inhibition of the Cys-less CTP mediated by PLP and the inhibition constants were determined as follows. Proteoliposomes (585μl) were incubated with 45μl of either buffer (experimental incubation) or 1,2,3-benzenetricarboxylic acid (BTC; Sigma Aldrich) (control incubation) for 10 min at 21°C. Transport reactions (40-75 sec) were then triggered by adding 49 μl of the above reaction mix to 25 μl of [1, 5-14C] citrate (GE Biosciences; 0.1 - 4.0 mM; specific radioactivity = 12.5 - 60 × 103 cpm/nmol) plus varying concentrations of PLP (0 - 40 mM; Sigma Aldrich). Note: For all experiments PLP was prepared fresh each day in sodium hydroxide and adjusted to pH 8.0. Experimental incubations were quenched by the addition of 3.5μl of 200mM BTC, whereas control incubations received an equal volume of buffer. All transport incubations contained a final concentration of 9 mM BTC. Following transport, intraliposomal radiolabeled citrate was separated from the external radiolabel via chromatography on 2.6ml of Dowex resin in Biorad support columns. The eluted intraliposomal radiolabel was quantified via liquid scintillation counting. The BTC-sensitive transport rate was calculated by subtracting the control value from the experimental value. The data were then analyzed via Dixon (1/v versus I), Cornish-Bowden (S/v versus I), and Lineweaver-Burk (1/v versus 1/S) plots, in which v corresponds to the rate of a given transport reaction. Both the Kic (the competitive inhibition constant) and Kiu (the uncompetitive inhibition constant) values were calculated using the global fit method in the Grafit program (Erithacus Software Ltd.). In this method the observed rate vs [S] data (in the presence and absence of inhibitors) can be fitted simultaneously according to the full non-linear expression for mixed inhibition
| (Equation 1) |
Effect of Pyridoxal 5′-phosphate on Reconstituted [14C]citrate/citrate Exchange Catalyzed by the Cys-less and the Single-Cys CTP Variants
Proteoliposomes (45μl) were pre-incubated with 3.5 μl of either water or varying concentrations of PLP for 10 min. 3.5 μl of either water or sodium borohydride (note: the latter was added at a concentration of 1.0 - 1.5 times that of PLP) was then added and allowed to react for 10 min. The reaction mix was then incubated with 7.0 μl of either 1M BTC (control sample) or with buffer (experimental sample) for 10 min. Transport was initiated by the addition of 21.5μl of 34.2mM of [1, 5-14C] citrate. It should be noted that prior to the transport experiments, the [14C]citrate source had been concentrated approximately 22-fold by evaporation under vacuum in order to enable construction of a stock solution with increased specific radioactivity. Transport reaction times ranged from 40s to 2.5h depending upon the intrinsic activity of a given mutant. Experimental incubations were quenched by the addition of 7.0 μl of 1M BTC, whereas the control incubation received an equal volume of buffer. All reactions were conducted at room temperature (21°C) and contained a final BTC concentration of 80 mM. Sodium borohydride was prepared fresh daily in deionized water. The BTC-sensitive transport rate was calculated by subtracting the control value from the experimental value. The effect of PLP on citrate transport is expressed as the percentage of the initial BTC-sensitive citrate uptake (measured in the presence of water) that remained in the presence of a given PLP concentration. This was calculated by: i) determining the BTC-sensitive transport rate via subtraction of the control value from the experimental value, in the absence of PLP; ii) subtracting the control value from the experimental value obtained in the presence of varying concentrations of PLP; iii) determining the ratio of this difference to the uninhibited BTC-sensitive citrate transport rate; and iv) application of the formula (1-ratio) × 100. The percentage inhibition versus PLP concentration was fitted to a one site binding equation,
| (Equation 2) |
using a non-linear least squares curve fit in Graph Pad Prism.
Effect of MTSEA Labeling of Lysine to Cysteine Substitution Mutants on the Pyridoxal 5′-phosphate-mediated Inhibition of [14C]citrate/citrate Exchange
Experiments probing the effect of MTSEA (Toronto Research Chemicals) on the pyridoxal 5′-phosphate-mediated inhibition of the [14C]citrate/citrate exchange catalyzed by substrate binding site single-Cys substitution mutants were conducted in a manner very similar to that described above. However, there were two methodological differences. First, proteoliposomes were preincubated with MTSEA (0.05 - 0.06 mM) for 10 min prior to incubation with either PLP or water. Second, NaBH4 was excluded from these incubations in order to avoid reduction of the Cys-MTS reagent disulfide bond.
Docking of Pyridoxal 5-phosphate into the CTP Substrate Binding Sites
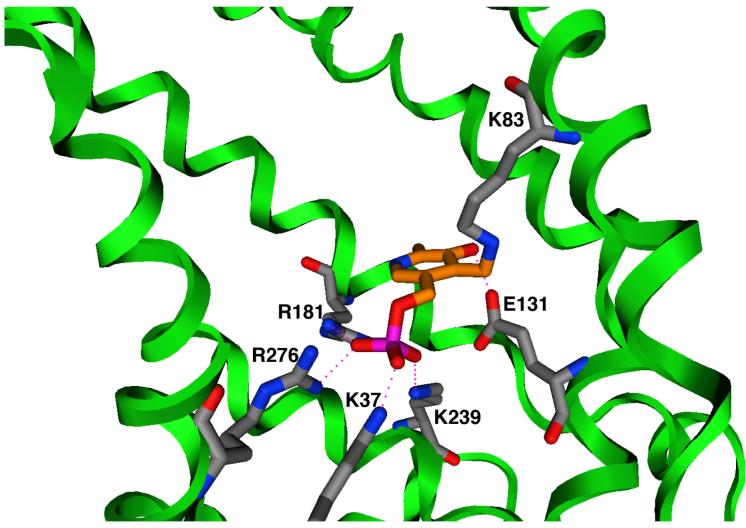
Molecular modeling was carried out using MOE (version 2007.09). CTP was homology modeled as described previously (Walters & Kaplan, 2004). PLP was modeled using the MMFF parameter set (Halgren, 1999). Initial docking orientations for PLP were generated using the Dock module of MOE. Those orientations that were consistent with the experimental data, i.e., interacting with Lys-83, Lys-37, and Lys-239, were investigated as follows. First, since the K83C mutant has, by far, the largest effect on PLP inhibition, we assumed that this lysine forms a Schiff base with the PLP, and following reduction with sodium borohydride, we modeled the covalent bond accordingly. Then we manually adjusted PLP and nearby side-chain conformations to maximize attractive interactions. Finally we fixed the positions of the protein backbone atoms and allowed the remainder of the structure to minimize. After several cycles of conformation adjustment and minimization were carried out, minimization consistently arrived at one orientation shown in Figure 5.
Figure 5. Docking of PLP within the CTP substrate binding sites.
Pyridoxal 5′-phosphate is covalently bonded to the side-chain of Lys-83. The phosphate forms ionic hydrogen bonds with Lys-37, Lys-239, Arg-181, and Arg-276. The hydroxyl group of PLP forms an ionic hydrogen bond with Glu-131. The protein backbone is depicted as a green ribbon; blue = nitrogen, red = oxygen, magenta = phosphorus, gray = protein carbon atoms, orange = PLP carbon atoms. Hydrogen bonds are shown as dotted lines.
RESULTS
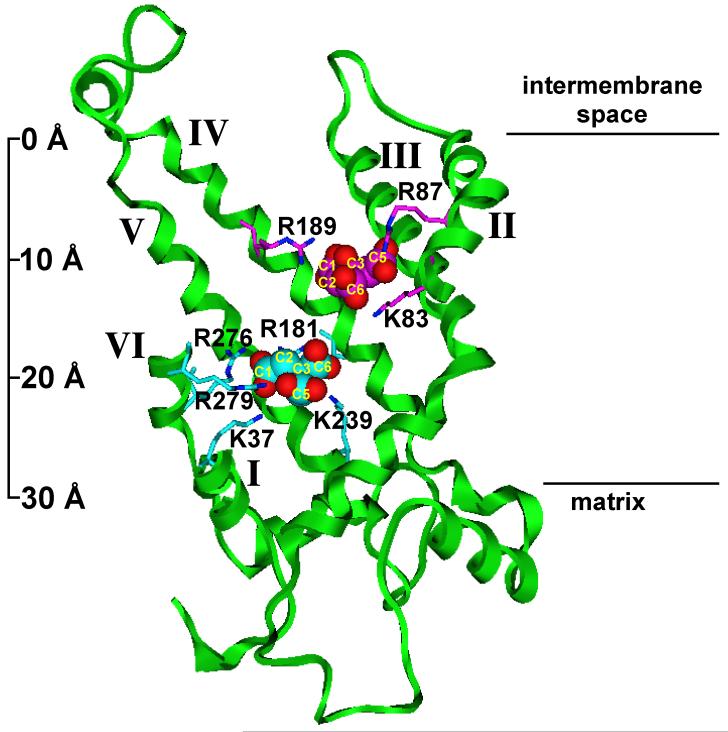
We previously demonstrated that the lysine-selective reagent, pyridoxal 5′-phosphate, inhibits the function of the purified CTP from rat liver mitochondria in a manner that could be protected by substrate, suggesting that the reagent interacted with a lysine(s) residue that resided at or near the substrate binding site(s) (Gremse et al., 1995). This finding elicited renewed interest in light of our recent report (Ma et al., 2007) wherein we clearly identified two substrate binding sites within the yeast mitochondrial CTP that reside at different depths within the membrane bilayer (see Figure 1). The residues comprising these binding sites were identified via kinetic characterization of single-Cys substitution mutants in combination with molecular modeling approaches. As depicted in Figure 1, each substrate binding site contains at least one lysine residue. Thus, in the present studies, we sought to identify the specific PLP-lysine interaction(s) that results in inhibition of citrate transport.
Figure 1. Two citrate binding sites within the CTP transport pathway viewed in the plane of the membrane bilayer.
Two citrate binding sites, viewed in the plane of the membrane bilayer (i.e., a side view) are presented. The backbone of the CTP is represented as a green ribbon. Important side chains are shown as stick structures, and citrate molecules are shown as space filling structures. Citrate oxygens are red. In site one, the citrate carbons and the directly interacting side chains are magenta; in site two, the citrate carbons and the directly interacting side chains are cyan. The distance between the two sites (measuring at the central carbon atoms of the two citrates) is 9.2 Å. Portions of TMDs I, II, and VI have been cut away for clarity. Black horizontal lines at right indicate the approximate boundaries of the bilayer and the atomic ruler at the left indicates approximate dimensions. Reproduced from Ma et al. (2007) with permission from the J. Biol. Chem.
Characterization of the Kinetic Mechanism of the PLP-Mediated Inhibition of the Cys-less CTP
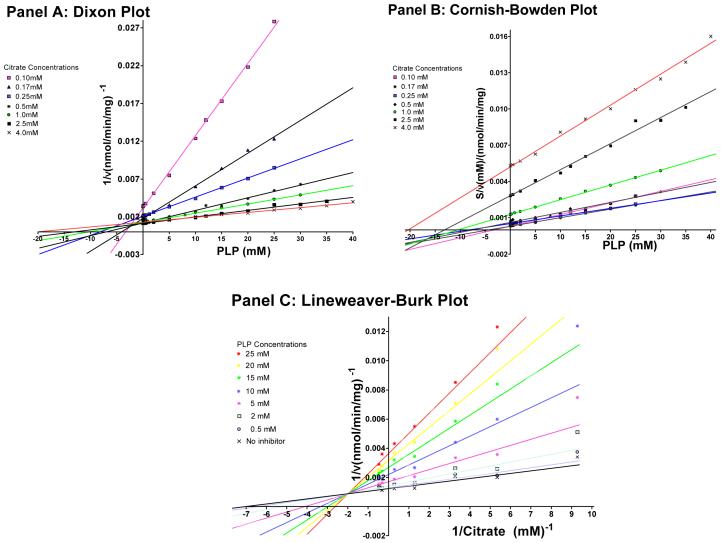
Our initial studies focused on characterizing the PLP-mediated inhibition of the Cys-less CTP variant. For these studies the transport rate was measured at a variety of different citrate and PLP concentrations. It is important to note that these studies were conducted under conditions in which the condensation of PLP with a lysine amino group to form a Schiff base is reversible (i.e., in the absence of sodium borohydride) (Rippa et al., 1967; Lundblad, 1991; Dierks et al., 1992). Figure 2 depicts these results displayed in three different types of plots in order to determine the most appropriate inhibition model. Panel A depicts a Dixon plot (1/v vs. [I]) of the data, in which the pattern of intersecting lines indicates either a competitive or a mixed inhibition mode (Cornish-Bowden, 1974). However, since this plot does not distinguish competitive from mixed inhibition, the data were also plotted via a Cornish-Bowden plot (S/v vs. [I]). The fact that the Cornish-Bowden Plot does not reveal parallel lines indicates that the inhibition is not strictly competitive. Rather, the pattern of intersecting lines is most consistent with a mixed type of inhibition (Cornish-Bowden, 1974). However, since this plot is not always able to distinguish between uncompetitive and mixed inhibition, the data were also plotted via the classical Lineweaver-Burk plot (1/v vs. 1/S). In this plot we observed a family of lines that do not intersect on either the X- or the Y-axes, thereby indicating changes in both the Km and the Vmax values. Based on the patterns of inhibition seen in these three plots, a model of mixed inhibition was chosen to describe the PLP-mediated inhibition of the CTP and a global fit of the data using the Marquart algorithm as implemented by the program GRAFIT was conducted to determine the parameters Vmax, Km, Kic (the competitive inhibition constant), and Kiu (the uncompetitive inhibition constant) in Equation 1 (see “Experimental Procedures”). The competitive inhibition constant for PLP (Kic = 3.6 ± 0.8 mM) was found to be approximately 5 times less than the uncompetitive inhibition constant (Kiu = 17.7 ± 2.3 mM) indicating that PLP acts predominantly by a competitive mechanism. The competitive aspect of this inhibition arises due to PLP binding to the same location as citrate.
Figure 2. Determination of the kinetic type of PLP-mediated inhibition of the Cys-less CTP.
The rate of BTC-sensitive [14C]citrate/citrate exchange was determined in the presence of varying concentrations (i.e., 0 - 40 mM) of PLP. These inhibition plots were determined at multiple citrate concentrations ranging from 0.1 mM - 4.0 mM. Best fit lines in Panels A and B were constructed according to linear regression analysis by the methods of least squares. In Panel C the lines were derived from a Global Fit to Equation 1 at each PLP concentration. Other conditions were as described under “Experimental Procedures”.
PLP-Mediated Inhibition of CTP Lysine to Cysteine Substitution Mutants
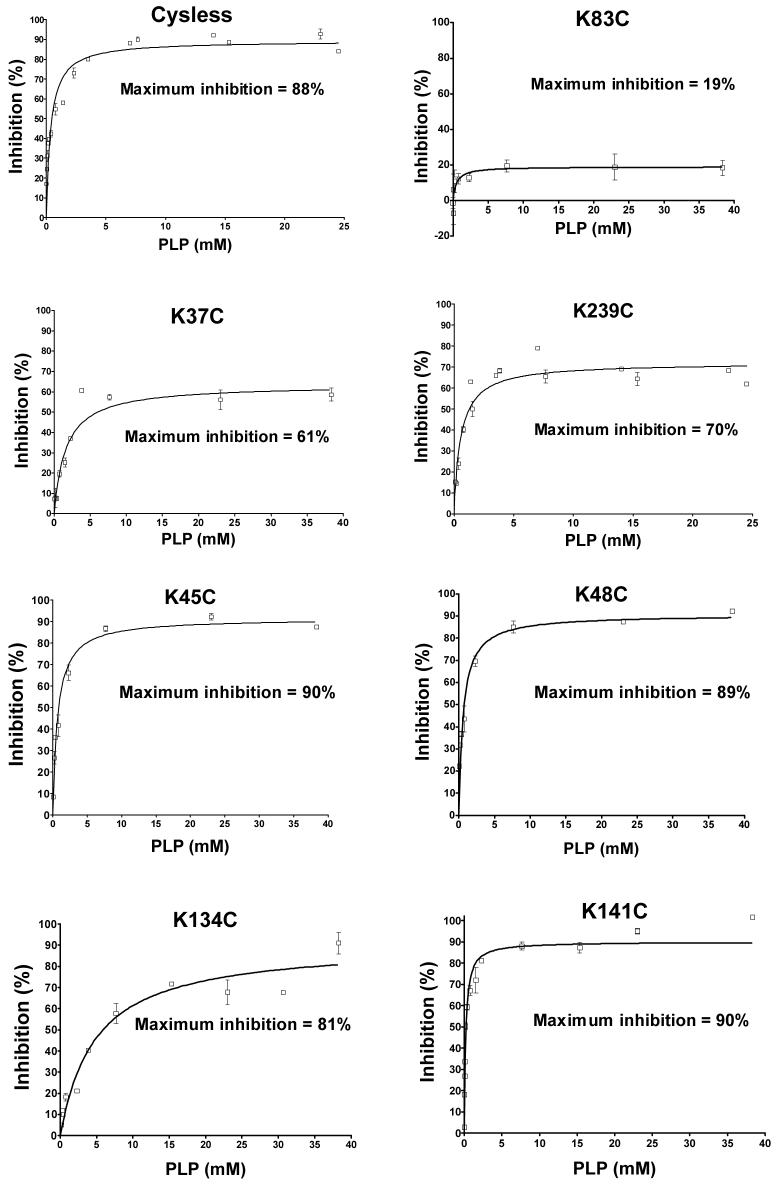
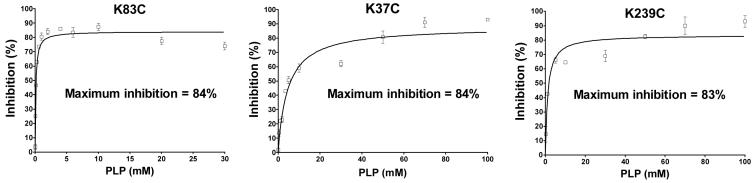
We have quantified the ability of varying concentrations of PLP to inhibit the Cys-less and seven single-Cys substitution mutants. As depicted in Figure 3, PLP causes an 88% inhibition of the Cys-less CTP. In contrast, with the Lys83Cys substitution mutant, only 19% inhibition is observed. Thus Lys-83, located in binding site one, is responsible for most of the PLP effect. Upon substitution of site two residues Lys-37 and Lys-239 with Cys, the PLP inhibition was only slightly reduced (i.e., from 88% to 61% and 70%, respectively). In contrast, upon substitution of cysteines for lysines located in non-binding site regions of the protein (Lys-45: Matrix Loop A; Lys-48: Matrix Loop A; Lys-134: Transmembrane Domain III, near binding site two; and Lys-141:Matrix Loop C), little reduction in the PLP-mediated inhibition was observed. Finally, replacement of Glu-131 or Arg-181 (i.e., two residues that are thought to form ionic hydrogen bonds with the PLP; see Figure 5) with Cys caused little change in the PLP inhibition (i.e., from 88% to 80% and 76%, respectively; data not shown).
Figure 3. Inhibition of yeast mitochondrial CTP variants by PLP.
The Cys-less CTP, as well as single lysine to cysteine substitution mutants were incubated with varying concentrations of PLP for 10 min followed by incubation with sodium borohydride (1.0-1.5-fold excess relative to PLP concentration) for 10 min. BTC-sensitive citrate transport was then measured at 21°C. Data represent means ± S.E. of incubations repeated 2-6 times. Percent maximum inhibition was obtained from the best fit curve generated via non-linear regression analysis using Equation 2 in “Experimental Procedures”. Other conditions were as described under “Experimental Procedures”.
Influence of Chemical Modification of Substrate Binding Site Single-Cys CTP Variants on PLP-Mediated Inhibition
Figure 4 depicts the effect of chemical modification of lysine to cysteine substitution mutants, located within the substrate binding sites, with MTSEA. This reagent is highly polar, cysteine-specific, and forms a mixed disulfide by covalent attachment of SCH2CH2NH3+ to the cysteine sulfhydryl, producing a lysine-like side-chain (Ma et al., 2007). The data show that modification of cysteines substituted at binding site locations 83, 37, 239 with MTSEA, causes a near total rescue of the PLP-mediated inhibition to the value obtained with the Cys-less CTP, in which the native lysines are present at each site (compare Figures 3 and 4).
Figure 4. Effect of MTSEA labeling of binding site cysteine substitution mutants on PLP-mediated inhibition of CTP function.
Proteoliposomes were preincubated with 0.05-0.06 mM MTSEA for 10 min prior to the addition of PLP. Sodium borohydride was excluded from these incubations in order to avoid reduction of the Cys-MTS reagent disulfide bond. Data represent means ± S.E. of incubations repeated 2-3 times. Other conditions were as described under “Experimental Procedures”.
Docking of Pyridoxal 5′-phosphate into the Homology-Modeled Structure of the CTP
Pyridoxal 5′-phosphate was docked into the homology modeled structure of the CTP using the Dock module of MOE as described under “Experimental Procedures”. As depicted in Figure 5, PLP is hypothesized to block the CTP transport pathway via covalently binding to site one residue Lys-83, and then extending into site two where its phosphate group can form ionic hydrogen bonds with residues Lys-37, Lys-239, Arg-276, and Arg-181. Furthermore, the hydroxyl group of PLP is thought to hydrogen bond with Glu-131, a residue that is near binding site one. The experimental data suggest that the ionic interactions have a minor role in giving rise to the PLP effect.
DISCUSSION
The present studies provide the first clear-cut characterization of the molecular origin for the inhibitory effect mediated by the lysine-selective reagent pyridoxal 5′-phosphate with any member of the mitochondrial carrier family. Our data with seven CTP lysine to cysteine substitution mutants indicate that: i) replacement of Lys83 (a residue located within binding site one) with cysteine results in a loss of 78% of the PLP-mediated inhibition of CTP function; ii) replacement of Lys-37 or Lys-239 (binding site two residues) with cysteine caused a modest reduction in the PLP effect (i.e., 31% and 20%, respectively); iii) replacement of non-binding site lysines (i.e., Lys-45, Lys-48, Lys-134, Lys-141) with cysteine resulted in little change in the PLP-mediated inhibition; and iv) replacement of Glu-131 or Arg-181 (i.e., two residues that are thought to form ionic hydrogen bonds with the PLP) with cysteine caused little change in the PLP inhibition. In combination, our data point to Lys-83 as being the primary determinant of the total PLP effect since the replacement of this single lysine abolishes nearly all of the observed inhibition by PLP. Site two residues Lys-37 and Lys-239 appear to be secondary determinants of the PLP effect, since their replacement with cysteine causes a modest relief of the PLP-mediated inhibition. Based on these results, in combination with docking calculations, we developed a model for PLP binding in which the reagent is tethered to binding site one via formation of a covalent bond with Lys-83. PLP then extends further into the translocation pathway and forms ionic interactions with several binding site two residues (i.e., Lys-239, Lys-37, Arg-181, and Arg-276). This model supports the conclusion that PLP inhibits CTP function via direct steric blockage of the substrate binding sites within the translocation pathway.
Several points merit further comment. First, the observation that most of the PLP-mediated inhibition of CTP function can be accounted for by interaction of the reagent with substrate binding site residues, is consistent with the predominantly competitive mode of inhibition that we observed in our kinetic studies (Figure 2). However, we cannot rule out possible additional minor effects of PLP that may arise by interaction with select lysine(s) located in other regions of the protein. Second, the present findings are entirely consistent with our earlier studies that focused on the reconstitutively active CTP purified from rat liver mitochondria (Gremse et al., 1995). In those studies we demonstrated that substrate could protect against PLP-mediated inhibition in a specific manner, thereby suggesting that PLP interacted with a residue at or near the substrate binding site. Third, our model suggests that the mode of PLP binding to lysine at position 83 within site one differs significantly from the binding of citrate to this site (compare Figures 1 and 5). Thus, upon interaction of the C-4 aldehyde group within PLP with the lysine 83 amine moiety to form a protonated Schiff base followed by its subsequent reduction, we propose that PLP is able to extend further into the transport pathway whereupon it forms ionic hydrogen bonds with several residues within site two. In contrast, when citrate reacts at site one, it adopts a conformation enabling it to form high affinity interactions with three residues within this binding site (i.e., Lys-83, Arg-87, and Arg-189). In this conformation, citrate is not able to simultaneously interact with residues in both sites one and two (Ma et al., 2007).
The identification of lysine 83 both as a substrate binding site residue (Ma et al., 2007), and as being predominantly responsible for the PLP-mediated inhibition of the CTP (the present investigation), has important ramifications with regard to other mitochondrial transporters. For example, based on sequence alignment, we note that this lysine is conserved amongst mitochondrial CTPs of widely divergent evolutionary origin ranging from yeast to human (Kaplan et al., 1995; and unpublished observations). This conservation is consistent with and supportive of its important mechanistic role in CTP function. Furthermore, the observation that a lysine at this location is conserved amongst other types of mitochondrial anion carriers (i.e., the α-ketoglutarate and the succinate carriers) (Robinson and Kunji, 2006) suggests a possible role for this residue: i) in the substrate binding sites of these transporters and thus in the possible conservation of binding site architecture across several members of this carrier family (Robinson and Kunji, 2006); and ii) in the PLP-mediated inhibition observed with these carriers (Palmieri et al., 1997a, 2001). Conversely, the observation that a lysine at this location is not preserved in numerous other mitochondrial carriers with which PLP inhibition has been observed (Palmieri et al., 1997b; Robinson and Kunji, 2006; Todisco et al., 2006), indicates that in these cases the reagent reacts with “other” lysine residues that are both accessible to the external environment and are critical for the carrier function. It will be of interest to ascertain whether these PLP-reactive lysines also reside within substrate binding sites as observed with the CTP.
Finally, in the present investigation, chemical modification of substrate binding site lysine to cysteine substitution mutants with MTSEA resulted in a near-total restoration of the PLP inhibitory effect. Since formation of a mixed disulfide between SCH2CH2NH3+ and the cysteine sulfhydryl group results in the addition of a group with a similar distribution of positive charge, length, and molecular volume to that observed with the native lysine side-chain (Ma et al., 2007), a chemical rescue of the inhibitory effect of PLP on CTP function was both predicted and observed.
In conclusion, the present investigation has provided direct evidence demonstrating that a primarily competitive type of PLP-mediated inhibition of the CTP arises from PLP interaction with lysine residues located in the previously identified substrate binding sites within this transporter. These findings lend further credence to our identification of the binding site composition and represent a continuation of our efforts to merge data from functional and structural approaches in order to understand the details of the CTP mechanism at the molecular and atomic levels.
Acknowledgments
This work was supported by National Institutes of Health Grant GM-054642 to R.S.K.
Footnotes
- BTC
- 1,2,3-benzenetricarboxylate
- CTP
- citrate transport protein
- MOE
- molecular operating environment
- MTS
- methanethiosulfonate
- MTSEA
- (2-aminoethyl)methanethiosulfonate hydrobromide
- PLP
- pyridoxal 5′-phosphate
- sarkosyl
- sodium N-lauroylsarcosinate
- S.E.
- standard error
- TMD
- transmembrane domain
REFERENCES
- Brunengraber H, Lowenstein JM. FEBS Lett. 1973;36:130–132. doi: 10.1016/0014-5793(73)80353-9. [DOI] [PubMed] [Google Scholar]
- Conover TE. Trends Biochem. Sci. 1987;12:88–89. [Google Scholar]
- Cornish-Bowden A. Biochem. J. 1974;137:143–144. doi: 10.1042/bj1370143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes A, Cascante M, Cardenas ML, Cornish-Bowden A. Biochem. J. 2001;357:263–268. doi: 10.1042/0264-6021:3570263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierks T, Stappen R, Salentin A, Kramer R. Biochim. Biophys. Acta. 1992;1103:13–24. doi: 10.1016/0005-2736(92)90052-n. [DOI] [PubMed] [Google Scholar]
- Endemann G, Goetz PG, Edmond J, Brunengraber H. J. Biol. Chem. 1982;257:3434–3440. [PubMed] [Google Scholar]
- Gremse DA, Dean B, Kaplan RS. Arch. Biochem. Biophys. 1995;316:215–219. doi: 10.1006/abbi.1995.1030. [DOI] [PubMed] [Google Scholar]
- Halgren TA. J. Comput. Chem. 1999;20:616–641. doi: 10.1002/(SICI)1096-987X(199905)20:7<720::AID-JCC7>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Kaplan RS, Morris HP, Coleman PS. Cancer Res. 1982;42:4399–4407. [PubMed] [Google Scholar]
- Kaplan RS, Pedersen PL. Anal. Biochem. 1985;150:97–104. doi: 10.1016/0003-2697(85)90445-2. [DOI] [PubMed] [Google Scholar]
- Kaplan RS, Mayor JA, Johnston N, Oliveira DL. J. Biol. Chem. 1990a;265:13379–13385. [PubMed] [Google Scholar]
- Kaplan RS, Oliveira DL, Wilson GL. Arch. Biochem. Biophys. 1990b;280:181–191. doi: 10.1016/0003-9861(90)90534-6. [DOI] [PubMed] [Google Scholar]
- Kaplan RS, Mayor JA, Wood DO. J. Biol. Chem. 1993;268:13682–13690. [PubMed] [Google Scholar]
- Kaplan RS, Mayor JA, Gremse DA, Wood DO. J. Biol. Chem. 1995;270:4108–4114. doi: 10.1074/jbc.270.8.4108. [DOI] [PubMed] [Google Scholar]
- Kaplan RS, Mayor JA, Brauer D, Kotaria R, Walters DE, Dean AM. J. Biol. Chem. 2000;275:12009–12016. doi: 10.1074/jbc.275.16.12009. [DOI] [PubMed] [Google Scholar]
- Lundblad RL. Chemical Reagents for Protein Modification. CRC Press; Boca Raton, FL: 1991. [Google Scholar]
- Ma C, Kotaria R, Mayor JA, Eriks LR, Dean AM, Walters DE, Kaplan RS. J. Biol. Chem. 2004;279:1533–1540. doi: 10.1074/jbc.M310866200. [DOI] [PubMed] [Google Scholar]
- Ma C, Kotaria R, Mayor JA, Remani S, Walters DE, Kaplan RS. J. Biol. Chem. 2005;280:2331–2340. doi: 10.1074/jbc.M411474200. [DOI] [PubMed] [Google Scholar]
- Ma C, Remani S, Sun J, Kotaria R, Mayor JA, Walters DE, Kaplan RS. J. Biol. Chem. 2007;282:17210–17220. doi: 10.1074/jbc.M611268200. [DOI] [PubMed] [Google Scholar]
- Mannella CA. J. Bioenerg. Biomembr. 1997;29:525–531. doi: 10.1023/a:1022489832594. [DOI] [PubMed] [Google Scholar]
- Palmieri F, Stipani I, Quagliariello E, Klingenberg M. Eur. J. Biochem. 1972;26:587–594. [Google Scholar]
- Palmieri L, Lasorsa FM, De-Palma A, Palmieri F, Runswick MJ, Walker JE. FEBS Lett. 1997a;417:114–118. doi: 10.1016/s0014-5793(97)01269-6. [DOI] [PubMed] [Google Scholar]
- Palmieri L, Marco VD, Iacobazzi V, Palmieri F, Runswick MJ, Walker JE. FEBS Lett. 1997b;410:447–451. doi: 10.1016/s0014-5793(97)00630-3. [DOI] [PubMed] [Google Scholar]
- Palmieri L, Agrimi G, Runswick MJ, Fearnley IM, Palmieri F, Walker JE. J. Biol. Chem. 2001;276:1916–1922. doi: 10.1074/jbc.M004332200. [DOI] [PubMed] [Google Scholar]
- Rippa M, Spanio L, Pontremoli S. Arch. Biochem. Biophys. 1967;118:48–57. [Google Scholar]
- Robinson AJ, Kunji ERS. Proc. Nat. Acad. Sci. USA. 2006;103:2617–2622. doi: 10.1073/pnas.0509994103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todisco S, Agrimi G, Castegna A, Palmieri F. J. Biol. Chem. 2006;281:1524–1531. doi: 10.1074/jbc.M510425200. [DOI] [PubMed] [Google Scholar]
- Walters DE, Kaplan RS. Biophys. J. 2004;87:907–911. doi: 10.1529/biophysj.104.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JA, Lowenstein JM. J. Biol. Chem. 1970;245:5993–6002. [PubMed] [Google Scholar]
- Xu Y, Mayor JA, Gremse D, Wood DO, Kaplan RS. Biochem. Biophys. Res. Commun. 1995;207:783–789. doi: 10.1006/bbrc.1995.1255. [DOI] [PubMed] [Google Scholar]
- Xu Y, Kakhniashvili DA, Gremse DA, Wood DO, Mayor JA, Walters DE, Kaplan RS. J. Biol. Chem. 2000;275:7117–7124. doi: 10.1074/jbc.275.10.7117. [DOI] [PubMed] [Google Scholar]