Abstract
RNA editing provides a post-transcriptional mechanism to increase structural heterogeneity of gene products. Recently, the α3 subunit of the GABAA receptors has been shown to undergo RNA editing. As a result, a highly conserved isoleucine residue in the third transmembrane domain is replaced with a methionine. To determine the effect of this structural change on receptor function, we compared the GABA sensitivity, pharmacological properties and macroscopic kinetics of recombinant receptors containing either the edited or unedited forms of the α3 subunit along with β3 and γ2L. Editing substantially altered the GABA sensitivity and deactivation rate of the receptors, with the unedited form showing a lower GABA EC50 and slower decay. Comparable effects were observed with a mutation at the homologous location in the α1 subunit, suggesting a common role for this site in regulation of channel gating. Except for the response to GABA, the pharmacological properties of the receptor were unaffected by editing, with similar enhancement by a variety of modulators. Since RNA editing of the α3 subunit increases through development, our findings suggest that GABAergic neurotransmission may be more effective early in development, with greater GABA sensitivity and slower decay rates conferred by the unedited α3 subunit.
Keywords: GABA, development, gating, mutation, patch clamp, channel kinetics
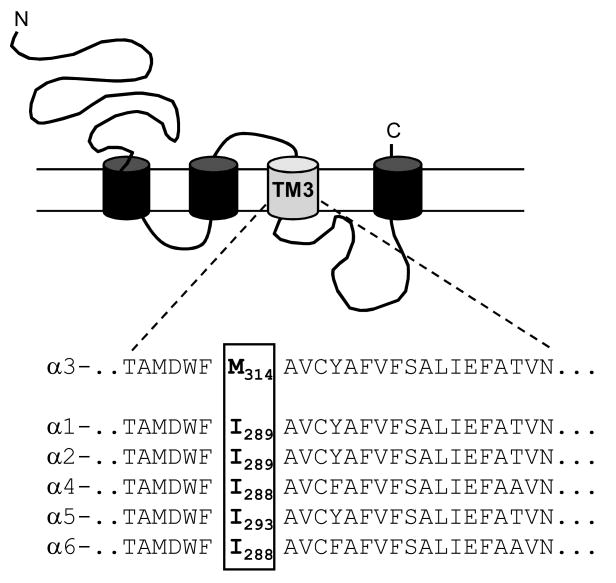
The process of RNA editing is very common in the mammalian nervous system and provides a post-transcriptional mechanism for altering protein structure. Many proteins important for development and function in the nervous system are regulated through the action of RNA-specific adenosine deaminases(ADARs) (Mattick and Mehler, 2008). Several of these gene products are ion channels, including the ligand-gated glutamate receptors. The editing of the AMPA and kainate subtypes of glutamate receptors causes a genomically-encoded glutamine residue to be replaced by an arginine within the second transmembrane domain (Sommer et al., 1991). This structural change results in reduced channel permeability to divalent cations (Burnashev et al., 1992). Editing of the glutamate receptor subunits is vital for normal brain development and function, as mice engineered to produce only the unedited GluR2 subunit do not survive into adulthood (Brusa et al., 1995). Recently it was reported that the α3 subunit of the GABAA receptor (GABAR) also undergoes RNA editing by ADARs (Ohlson et al., 2007; Rula et al., 2008). The result is a change in amino acid sequence within the third transmembrane domain (TM3) that replaces a highly conserved isoleucine residue with a methionine that is found only in the α3 subunit (Figure 1). The extent of this editing is developmentally regulated, with about 50% of the sites edited in the newborn rat brain increasing to nearly 100% edited in the adult. RNA editing has been shown to occur at four sites within the homologous RDL subunit of Drosophila (Hoopengardner et al., 2003; Es-Salah et al., 2008), but had not previously been shown in the vertebrate GABARs.
Figure 1. Mutation site.
The amino acid sequence of the 3rd transmembrane domain (TM3) for each of the α subunits is shown beneath the schematic of the subunit structure. The isoleucine/methionine residue affected by RNA editing (number 314 of the mature α3 rat peptide) is boxed and indicated in bold. Sequence alignment from Tyndale et al., 1995.
The GABARs are ligand-gated, chloride-permeable ion channels responsible for fast inhibitory neurotransmission. The GABARs exhibit substantial structural heterogeneity through the expression of at least 16 different subunits in the mammalian brain (Whiting et al., 1999). There are six different subtypes within the α subunit family, each of which has a distinct, developmentally regulated, pattern of expression (Laurie et al., 1992a, 1992b; Wisden et al., 1992). The α3 subunit is one of the predominant α subunits in the embryonic brain, where it is widely and highly expressed. As development progresses, the α3 subunit is largely replaced by the α1 subunit, and its expression is restricted primarily to cortical neurons in the adult (Laurie et al., 1992a, 1992b; Wisden et al., 1992). Production of the α3 subunit can be influenced by pathological changes in the brain. An increase in α3 mRNA is observed during epileptogenesis (Brooks-Kayal et al,. 1998) while a reduction is often observed following seizure onset (Poulter et al., 1999; Loup et al., 2006). Animals lacking the α3 subunit exhibit abnormalities in sensorimotor processing similar to those observed in schizophrenic patients (Yee et al., 2005). Drugs selectively targeting the α3-containing receptors are under investigation for treatment of anxiety and chronic pain (Dias et al, 2005; Knabl et al., 2008).
A recent report by Rula et al (2008) showed that editing of the α3 subunit alters some of its kinetic properties. To further examine how the change in amino acid sequence created by RNA editing alters the function of α3-containing GABARs, we compared the pharmacological and electrophysiological properties of receptors containing either the unedited (Iso) or the edited (Met) forms of the subunit. In addition, we made the same residue change within the α1 subunit to determine if this site plays a general role in controlling GABAR function. The subunits were transiently transfected into HEK-293T cells and the chloride currents in response to GABA measured through whole-cell and excised patch recordings.
Methods
Transfection of mammalian cells
Full-length cDNAs in pCMVNeo (Dr. Robert Macdonald, Vanderbilt University) expression vectors were transfected into the human embryonic kidney cell line HEK-293T (GenHunter, Nashville, TN). For selection of transfected cells, the plasmid pHookTM-1 (Invitrogen) containing cDNA encoding the surface antibody sFv was also transfected into the cells (Chesnut et al., 1996). Cells were maintained in Dulbecco's modified Eagle medium (DMEM) plus 10% fetal bovine serum, 100 IU/ml penicillin and 100 μg/ml streptomycin. Cells were passaged by a 5 min. incubation with 0.05% trypsin/0.02% EDTA solution in phosphate buffered saline (10 mM Na2HPO4, 150 mM NaCl, pH=7.3).
The cells were transfected using calcium phosphate precipitation. Plasmids encoding GABAR subunit cDNAs were added to the cells in 1:1:1 ratios of 2 μg each. 1 μg of a plasmid encoding a surface antibody (pHook) was also transfected as a marker for transfection. Following a 4-6 hr. incubation at 3% CO2, the cells were treated with a 15% glycerol solution in BBS buffer (50 mM BES(N,N-bis[2-hydroxyethyl]-2-aminoethanesulfonic acid), 280 mM NaCl, 1.5 mM Na2HPO4) for 30 sec. The selection procedure for pHook expression was performed 44-52 hrs later. The cells were passaged and mixed with 3-5 μl of magnetic beads coated with antigen for the pHook antibody (approximately 6 × 105 beads) (Chesnut et al., 1996). Following a 30-60 min. incubation to allow the beads to bind to positively transfected cells, the beads and bead-coated cells were isolated using a magnetic stand. The selected cells were resuspended into DMEM, plated onto glass coverslips treated with poly L-lysine and coated with collagen and used for recordings 18-28 hrs. later.
Electrophysiological recording solutions and techniques
For all recordings the external solution consisted of (in mM): 142 NaCl, 8.1 KCl, 6 MgCl2, 1 CaCl2, and 10 HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) with pH = 7.4 and osmolarity adjusted to 295-305 mOsm. Recording electrodes were filled with an internal solution of (in mM); 153 KCl, 1 MgCl2, 5 K-EGTA (ethylene glycol-bis (β-aminoethyl ether N,N,N′N′-tetraacetate), 2 MgATP and 10 HEPES with pH = 7.4 and osmolarity adjusted to 295-305 mOsm. GABA was diluted into external solution from freshly made or frozen stocks in water. Drugs were diluted from freshly made stocks in DMSO and were obtained from commercial sources. Patch pipettes were pulled from borosilicate glass with an internal filament (World Precision Instruments, Sarasota FL) on a two-stage puller (Narishige, Japan) to a resistance of 5-10 MΩ. For whole-cell recordings GABA was applied to cells using a stepper solution exchanger with a complete exchange time of <50 msec (open tip, SF-77B, Warner Instruments, Hamden CT). For macropatch recordings the 3-barrel square glass was pulled to a final size near 200 μm. 10-90% rise times of the junction potential at the open tip were consistently faster than 400 μsec and were tested using a diluted external solution. There was a continuous flow of external solution through the chamber. Currents were recorded with an Axon 200B (Foster City, CA) patch clamp amplifier.
Construction of mutated subunit cDNAs
Point mutations were generated using the QuikChange procedure and products (Stratagene, La Jolla, CA). Oligonucleotide primers were synthesized and DNA sequencing was performed by the University of South Carolina DNA core facility (Columbia, SC).
Analysis of whole-cell and macropatch currents
Whole-cell currents were analyzed using the programs Clampfit (pClamp8 suite, Axon Instruments, Foster City CA) and Prism (Graphpad, San Diego, CA). Concentration-response data was fit with a four-parameter logistic equation. All fits were made to normalized data with current expressed as a percentage of the maximum peak response to GABA for each cell.
Macropatch currents were digitized at 10 kHz and analyzed with the pClamp8.0 suite of programs (Axon Instruments). The deactivation or desensitization rate was determined by fitting the decay current with the Levenberg-Marquardt least squares method with increasing numbers of exponential functions until additional components did not significantly improvement of the fit (F test of the sum of squared residuals). In all cases the decay was best fit with the sum of two components with a correlation coefficient for the fit greater than 0.90. Student's paired or unpaired t-tests were performed using the Instat program (Graphpad) with a significance level of p<0.05. The logs of the GABA EC50 measurements were used for statistical comparison.
Results
Exchanging Isoleucine314 for Methionine in the α3 subunit increases GABA sensitivity
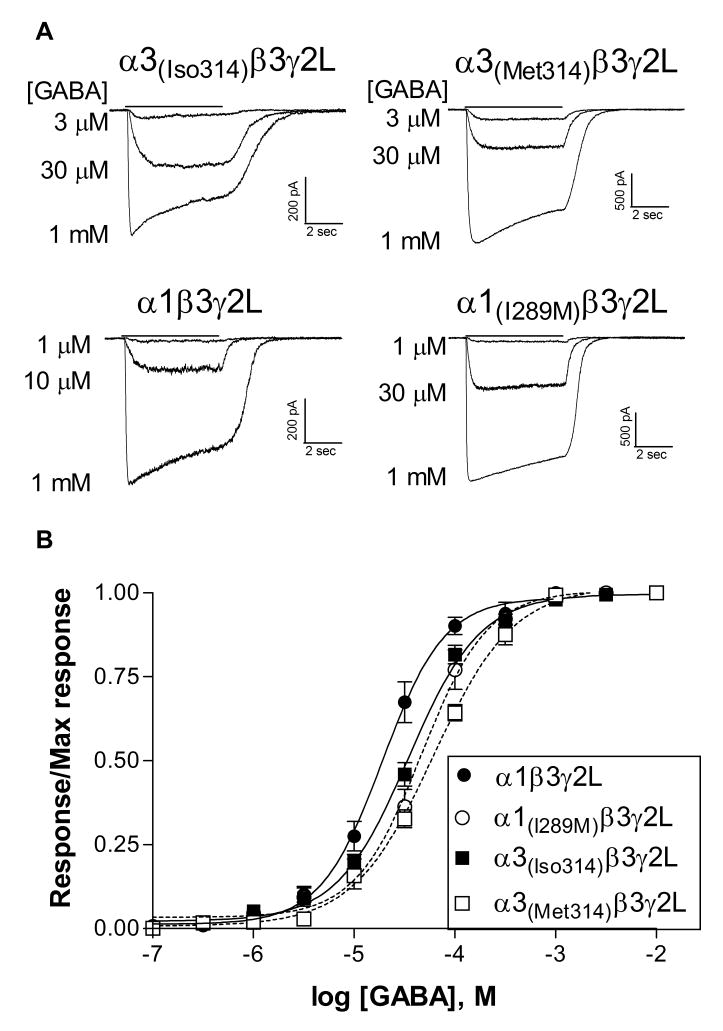
Receptors containing the edited α3 subtype are characterized by a low GABA sensitivity compared to those containing any of the other α subtypes (Dučić et al., 1995; Böhme et al., 2004; Picton et al., 2007). To determine if RNA editing affects the GABA EC50 of the receptor, α3 or α1 subunits containing either an isoleucine or methionine residue at the edit site were transfected into HEK-293T cells. All constructs produced GABA-sensitive currents when combined with wild-type β3 and γ2L subunits. For both α1 and α3 subunits, the isoleucine residue was associated with a greater sensitivity to GABA compared to subunits carrying the methionine residue (Figure 2). For the α3 subunit, the average GABA EC50 for the α3(Iso314)β3γ2L isoform was 35.7 ± 3.1 μM (N=6), significantly different (p≤0.001) from receptors with the α3(Met314) subunit, which had an average EC50 of 62.0 ± 5.0 μM (N=6). Similarly, the average GABA EC50 of the wild-type α1 subunit, containing an isoleucine residue, was 20.5 ± 3.2 μM (N=5), significantly less (p≤0.01) than that of α1(I289M)-containing receptors, which averaged 50.1 ± 9.5 μM (N=5). This suggests that RNA editing of the α3 subunit is one contributor to its characteristic low sensitivity to the endogenous agonist GABA and that GABA sensitivity of these receptors will decrease through development as editing increases.
Figure 2. GABA sensitivity.
A. Representative whole-cell current traces from cells transfected with the α subunit indicated in combination with β3 and γ2L. GABA was applied for 5 sec (solid line) to cells voltage clamped at -50 mV.
B. Normalized concentration-response relationships were constructed by dividing the peak response to each concentration of GABA by the maximum current response for each cell. Points represent the mean ± SEM. Data were fit with a four-parameter logistic equation. EC50's of the fits shown to averaged data were 19.1 μM (α1 wild-type, N=5), 47.0 μM (α1(I289M), N=5), 34.7 μM (α3(Iso314), N=6), and 61.3 μM (α1(Met314), N=6).
One concern with heterologous expression systems is the incorporation of all transfected subunits. Expression of α3β3 heterodimers might influence the characteristics of the functional response. We and others have found that α3(Met314)β1 (Ranna et al., 2006) or α3(Met314)β3 (GABA EC50 = 2.7 ± 1.1 μM, N=3, data not shown) have substantially higher sensitivity to GABA compared to γ2-containing receptors. We did not observe a higher affinity component in any GABA concentration response relationships, suggesting that αβ heteromers did not substantially contribute to the receptor population. The pharmacological properties of the receptors, described below, also confirm the presence of the γ2 subunit.
Editing of the α3 subunit does not alter other pharmacological properties
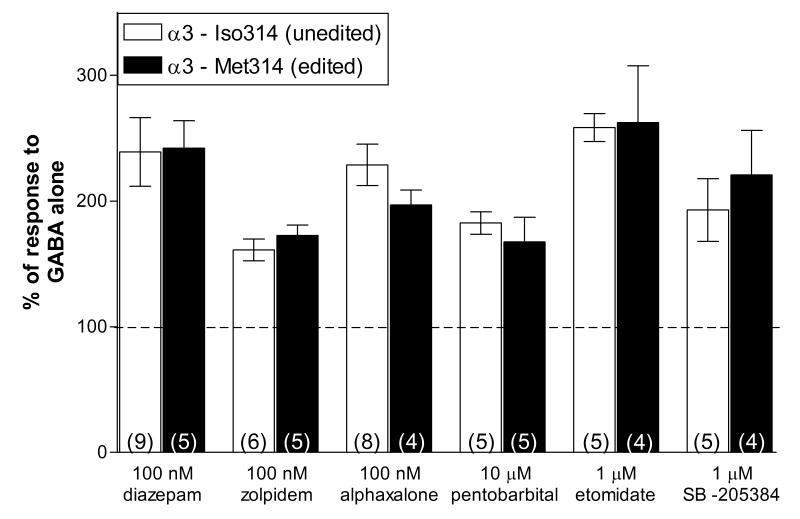
A wide variety of different allosteric modulators act upon the GABARs in a subunit-dependent manner and many of these are used clinically as sedatives, anxiolytics, anesthetics and anticonvulsants (Korpi et al., 2002). A change in pharmacological properties by RNA editing might have important implications for the treatment of disorders in the developing brain. Therefore, we compared the response of receptors with the unedited (α3-Iso314) or the edited (α3-Met314) subunit to a variety of different classes of positive allosteric modulators (Figure 3). We examined potentiation by benzodiazepine-site agonists (diazepam and zolpidem), a neurosteroid (alphaxalone), two intravenous anaesthetics (pentobarbital and etomidate) and an α3-selective modulator (SB-205384, Meadows et al., 1998). We have previously shown that editing at position 314 had no effect on another α3-preferring anticonvulsant, stiripentol (Fisher, in press). The drugs were tested at sub-maximal concentrations in order to detect any shift in sensitivity to the modulator. All drugs were co-applied with a GABA concentration near the EC10-20 for each receptor isoform. The presence of a methionine or isoleucine residue at the edit site had no significant effect on the response to any of the drugs tested (Figure 3). Sensitivity to diazepam and zolpidem provide further evidence of incorporation of the γ2L subunit in both receptor populations. Since only a single concentration of each modulator was examined, it is possible that differences in maximum potentiation or other concentration-dependent effects may be influenced by editing. However, these findings suggest that while RNA editing alters the receptor's response to its endogenous agonist, it would not be expected to influence the effectiveness of many clinically relevant GABAR modulators.
Figure 3. Editing does not alter other pharmacological properties of the α3 subunit.
The allosteric modulators indicated were co-applied for 5 sec with either 3 μM (α3-Iso314) or 6 μM GABA (α3-Met314). The peak current amplitude in the presence of the modulator was divided by the response to GABA alone and multiplied by 100. The dashed line represents the response to GABA alone (100%). None of the responses were significantly altered by the point mutation.
Macroscopic kinetic properties
The rates of channel deactivation and desensitization are major contributors to the characteristics of the post-synaptic current. The α3 subtype is associated with slower rates of deactivation and onset of desensitization than most other α subtypes, including the α1, in both recombinant studies using the edited form of α3 (Gingrich et al., 1995; Picton and Fisher, 2007) and in neuronal studies in which the RNA editing status is unknown (Huntsman and Huguenard, 2006). To provide a more accurate prediction of the potential effect of RNA editing on post-synaptic currents mediated by α3-containing receptors, we rapidly applied 3 mM GABA to excised macropatches and measured the rates of current deactivation and desensitization.
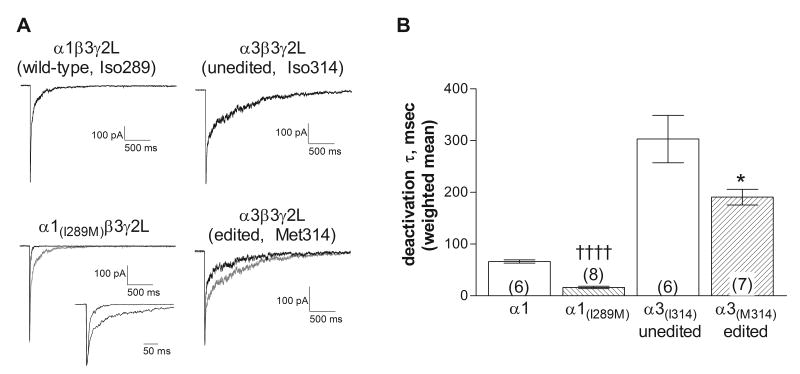
Deactivation
The deactivation rate for all receptors in response to a 5 msec application of GABA was fit with the sum of two exponential components (Table 1). Replacement of the isoleucine residue with methionine in either the α1 or the α3 subunit caused faster deactivation (Figure 4). For the α3 subunit, this was due primarily to a significant increase in the relative contribution from the fast component, rather than a change in the time constants (Table 1). These findings are generally consistent with Rula et al. (2008), who also found faster deactivation with the edited form. Quantitative differences in the measured rates are likely due to the slower agonist application rates associated with the lifted cells used in their study. More rapid deactivation is consistent with the reduced GABA sensitivity associated with the presence of a methionine (Figure 2). This finding suggests that the post-synaptic current at α3-expressing neurons would decay more slowly in response to GABA release early in development, when the site is unedited. GABAergic neurotransmission at neurons expressing the unedited α3 subunit might therefore be expected to be more effective, due to a longer-lasting chloride current, compared to neurons expressing the edited subunit.
Table 1.
Deactivation and desensitization kinetics
| Deactivation kinetics – 5 msec application | |||
|---|---|---|---|
| α subunit | τfast (msec) | areafast | τslow (msec) |
| α1-wildtype (N=6) |
15.9 ± 1.8 | 67.6 ± 4.2 % | 183.0 ± 20.8 |
| α1(I289M) (N=8) |
8.3 ± 0.7*** | 88.2 ± 1.7 %*** | 75.3 ± 11.0*** |
| α3-Iso314 (N=6) |
24.8 ± 2.4 | 45.3 ± 3.2 % | 549.6 ± 96.8 |
| α3-Met314 (N=7) |
25.4 ± 2.4 | 67.7 ± 4.2 %†† | 554.0 ± 37.3 |
| Desensitization kinetics – 400 msec application | |||
| α1-wildtype (N=8) |
11.5 ± 1.2 | 63.3 ± 4.0 % | 149.5 ± 22.8 |
| α1(I289M) (N=12) |
14.7 ± 1.8 | 54.5 ± 2.4% | 192.3 ± 23.3 |
| α3-Iso314 (N=6) |
19.2 ± 3.5 | 59.5 ± 3.6% | 198.6 ± 29.5 |
| α3-Met314 (N=6) |
24.3 ± 1.9 | 40.8 ± 4.9%† | 266.0 ± 50.6 |
(p≤0.05),
(p≤0.01) compared to α3-Iso314, unpaired, 2-tailed t-test.
(p≤0.001) compared to α1 – wild-type, unpaired 2-tailed t test.
Figure 4. Deactivation rate.
A. Representative macropatch current traces from receptors containing the subunits indicated in response to a 5 ms application of 3 mM GABA. The gray traces show the isoleucine-containing α subunits, scaled to overlay the response of the methionine-containing subunits. Inset shows overlaid traces from α1-containing receptors at an expanded time scale. For the α1-containing receptors, the components for the traces shown had time constants (and relative areas) of 15.2 msec (65.6%) and 152.8 msec for the wild-type subunit and 7.3 msec (83.7%) and 31.3 msec for the α1(I289M). For the α3-containing receptors, the components for the traces shown had time constants (and relative areas) of 30.2 msec (48.0%) and 852.6 msec for the unedited form (Iso314) and 25.3 msec (70.8%) and 558.7 msec for the edited form (Met314). Correlation coefficients for all fits were greater than 0.95.
B. The current deactivation was fit with the sum of two exponential components. Symbols and bars represent the weighted average ± SEM and the number in parentheses indicates number of patches. The average decay rate of the methionine-containing subunits was significantly faster than those with the isoleucine residue for both the α1 and α3 subtypes. †††† (p≤0.0001, compared to α1-wildtype), * (p ≤0.05 compared to α3-isoleucine) using a 2-tailed unpaired t-test.
Desensitization
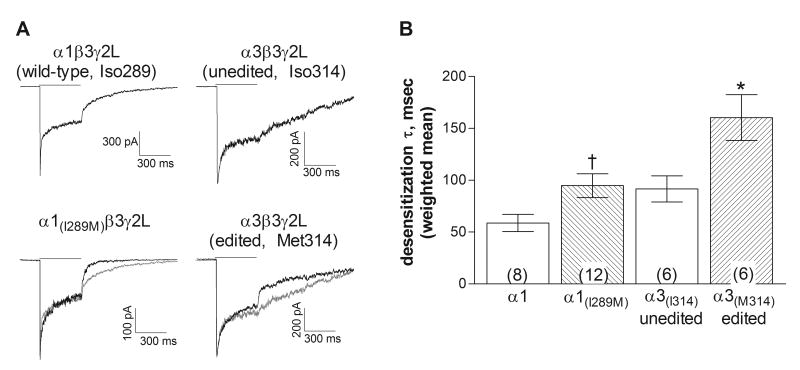
To examine entry into desensitized states, 3 mM GABA was applied to macropatches for 400 msec (Figure 5A). The onset of desensitization was fit with the sum of two exponential components, (Table 1). Receptors composed of the α1 or α3 subunit carrying the methionine residue showed a slightly slower mean rate of desensitization compared to those with the isoleucine at the edit site (Figure 5B). Similar to the effect on deactivation, the only significant difference in desensitization between the two α3 forms was found in the relative area of the time constants (Table 1). Entry into long-lived desensitized states would be expected to influence the duration of the synaptic current, the spread of the synaptic signal and the response to repetitive stimulation (Jones and Westbrook, 1995; Overstreet et al., 2000).
Figure 5. Desensitization rate.
A. Representative macropatch current traces from receptors containing the subunits indicated in response to a 400 ms application (solid line) of 3 mM GABA. The gray traces show the currents from the isoleucine-containing α subunits, scaled to overlay the response of the methionine-containing subunits. For the α1-containing receptors, the components for the traces shown had time constants (and relative areas) of 7.0 msec (67.7%) and 155.8 msec for the wild-type subunit and 12.8 msec (48.7%) and 171.0 msec for the α1(I289M). For the α3-containing receptors, the components for the traces shown had time constants (and relative areas) of 24.3 msec (69.1%) and 196.8 msec for the unedited form (Iso314) and 23.4 msec (40.6%) and 179.2 msec for the edited form (Met314). Correlation coefficients for all fits were greater than 0.95.
B. Bars represent the weighted average (±SEM) of the rate of onset of desensitization. The current decay was fit with the sum of two exponential components. The desensitization rate of the methionine-containing subunits was significantly slower than that of its isoleucine-containing counterpart for both the α1 and α3 subtypes (p≤0.05, compared to α1-wildtype(†) or α3-isoleucine(*) using a 2-tailed unpaired t-test). Number of patches is given by the number in parentheses.
Discussion
The process of RNA editing can often have dramatic effects on the function of proteins (Mattick and Mehler, 2008). While many neuronally expressed gene products have been shown to be post-transcriptionally regulated in this manner, within the large GABAR subunit family only the α3 subunit has been reported to be RNA edited in mammals. The closely related RDL subunits also undergo RNA editing at four sites, at least one of which influences the pharmacological characteristics of the receptor (Hoopengardner et al., 2003; Es-Salah et al., 2008). We showed that exchanging the isoleucine residue encoded by the DNA sequence for the methionine residue produced by RNA editing has functional consequences for the receptor. The most dramatic effects were on GABA sensitivity, which was significantly reduced by editing, and deactivation rate, which was faster with the edited form. A modest effect on desensitization rate was also observed, with editing producing slower onset of desensitization. Similar effects were observed with the α1 subunit with mutation from the wild-type isoleucine to methionine. These results are generally consistent with those recently reported by Rula et al., (2008) who also found higher GABA sensitivity, faster activation, slower deactivation, and greater outward rectification associated with the non-edited α3 subunit using lifted cell recordings. We also examined whether editing might alter the response of the GABARs to commonly used sedatives, anxiolytics and anticonvulsants. We found that, except for the response to the agonist GABA, the pharmacological properties do not appear to be influenced by editing. Many of these modulators have been shown to act through sites distinct from the TM3 (Korpi et al., 2002; Hosie et al., 2007), and our results suggest that even those that have selective actions at α3-containing receptors are not affected by structural heterogeneity within this region.
Role of the 3rd transmembrane domain in channel function
These findings add to growing evidence that changes in amino acid sequence within the TM3 of the GABAR can alter channel gating. A mutation in this area of the α1 subunit has been linked to inheritance of juvenile myoclonic epilepsy and found to reduce GABA sensitivity and channel open time (Cossette et al., 2002; Fisher, 2004a). We previously suggested that heterogeneity in this region may contribute to differences in channel gating conferred by the α6 subunit (Fisher, 2004b). GABA binding causes changes in the secondary structure of TM3, consistent with a role in channel activation (Williams et al., 1999) and residues within this region contribute to a binding pocket for volatile anesthetics and ethanol (Bonin and Orser, 2008). The edited residue is located near the extracellular end of TM3 and is believed to lie within the region of the helix that faces the TM2 domain. It is structurally adjacent to residues that can cross-link to TM2 and may influence conformational stability (Jansen and Akabas, 2006). Its location is therefore consistent with a role in channel gating. We found that the functional effects of structural changes at the editing site were not specific for the α3 subunit, but were shared by the α1 subunit, suggesting a common role for this region for the α subunits. Together this evidence indicates an important regulatory role for the TM3 in controlling the activation and channel gating properties of the GABAR.
Potential effects of RNA editing on GABAergic neurotransmission
Many changes occur in neurotransmitter systems with development. Typically these processes lead to less excitation and more inhibition as the brain matures (Raol et al., 2001; Ben-Ari et al., 2007). The effect of RNA editing on the α3 GABAR subunit would therefore appear to be distinctive in that the immature, unedited, form of the GABAR would be expected to produce more effective GABAergic neurotransmission. However, in the fetal brain, activation of GABARs may be depolarizing and excitatory because of the difference in chloride equilibrium potential compared to mature neurons (Ben-Ari et al., 2007). Therefore, a post-synaptic receptor that is more responsive and decays more slowly in response to GABA release may well contribute to hyper-excitability, providing the important depolarizing signals that regulate cortical development.
It is likely that neurons will produce a heterogeneous population of α3 editing forms, especially early in development. This raises the possibility that mixed receptors will be formed carrying one edited and one unedited subunit. Most studies show that receptors containing two different α subtypes or subunits with different properties have characteristics intermediate to the homogeneous receptor populations (Verdoorn, 1994; Sigel et al., 2006). As editing becomes more complete with development, the adult neurons would be expected to produce a nearly pure population of edited subunits. Further studies in neurons will be necessary to verify this predicted functional impact of the RNA editing of the α3 subunit.
Acknowledgments
This work was supported by funds from NIH-NINDS (RO1-NS045950) and the Epilepsy Foundation. Thanks to Amber Picton and Matt Fisher for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brooks-Kayal AR, Shumate MD, Jin H, Rikhter TY, Coulter DA. Selective changes in single cell GABAA receptor subunit expression and function in temporal lobe epilepsy. Nature Med. 1998;4:1166–1172. doi: 10.1038/2661. [DOI] [PubMed] [Google Scholar]
- Ben-Ari Y, Gaiarsa JL, Tyzio R, Khazipov R. GABA: a pioneer transmitter that excites immature neurons and generates primitive oscillations. Physiol Rev. 2007;87:1215–1284. doi: 10.1152/physrev.00017.2006. [DOI] [PubMed] [Google Scholar]
- Böhme I, Rabe H, Lüddens H. Four amino acids in the α subunits determine the γ-aminobutyric acid sensitivities of GABAA receptor subtypes. J Biol Chem. 2004;279:35193–35200. doi: 10.1074/jbc.M405653200. [DOI] [PubMed] [Google Scholar]
- Bonin RP, Orser BA. GABAA receptor subtypes underlying general anesthesia. Pharm Biochem Behav. 2008;90:105–112. doi: 10.1016/j.pbb.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Brusa R, Zimmermann F, Koh DS, Feldmeyer D, Gass P, Seeburg PH, Sprengel R. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science. 1995;270:1677–1680. doi: 10.1126/science.270.5242.1677. [DOI] [PubMed] [Google Scholar]
- Burnashev N, Monyer H, Seeburg H, Sakmann B. Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron. 1992;8:189–198. doi: 10.1016/0896-6273(92)90120-3. [DOI] [PubMed] [Google Scholar]
- Chesnut JD, Baytan AR, Russell M, Chang MP, Bernard A, Maxwell IH, Hoeffler JP. Selective isolation of transiently transfected cells from a mammalian cell population with vectors expressing a membrane anchored single-chain antibody. J Immunol Methods. 1996;193:17–27. doi: 10.1016/0022-1759(96)00032-4. [DOI] [PubMed] [Google Scholar]
- Cossette P, Liu L, Brisebois K, Dong H, Lortie A, Vanasse M, Saint-Hilaire JM, Carmant L, Verner A, Lu WY, Wang YT, Rouleau GA. Mutation of GABRA1 in an autosomal dominant form of juvenile myoclonic epilepsy. Nature Genetics. 2002;31:184–189. doi: 10.1038/ng885. [DOI] [PubMed] [Google Scholar]
- Dias R, Sheppard WFA, Fradley RL, Garrett EM, Stanley JL, Tye SJ, Goodacre S, Lincoln RJ, Cook SM, Conley R, Hallett D, Humphries AC, Thompson SA, Wafford KA, Street LJ, Castro JL, Whiting PJ, Rosahl TW, Atack JR, McKernan RM, Dawson GR, Reynolds DS. Evidence for a significant role of α3-containing GABAA receptors in mediating the anxiolytic effects of benzodiazepines. J Neurosci. 2005;25:10682–10688. doi: 10.1523/JNEUROSCI.1166-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dučić I, Caruncho HJ, Zhu WJ, Vicini S, Costa E. γ-Aminobutyric acid gating of Cl- channels in recombinant GABAA receptors. J Pharm Exp Ther. 1995;272:438–445. [PubMed] [Google Scholar]
- Es-Salah Z, Lapied B, Le Goff G, Hamon A. RNA editing regulates gamma-amino butyric acid receptor function and insecticide sensitivity. Neuroreport. 2008;19:939–943. doi: 10.1097/WNR.0b013e32830216c7. [DOI] [PubMed] [Google Scholar]
- Fisher JL. A mutation in the GABAA receptor α1 subunit linked to human epilepsy affects channel gating properties. Neuropharmacology. 2004a;46:629–637. doi: 10.1016/j.neuropharm.2003.11.015. [DOI] [PubMed] [Google Scholar]
- Fisher JL. The α1 and α6 subunit subtypes of the mammalian GABAA receptor confer distinct channel gating kinetics. J Physiol. 2004b;561:433–448. doi: 10.1113/jphysiol.2003.051839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher JL. The anticonvulsant stiripentol acts directly on the GABAA receptor as a positive allosteric modulator. Neuropharmacology. doi: 10.1016/j.neuropharm.2008.06.004. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingrich KJ, Roberts WA, Kass RS. Dependence of the GABAA receptor gating kinetics on the α-subunit isoform: Implications for structure-function relations and synaptic transmission. J Physiol. 1995;489:529–543. doi: 10.1113/jphysiol.1995.sp021070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopengardner B, Bhalla T, Staber C, Reenan R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301:832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- Hosie AM, Wilkins ME, Smart TG. Neurosteroid binding sites on GABAA receptors. Pharm Ther. 2007;116:7–19. doi: 10.1016/j.pharmthera.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Huntsman MM, Huguenard JR. Fast IPSCs in rat thalamic reticular nucleus require the GABAA receptor β1 subunit. J Physiol. 2006;572:459–475. doi: 10.1113/jphysiol.2006.106617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen M, Akabas MH. State-dependent cross-linking of the M2 and M3 segments: functional basis for the alignment of GABAA and acetylcholine receptor M3 segments. J Neurosci. 2006;26:4492–4499. doi: 10.1523/JNEUROSCI.0224-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MV, Westbrook GL. Desensitized states prolong GABAA channel responses to brief agonist pulses. Neuron. 1995;15:181–191. doi: 10.1016/0896-6273(95)90075-6. [DOI] [PubMed] [Google Scholar]
- Knabl J, Witschi R, Hösl K, Reinold H, Zeilhofer UB, Ahmadi S, Brockhaus J, Sergejeva M, Hess A, Brune K, Fritschy JM, Rudolph U, Möhler H, Zeilhofer HU. Reversal of pathological pain through specific spinal GABAA receptor subtypes. Nature. 2008;451:330–334. doi: 10.1038/nature06493. [DOI] [PubMed] [Google Scholar]
- Korpi ER, Gründer G, Lüddens H. Drug interactions at GABAA receptors. Prog Neurobiol. 2002;67:113–159. doi: 10.1016/s0301-0082(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Laurie DJ, Wisden W, Seeburg PH. The distribution of thirteen GABAA receptor subunit mRNAs in the rat brain. III. Embryonic and postnatal development. J Neurosci. 1992a;12:4151–4172. doi: 10.1523/JNEUROSCI.12-11-04151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie DJ, Seeburg PH, Wisden W. The distribution of 13 GABAA receptor subunit mRNAs in the rat brain. III. Olfactory bulb and cerebellum. J Neurosci. 1992b;12:1063–1076. doi: 10.1523/JNEUROSCI.12-03-01063.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loup F, Picard F, André VM, Kehrli P, Yonekawa Y, Wieser HG, Fritschy JM. Altered expression of α3-containing GABAA receptors in the neocortex of patients with focal epilepsy. Brain. 2006;129:3277–3289. doi: 10.1093/brain/awl287. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Mehler MF. RNA editing, DNA recoding and the evolution of human cognition. Trends Neurosci. 2008;31:227–233. doi: 10.1016/j.tins.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Meadows HJ, Kumar CS, Pritchett DB, Blackburn TP, Benham CD. SB-205384: a GABAA receptor modulator with novel mechanism of action that shows subunit selectivity. Br J Pharmacol. 1998;123:1253–1259. doi: 10.1038/sj.bjp.0701721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlson J, Pedersen JS, Haussler D, Öhman M. Editing modifies the GABAA receptor subunit α3. RNA. 2007;13:698–703. doi: 10.1261/rna.349107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overstreet LS, Jones MV, Westbrook GL. Slow desensitization regulates the availability of synaptic GABAA receptors. J Neurosci. 2000;20:7914–7921. doi: 10.1523/JNEUROSCI.20-21-07914.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picton AJ, Fisher JL. Effect of the α subunit subtype on the macroscopic kinetic properties of recombinant GABAA receptors. Brain Res. 2007;1165:40–49. doi: 10.1016/j.brainres.2007.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter MO, Brown LA, Tynan S, Willick G, William R, McIntyre DC. Differential expression of α1, α2, α3, and α5 GABAA receptor subunits in seizure-prone and seizure-resistant rat models of temporal lobe epilepsy. J Neurosci. 1999;19:4654–4661. doi: 10.1523/JNEUROSCI.19-11-04654.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranna M, Sinkkonen ST, Möykkynen T, Uusi-Oukari M, Korpi ER. Impact of ε and θ subunits on pharmacological properties of α3β1 GABAA receptors expressed in Xenopus oocytes. BMC Pharmacol. 2006;6:1. doi: 10.1186/1471-2210-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raol YH, Lynch DR, Brooks-Kayal AR. Role of excitatory amino acids in developmental epilepsies. Mental Retardation Dev Disab Res Rev. 2001;7:254–260. doi: 10.1002/mrdd.1035. [DOI] [PubMed] [Google Scholar]
- Rula EY, Lagrange AH, Jacobs MM, Hu N, Macdonald RL, Emeson RB. Developmental modulation of GABAA receptor function by RNA editing. J Neurosci. 2008;28:6196–6201. doi: 10.1523/JNEUROSCI.0443-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigel E, Baur R, Boulineau N, Minier F. Impact of subunit positioning on GABAA receptor function. Biochem Soc Trans. 2006;34:868–871. doi: 10.1042/BST0340868. [DOI] [PubMed] [Google Scholar]
- Sommer B, Köhler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;87:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- Tyndale RF, Olsen RW, Tobin AJ. GABAA receptors. In: North RA, editor. Ligand- and voltage-gated ion channels. CRC Press; Boca Raton: 1995. pp. 265–290. [Google Scholar]
- Verdoorn TA. Formation of heteromeric γ-aminobutyric acid type A receptors containing two different α subunits. Mol Pharmacol. 1994;45:475–480. [PubMed] [Google Scholar]
- Whiting PJ, Bonnert TP, McKernan RM, Farrar S, Le Bourdelles B, Heavens RP, Smith DW, Hewson L, Rigby MR, Sirinathsinghji JS, Thompson SA, Wafford KA. Molecular and functional diversity of the expanding GABAA receptor gene family. Ann N Y Acad Sci. 1999;868:645–653. doi: 10.1111/j.1749-6632.1999.tb11341.x. [DOI] [PubMed] [Google Scholar]
- Williams DB, Akabas MH. γ-aminobutyric acid increases the water accessibility of M3 membrane spanning segment residues in γ-aminobutyric acid type A receptors. Biophys J. 1999;77:2563–2574. doi: 10.1016/s0006-3495(99)77091-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisden W, Laurie DJ, Monyer H, Seeburg PH. The distribution of thirteen GABAA receptor subunits mRNAs in the rat brain. I. Telencephalon, diencephalons, mesencephalon. J Neurosci. 1992;12:1040–1062. doi: 10.1523/JNEUROSCI.12-03-01040.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee BK, Keist R, von Bohmer L, Studer R, Benke D, Hagenbuch N, Dong Y, Malenka RC, Fritschy JM, Bluethmann H, Feldon J, Möhler H, Rudolph U. A schizophrenia-related sensorimotor deficit links α3-containing GABAA receptors to a dopamine hyperfunction. Proc Natl Acad Sci. 2005;102:17154–17159. doi: 10.1073/pnas.0508752102. [DOI] [PMC free article] [PubMed] [Google Scholar]